Abstract
Asian rice (Oryza sativa) was domesticated from O. rufipogon, and reduced seed-shattering behaviour was selected to increase yields. Two seed-shattering loci, qSH3 and sh4, are involved in reducing seed shattering in both japonica and indica rice cultivars, while qSH1 and qCSS3 are likely specific to japonica cultivars. In indica cultivars, qSH3 and sh4 fail to explain the degree of seed shattering, as an introgression line (IL) of O. rufipogon W630 carrying domesticated alleles at qSH3 and sh4 still showed seed shattering. Here we analysed differences in seed-shattering degree between the IL and the indica cultivar IR36. The values for grain detachment in the segregating population between the IL and IR36 were continuous. Based on QTL-seq analysis using the BC1F2 population between the IL and IR36, we detected two novel loci, qCSS2 and qCSS7 (QTLs for the Control of Seed Shattering in rice on chromosomes 2 and 7), which contributed to the reduced seed shattering in IR36. We further investigated the genetic interaction of qCSS2 and qCSS7 under the presence of qSH3 and sh4 mutations in O. rufipogon W630 and found that IL carrying IR36 chromosomal segments covering all four loci are required to explain seed-shattering degree in IR36. Since qCSS2 and qCSS7 were not detected in previous studies on seed shattering in japonica, their control may be specific to indica cultivars. Therefore, they are important to understanding the history of rice domestication as well as to adjusting the seed-shattering degree of indica cultivars to maximise their yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A reduction in seed shattering is a well-known phenotypic change observed in many crops (Doebley et al. 2006). Wild plants shatter their seeds soon after maturation for more efficient propagation, but early farmers selected plants with reduced seed shattering to improve harvest efficiency. The more stable yield provided by the loss of seed shattering allowed for more efficiently organised agricultural systems, an important factor leading to the development of human civilisation. Rice (Oryza sativa L.) is a vital staple crop worldwide, together with wheat and maize, and is domesticated from the wild rice O. rufipogon, which is widely distributed in tropical Asian countries (Khush 1997). During its domestication, several important characteristics were altered to make it more convenient for human use (Ishikawa et al. 2020).
Notably, inhibition of abscission layer formation is associated with reduced seed-shattering behaviour, and genetic analyses have revealed several loci involved in a loss of seed shattering based on natural variations in the degree of seed shattering in wild and cultivated rice species. A major locus of sh4 was identified by a quantitative trait locus (QTL) analysis of the degree of seed shattering of the F2 segregating population between the wild rice O. nivara (an annual form of O. rufipogon) and O. sativa indica rice (Li et al. 2006). The mutation at sh4 was caused by a single-nucleotide polymorphism (SNP) in the gene encoding the MYB transcription factor. All the cultivated rice investigated for their genotype at sh4 carried the mutation, indicating it played an important role in reducing seed shattering during rice domestication. Another locus of qSH1 was identified by QTL analysis using the F2 segregating population between two cultivated rice varieties, O. sativa Nipponbare and Kasalath (Konishi et al. 2006). The mutation was found to be an SNP in the regulatory region of the gene encoding a transcription factor, similar to Arabidopsis REPLUMLESS, and the mutation was found among japonica rice cultivars. The seed-shattering locus qSH3 was originally detected by QTL analysis (Onishi et al. 2007) and was further confirmed by the QTL analysis of the degree of seed shattering using the segregating population between O. sativa Nipponbare and O. rufipogon (Htun et al. 2014). The causal mutation at qSH3 was revealed to be an SNP in the previously identified gene of OsSh1, a homologue of the sorghum seed-shattering gene (Lin et al. 2012; Ishikawa et al. 2022). Interestingly, introgression lines (IL) carrying the domesticated allele at qSH3 in the wild genetic background showed complete seed-shattering behaviour as the wild rice O. rufipogon, suggesting that the single mutation at qSH3 in wild rice was insufficient to reduce seed shattering, and which is also observed for sh4 (Ishikawa et al. 2022). However, the ILs having mutations at both qSH3 and sh4 slightly inhibited the abscission layer formation, resulting in reduced seed-shattering behaviour (Inoue et al. 2015; Ishikawa et al. 2022). In our previous study, we reported qCSS3, a novel locus involved in reducing seed shattering in the japonica cultivar Nipponbare (Tsujimura et al. 2019). Studies of these loci suggested that a loss of seed shattering in cultivated rice can be established by the combination of variations at several loci with specific and common mutations among cultivars. Furthermore, we previously investigated the non-seed-shattering behaviour of the indica cultivar O. sativa IR36, one of the leading varieties grown in tropical Asia with semi-dwarf morphology and high resistance to major insects and disease (Spielmeyer et al. 2002; Ballini et al. 2007), and showed that qSH3 and sh4 were responsible for the reduced shattering phenotype (Ishikawa et al. 2017).
In this study, we aimed to understand the genetic loci involved in the reduced seed-shattering behaviour of IR36. We performed a QTL analysis for the degree of seed shattering on the segregating population between IR36 and wild rice IL carrying domesticated alleles at qSH3 and sh4. The loci identified in this study were further evaluated to confirm their roles in regulating rice seed shattering.
Material and methods
Plant materials
An indica rice cultivar O. sativa cv. IR36, a japonica rice cultivar O. sativa cv. Nipponbare, and the wild rice accession of O. rufipogon acc. W630 originated from Myanmar were used in this study. By backcrossing with the W630, an IL carrying the Nipponbare chromosomal segments covering the qSH3 and sh4 loci in the genetic background of wild rice was produced and named as IL(qSH3-Npb, sh4-Npb). The IL(qSH3-Npb, sh4-Npb) was crossed with IR36, and the resulting F1 plant was further backcrossed with IR36. Among 116 BC1F1 plants grown in 2016, No. 112 with higher seed-shattering behaviour was selected (Supplementary Table 2), and their self-pollinated BC1F2 seeds were obtained. A total of 166 BC1F2 individuals were grown in pots at Kobe University, Japan in 2017, with short-day treatments to induce similar flowering times, and their seed-shattering behaviour was evaluated. A progeny test was also conducted using two lines with heterozygous chromosomal constitution at the locus detected in this study.
Evaluation of the degree of seed shattering
To evaluate the degree of seed shattering, we measured the breaking tensile strength (BTS, gf: gram-force), which is the amount of force required to detach a grain from the pedicel, using a digital force gauge (FGP 0.5, Nidec-Shimpo Co., Japan). Approximately one month after heading, the BTS values of 75 seeds (25 randomly selected seeds from each of the three panicles) were measured, and their average BTS values were calculated.
DNA extraction, bulking, and library construction for next-generation sequencing analysis
Bulked DNA samples were prepared for QTL-seq analysis as described in previous studies (Abe et al. 2012; Takagi et al. 2013). DNA was extracted from 100 mg of fresh rice leaves using the DNeasy Plant Mini Kit (QIAGEN Sciences, Germany). Then their concentrations were quantified using the Qubit® 3.0 Fluorometer and Qubit® dsDNA BR Assay Kit (Life Technologies, Japan). Ten plants with the lowest and the highest BTS values were selected and genomic DNA from each plant was extracted and bulked in equal amounts into low- or high-BTS samples (L-bulk and H-bulk, respectively). The bulked samples and the IL(qSH3-Npb, sh4-Npb) as controls were subjected to whole-genome resequencing analysis using Illumina Hi-seq 2500 platform (paired-end 100).
Read processing
The Illumina short reads obtained from the bulked and control samples, IL(qSH3-Npb, sh4-Npb) and IR36, were processed as follows. Raw reads were first subjected to adapter and quality trimming using PEAT (Li et al. 2015) and Trimmomatic (Bolger et al. 2014), respectively. The parameters of Trimmomatic were: LEADING: 20, TRAILING: 20, SLIDINGWINDOW: 10: 20 and MINLEN: 20. Trimmed reads were aligned to the Nipponbare reference genome sequence obtained from the Rice Annotation Database Project (http://rapdb.dna.affrc.go.jp) using BWA-MEM (Li 2013) and transformed into BAM files using SAMtools (Danecek et al. 2021). Then, PCR duplicates were removed using Picard (http://broadinstitute.github.io/picard/). Resulting BAM files were used for the investigation of introgressed chromosomal segments and QTL-seq analysis.
Estimation of the chromosomal segments of Nipponbare and IR36
BAM files (PCR duplicates were removed) were processed into gVCFs using HaplotypeCaller function of GATK4 (Van der Auwera and O’Connor 2020). Each gVCF was combined into a VCF by GenomicsDBImport and GenotypeGVCFs functions of GATK4. SNP loci were selected, and quality filtered by VariantFiltration of GATK4 according to the following thresholds, QD < 5.0, QUAL < 30, SOR > 3.0, FS > 60.0, MQ < 50.0, MQRankSum < − 12.5 and ReadPosRankSum < − 8.0. SNP loci with missing data were further filtered using VCFtools (Danecek et al. 2011). Using the resulting VCF, we first estimated the chromosomal segments of Nipponbare that were introgressed in wild rice, O. rufipogon W630 genome using the Integrative Genomics Viewer (Robinson et al. 2011). We also investigated the IR36-homozygously fixed chromosomal regions in the BC1F2 population associated with backcrossing.
Detection of novel seed-shattering loci by QTL-seq analysis
The BAM files of the bulked samples and the IL(qSH3-Npb, sh4-Npb) were subjected to QTL-seq Pipeline (Takagi et al. 2013; Sugihara et al. 2022). The SNP-index was calculated for all the SNP positions. The Δ(SNP-index) was then calculated by subtracting the SNP-index values of the L-bulk from those of the H-bulk. A sliding window analysis was applied by averaging the Δ(SNP-index) values within a 500 kb window size and a 25 kb increment. Regions with averaged Δ(SNP-index) values exceeding the 95% confidence intervals were regarded as statistically significant loci under the null hypothesis of no QTL (P < 0.05). The 95% confidence intervals under the null hypothesis were preliminary defined for each read depth and are implemented on the QTL-seq pipeline (Takagi et al. 2013). Simple sequence repeat (SSR) or indel markers were used to survey the genotypes at the detected loci (Supplementary Table 1).
Morphological and histological analysis of abscission layer formation
A Leica Biosystems S6D microscope (Germany) was used to examine the abscission layer (axial images of detached spikelets), and MC170HD and the Leica Application Suite were used for photography (Leica Biosystems, Germany). Histological analysis of abscission layer formation was carried out using the pedicel tissue of grains before heading, as previously reported (Htun et al. 2014; Inoue et al. 2015). FAA solution (formaldehyde: acetic acid: 70% ethanol = 1:1:18 [volume ratio]) was used to fix the samples, followed by vacuum infiltration and preservation at 4 °C. They were dehydrated in an ethanol series (70%, 80%, 90% and 95% ethanol) for 2 days at each stage and then embedded in Technovit 7100 resin (Heraeus Kulzer, Germany), according to the manufacturer’s instructions. The samples were cut into 3-μm sections using an RM1215RT rotary microtome (Leica Biosystems, Germany), and toluidine blue O solution was used for staining. These sections were observed under a microscope and photographed with a digital camera using the imaging software, ToupView imaging software (× 86) (AmScope.com, USA).
Results
Characterisation of the seed-shattering behaviour of the IL(qSH3-Npb, sh4-Npb)
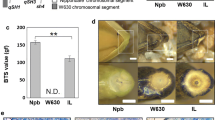
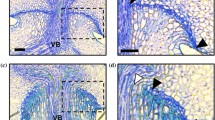
As reported previously, IR36 carries non-functional alleles at the causal SNPs of both qSH3 and sh4, and a functional allele at the causal SNP of qSH1 (Ishikawa et al. 2017). We previously developed an IL between O. sativa japonica Nipponbare and O. rufipogon W630. The IL(qSH3-Npb, sh4-Npb) carries O. sativa japonica Nipponbare chromosomal segments covering both qSH3 and sh4 in the genetic background of wild rice, O. rufipogon W630 (Ishikawa et al. 2022). Here, we confirmed that the IL carries domesticated alleles at qSH3 and sh4, with a functional allele at qSH1 (Fig. 1a). The appearance of the seed and panicle of the IL was similar to that of wild rice, O. rufipogon W630 (Fig. 1b and c). First, we investigated the seed-shattering behaviour of the IL. Comparison of the morphology of the spikelet base for IR36, W630, and IL(qSH3-Npb, sh4-Npb) revealed similar detachment at the abscission zone (Fig. 1d), but the degree of seed shattering was different; IR36 showed a BTS of 56.5 ± 4.3 gf, while W630 completely shed seeds at maturation. Comparatively, IL(qSH3-Npb, sh4-Npb) had a detectable BTS value of 10.7 ± 0.5 gf, which is lower than that of IR36 (Fig. 1e). We also confirmed that the differences in the degree of seed shattering can be attributed to the degree to which abscission layer formation was inhibited (Fig. 1f). In agreement with the findings of our previous study (Ishikawa et al. 2022), an increase in the BTS value was proportionally associated with an increase in abscission layer inhibition. Based on the analysis of the seed-shattering behaviour of the IL(qSH3-Npb, sh4-Npb), novel seed-shattering loci may still contribute to reduced seed shattering in IR36.
Characterisation of the introgression line (IL), IL(qSH3-Npb, sh4-Npb), carrying O. sativa japonica cv. Nipponbare (Npb) chromosomal segments covering qSH3 and sh4 loci in the genetic background of wild rice, O. rufipogon W630. a Genotyping at qSH1, sh4, and qSH3 for O. sativa japonica cv. Nipponbare, indica cv. IR36, O. rufipogon acc. W630, and the IL(qSH3-Npb, sh4-Npb) based on the causal single-nucleotide polymorphisms (SNPs) detected by derived cleaved amplified polymorphic sequence (dCAPS) markers (Supplementary Table 1). − and + represent PCR fragments undigested and digested PCR fragments with enzymes, respectively. b Seeds of Nipponbare, IR36, W630, and IL(qSH3-Npb, sh4-Npb). Scale bar = 5 mm. c Panicle morphologies of Nipponbare, IR36, W630, and IL(qSH3-Npb, sh4-Npb) approximately one month after flowering. Scale bar = 5 cm. d A close view of the spikelet base in IR36, W630, and IL (from left to right). Scale bars = 1 mm. e The breaking tensile strength (BTS) values for IR36, W630, and IL(qSH3-Npb, sh4-Npb). Data are mean ± S.D. (n = 3). ** indicates P < 0.01 by an unpaired Student’s t-test. N.D., not determined owing to complete seed shattering. f Longitudinal sections of the spikelet base after seed detachment in IR36 (left), W630 (centre), and IL(qSH3-Npb, sh4-Npb) (right). VB vascular bundle. AL abscission layer. Black triangles indicate both edges of the abscission layer. Scale bars = 100 μm
The degree of seed shattering in segregating populations obtained by crossing IR36 and IL(qSH3-Npb, sh4-Npb)
We previously observed segregation in the degree of seed shattering in an F2 population between IR36 and IL(qSH3-Npb, sh4-Npb). However, due to differing heading dates and morphological traits, proper evaluation of the degree of seed shattering was difficult (Tsujimura et al. 2017). To avoid segregation of other traits, we backcrossed an F1 plant with IR36 (Supplementary Fig. 1). A total of 116 BC1F1 plants were obtained with heading dates ranging from 1 to 18 August 2017 (Supplementary Table 2). Approximately, a month after heading, we evaluated the seed-shattering degree by measuring the BTS value for grain detachment of each BC1F1 plant, and the resulting values ranged from 18.8 gf to 37.9 gf (Fig. 2). Among these plants, we selected No. 112, as it had a low BTS value (19.7 gf). We expected the lower BTS value of the plant to be attributed to the W630-dominant allele(s) at the novel seed-shattering loci. The BTS values of the resulting BC1F2 plants were further evaluated for their seed-shattering degree. The BTS values of BC1F2 plants showed contentious segregation, with BTS values mostly falling between those of their parents: IL(qSH3-Npb, sh4-Npb), 12.2 ± 3.4 gf; and IR36, 45.6 ± 16.8 gf (Fig. 3; Supplementary Table 3). Because the qSH3 and sh4 alleles are fixed homozygously with domesticated alleles, the segregation of the degree of seed shattering in this population was likely caused by novel seed-shattering loci.
Frequency distribution of breaking tensile strength (BTS) values for 116 BC1F1 individuals between Oryza sativa IR36 and IL(qSH3-Npb, sh4-Npb). The average BTS values for the parent lines are indicated by black triangles. IR36: 34.3 ± 3.3 gf (n = 5) and the IL(qSH3-Npb, sh4-Npb), 11.6 ± 0.8 (n = 5) (mean ± SD). The BTS values for each BC1F1 individual are shown in Supplementary Table 2
Frequency distribution of breaking tensile strength (BTS) values for 166 BC1F2 individuals between Oryza sativa IR36 and IL(qSH3-Npb, sh4-Npb), used for QTL-seq analysis. The average BTS values for the parent lines and their F1 are shown with black triangles. IR36: 45.6 ± 16.8 gf (n = 4), the IL(qSH3-Npb, sh4-Npb): 12.2 ± 3.4 gf (n = 4), and F1: 19.9 ± 5.6 gf (n = 4) (mean ± SD). The DNA samples of BC1F2 plants with BTS values between 11.8 and 16.2 gf were selected as low (L) bulk and those with BTS values between 44.7 and 66.0 gf were selected as high (H) bulk for further studies. The BTS values for each BC1F2 individual are shown in Supplementary Table 3
Detection of novel loci involved in non-seed-shattering behaviour of IR36 by QTL-seq analysis
To estimate the genetic region(s) underlying differences in the seed-shattering degree between IR36 and IL(qSH3-Npb, sh4-Npb), we subjected their BC1F2 plants for QTL-seq analysis (Takagi et al. 2013; Sugihara et al. 2022). A total of 100.3, 105.7, 56.1, and 51.0 million sequence reads (each 100 bp) were obtained from the DNA of H-bulk, L-bulk, IL, and IR36, respectively. Using the Illumina short read data, we first estimated the chromosomal segments of Nipponbare that were introgressed in the wild rice, O. rufipogon W630 genome. In IL(qSH3-Npb, sh4-Npb), we found the chromosomal segments on chrs. 1, 3, 4, 5, 6, 7, 11, and 12 were derived from Nipponbare (Supplementary Table 4). We also investigated the IR36-homozygously fixed chromosomal regions in the BC1F2 population associated with backcrossing (Supplementary Table 5), suggesting that these regions are unlikely to be responsible for the difference in the seed-shattering degree of the BC1F2 population. According to the introgressed chromosomal segments of Nipponbare in IL(qSH3-Npb, sh4-Npb), those on chromosomes 1 and 12 were not transmitted because of IR36-backcrossing, while partial or whole segments on chromosomes 3, 4, 5, 6, 7 and 11 were transmitted to BC1F2 population (Supplementary Tables 3 and 4). Notably, the Nipponbare segment on chromosome 4 was present in BC1F2 population, but the sh4 genotype was fixed with domesticated allele of Nipponbare and IR36, that are identical (Fig. 1a). By examining the Δ(SNP-index) plot, we detected the two genomic regions with averaged Δ(SNP-index) values exceeding the 95% confidence interval by a sliding window analysis (statistical significance under the null hypothesis: P < 0.05): the regions on chr. 2 from 34.82 to 34.95 Mb with maximum Δ(SNP-index) = 0.43 (statistical significance under the null hypothesis: P < 0.05), and on chr. 7 from 19.43 to 23.15 Mb with maximum Δ(SNP-index) = 0.48 (Fig. 4). These two loci were named as QTL for the Control of Seed Shatteringin rice on chromosomes 2 and 7 (qCSS2 and qCSS7, respectively). Other regions exceeding the confidence interval were not detected in this analysis (Supplementary Fig. 2). Since the two regions were detected based on the segregation of IR36 and W630 chromosomal segments (Supplementary Tables 3 and 4), we estimated the qCSS2 and qCSS7 were likely to contribute to reduced seed-shattering behaviour in IR36.
Detection of novel loci for seed shattering on chromosomes 2 (qCSS2) and 7 (qCSS7) by QTL-seq analysis. qCSS2 and qCSS7 were detected in approximately the 0.13-Mb and 3.73-Mb regions on chromosomes 2 (a) and 7 (b), respectively. The Δ(SNP-index) plots with statistical intervals under the null hypothesis of no QTL (orange, P < 0.01; green; P < 0.05) are shown. The red line indicates the average Δ (SNP-index) calculated by a sliding window analysis
To further confirm the results of QTL-seq analysis, genotypes at qCSS2 and qCSS7 regions were surveyed for each BC1F2 plant using DNA markers flanking the regions on chromosomes 2 or 7, respectively (Supplementary Table 1). Regarding qCSS2, the region estimated by QTL-seq analysis was between Indel2-3 and RM208. Genotyping analysis showed that 40 (24.1%) and 39 (23.5%) of BC1F2 plants carried the IR36 and W630 homozygous chromosomal segments, respectively (Supplementary Table 3). A comparison of the BTS values of the two homozygous groups based on the chromosomal constitutions showed that plants with the IR36 chromosomal segment tended to have higher BTS values than those with W630 (Supplementary Fig. 3). Similarly, we analysed the qCSS7 region and found that 15 (9.0%) and 22 (13.3%) of BC1F2 plants carried IR36 and W630 homozygous chromosomal segments, respectively. We also found that plants with IR36 chromosomal segments at qCSS7 region tended to exhibit higher BTS values than those with W630 (Supplementary Fig. 3). These surveys of the BTS values of each BC1F2 plant confirmed that both qCSS2 and qCSS7 regions were likely to be involved in the non-seed-shattering behaviour of IR36.
Validation of the effect of qCSS2 and qCSS7 on seed-shattering degree
To further confirm the effect of the two loci on seed shattering, we carried out a progeny test for both the qCSS2 and qCSS7 loci. Two BC1F2 lines that carrying heterozygous chromosomal constitutions covering qCSS2 or qCSS7 candidate regions were selected (No. 95 for qCSS2 and No. 160 for qCSS7). Their self-pollinated seeds of BC1F3 generation were germinated and then their genotypes were confirmed to have plants as one of the two loci fixed with the IR36 chromosomal segment based on the DNA markers in the region (Supplementary Table 1). For qCSS2, a progeny test using BC1F3 plants from No. 95 showed that plants with the IR36 chromosomal segment had a BTS of 73.0 ± 14.2 gf, while those with the W630 chromosomal segment had a BTS of 45.3 ± 6.5 gf, with a significant difference in their BTS values (Fig. 5a and b). Similarly, the effect of the qCSS7 region on seed shattering was analysed using the progenies of two BC1F2 plants (No. 160). Difference in the BTS values was detected between plants carrying the IR36 and W630 chromosomal segments covering the qCSS7 region (Fig. 5c and d). Taken together, the progeny test for qCSS2 and qCSS7 indicated that these two loci play important roles in controlling the seed-shattering degree in rice.
Validation of the effect of qCSS2 and qCSS7 on seed shattering by subsequent progeny test. a Progeny test of qCSS2. Graphical genotype of BC1F3 plants (No. 95). b Comparison of the average breaking tensile strength (BTS) values for the BC1F3 plants are shown. c Progeny test of qCSS7. Graphical genotype of BC1F3 plants (No. 160). d Comparison of the average BTS values for the BC1F3 plants are shown. Data are mean ± SD (n = 3–5). Estimated regions of qCSS2 and qCSS7 are shown based on the result of QTL-seq analysis, with the 95% confidence intervals. Genotypes at qCSS2 and qCSS7 regions were estimated by the corresponding DNA markers, and the average of the BTS values were calculated. ** indicates P < 0.01 by unpaired Student’s t-test
Genetic interaction between qCSS2 and qCSS7 on reducing the degree of seed shattering in wild rice genetic background
To further confirm the effect of the qCSS2 and qCSS7 loci on reducing the degree of seed shattering during rice domestication, we produced ILs that carry IR36 chromosomal segments covering qCSS2 and qCSS7 under the presence of qSH3 and sh4 mutations in the genetic background of wild rice, O. rufipogon W630. We previously evaluated backcrossed recombinant inbred lines (BRILs) between IR36 and O. rufipogon W630 (Ishikawa et al. 2017). Among these, we found that two independent lines, namely BRIL-A4 and BRIL-H6, sustained more grains than the other lines (Supplementary Fig. 4). We previously genotyped them at known seed-shattering loci of qSH3 and sh4, finding that both carried IR36 domesticated alleles (Ishikawa et al. 2017). Interestingly, we noticed that the A4 and H6 lines contained IR36 chromosomal segments covering the qCSS2 and qCSS7 regions, respectively (Supplementary Fig. 4), suggesting that these two loci may contribute to further reductions in seed shattering of the two lines. To understand the genetic interaction between these two loci under the presence of qSH3 and sh4 mutations, we crossed these two lines, and the resulting F1 plants were self-pollinated to obtain F2 seeds. We employed two SSR markers for each of qCSS2 and qCSS7 to investigate the chromosomal constitutions covering the two loci. Based on the genotypes of the two loci, we produced four ILs: namely IL(qSH3-IR, sh4-IR), IL(qSH3-IR, sh4-IR, qCSS2-IR), IL(qSH3-IR, sh4-IR, qCSS7-IR), and IL(qSH3-IR, sh4-IR, qCSS2-IR, qCSS7-IR) in wild rice genetic background of O. rufipogon W630 (Supplementary Fig. 5). Seeds retained in the panicles of the four lines tended to be greater as the number of IR-chromosomal segments at qCSS2 and qCSS7 increased (Supplementary Fig. 6). We measured their BTS values together with the controls of O. rufipogon W630 and O. sativa IR36 (Fig. 6a). A proportional increase in the BTS values was observed depending on the number of seed-shattering loci with domesticated alleles (Fig. 6a). The BTS values for IL(qSH3-IR, sh4-IR) was 9.8 ± 1.4 gf. A slight increase in the BTS values of 11.7 ± 4.7 gf and 13.6 ± 4.7 gf were observed for IL(qSH3-IR, sh4-IR, qCSS2-IR) and IL(qSH3-IR, sh4-IR, qCSS7-IR), respectively, while significantly larger BTS values of 25.9 ± 1.6 gf were obtained for IL(qSH3-IR, sh4-IR, qCSS2-IR, qCSS7-IR). We then carried out a two-way analysis of variance (ANOVA) to investigate the relationship between qCSS2 and qCSS7 and found a significant digenic interaction (P < 0.05). Interestingly, no significant difference was detected between IL(qSH3-IR, sh4-IR, qCSS2-IR, qCSS7-IR) and IR36 (30.5 ± 4.4 gf) (Fig. 6a). We also analysed abscission layer formation for these ILs together with O. rufipogon W630 and O. sativa IR36. We found a proportional increase in the inhibition of abscission layer formation that was correlated with the BTS values (Fig. 6b). These results imply that the non-seed-shattering behaviour of IR36 can be controlled by the variations at least four seed-shattering loci: qSH3, sh4, qCSS2, and qCSS7.
Evaluation of the genetic effect of the IR36 alleles at qCSS2 and qCSS7 on seed-shattering degree in the genetic background of wild rice, O. rufipogon W630. a Evaluation of seed-shattering degree by BTS values in the ILs of qCSS2 and qCSS7 with qSH3 and sh4. ** and n.s. indicate significance (P < 0.01) and not significant (P ≥ 0.05) by unpaired Student’s t-test, respectively. N.D. Not determined owing to complete seed shattering. b Longitudinal sections of the spikelet base after seed detachment in W630, and the four ILs, and IR36. VB vascular bundle. AL abscission layer. Black triangles indicate both edges of the abscission layer. Scale bars = 50 μm. O. rufipogon W630 and O. sativa IR36 were used as the control
Discussion
Genetic dissection of quantitatively controlled non-seed-shattering behaviour of an indica rice cultivar, IR36
As Nipponbare and IR36 share the same alleles at qSH3 and sh4, the reduced seed-shattering characteristics of IR36 cannot be fully explained by these two alleles alone. Based on this observation, we conducted a genetic analysis to detect additional factor(s) contributing to the reduced seed-shattering behaviour of IR36. By backcrossing the F1 plant between IR36 and IL(qSH3-Npb, sh4-Npb), we first evaluated their BC1F1 population to screen the plants with the lowest BTS values. Based on the QTL-seq analysis using BTS values in the BC1F2 population, we identified two novel loci, qCSS2 and qCSS7. The effect of these loci on seed shattering was evaluated in each plant of the BC1F2 population and subsequent progeny tests (Supplementary Fig. 3, Fig. 5), confirming their roles in controlling seed shattering.
To investigate the genetic basis of the non-seed shattering behaviour of IR36, we produced ILs between IR36 and W630. Among the BRILs produced previously, we selected those carrying domesticated alleles at qSH3 and sh4, after screening their genotypes at these two loci (Ishikawa et al. 2017). Interestingly, the two lines exhibited relatively higher BTS values than those of the other lines. We noticed that these lines carried IR36 chromosomal segments in either the regions covering qCSS2 or qCSS7 (Supplementary Fig. 4). The high BTS values of these lines may be associated with sh4, qSH3, and qCSS2, or qCSS7. The BTS values of IL(qSH3-IR, sh4-IR, qCSS2-IR, qCSS7-IR) were significantly higher than those of IL(qSH3-IR, sh4-IR, qCSS2-IR) or IL(qSH3-IR, sh4-IR, qCSS7-IR), thereby confirming that the two loci play roles in reducing seed shattering (Fig. 6).
In the present study, we detected qCSS2 on chromosome 2 (Figs. 4 and 5). This region is closely linked to Sh13, which locates between RM208 and RM207 and was originally identified as a major locus responsible for the difference in the degree of seed shattering between cv. Oonari and cv. Takanari, moderate and easy-shattering rice cultivars, respectively (Li et al. 2019). The tendency toward seed shattering of Takanari was improved by gamma-ray irradiation-mediated mutagenesis, and the plants with reduced seed shattering were screened. Genetic analysis identified a responsible locus containing osa-mir172d miRNA, which was previously shown to control the grain threshability of wheat (Debernardi et al. 2017). Furthermore, upregulation of the mir172 miRNA is likely associated with gene duplication. The involvement of osa-mir172d miRNA is vital for understanding the regulation of seed shattering in rice, however, this mutation was obtained by artificially induced mutagenesis of gamma-ray irradiation (Li et al. 2019). A survey of the sequence of osa-mir172d in O. rufipogon W630, O. sativa Nipponbare, and IR36 showed that they share the same sequence for the primary transcript (Supplementary Fig. 7). Therefore, we posited here that osa-mir172d miRNA is unlikely to be involved in the difference of seed shattering degree in our segregating population, although further mapping experimentation is required to clarify the precise involvement of the osa-mir172d miRNA in qCSS2. Regarding qCSS7, a QTL named sh7.1 was previously detected near RM214 (approximately 12.78 Mb) on chromosome 7 using an advanced backcross population between O. rufipogon and O. sativa cv. Jefferson (Thomson et al. 2003). However, the position of qCSS7 (19.43 to 23.15 Mb by QTL-seq analysis) is far from sh7.1. As there are no previously known seed-shattering loci near qCSS7, novel factor(s) may underlie this region. Accordingly, further genetic dissections at the two loci are required to narrow down the candidate regions for identifying the causal mutations at qCSS2 and qCSS7. Taken together, qCSS2 and qCSS7 may encode novel genes or mutations distinct from those induced artificially.
Common and distinct loci involved in non-seed-shattering behaviour in japonica and indica rice cultivars
In this study, we detected two novel loci, qCSS2 and qCSS7, involved in reduced seed-shattering behaviour of an indica rice cultivar IR36 (Fig. 4). In contrast, we previously detected qCSS3, a novel locus involved in the reduced seed-shattering behaviour of the japonica rice cultivar O. sativa Nipponbare (Tsujimura et al. 2019). The qCSS3 was detected by QTL-seq analysis of F2 plants between Nipponbare and the IL(qSH1-Npb, sh4-Npb, qSH3-Npb), which carries Nipponbare chromosomal segments covering qSH1, sh4, and qSH3 regions in the genetic background of wild rice, O. rufipogon W630. In our QTL-seq analysis of reduced-seed-shattering behaviour in IR36, the qCSS3 region, located ~ 8.30 to 13.65 Mb, was not detected (Supplementary Fig. 2). This suggests that qCSS3 is unlikely to be involved in the reduced seed-shattering behaviour of IR36. In addition, qSH1 has also been detected as a locus involved in the reduction of seed shattering in some japonica rice cultivars (Konishi et al. 2006). As previously reported, the qSH1 malfunction allele is not observed in indica rice cultivars (Zhang et al. 2009), it is specific to japonica rice cultivars. Regarding our previous QTL-seq analysis of the non-seed-shattering behaviour of Nipponbare, no peaks associated with seed shattering were observed in either the qCSS2 or qCSS7 regions (Tsujimura et al. 2019), suggesting that the two loci detected in this study are unlikely to be involved in the control of seed shattering in japonica rice cultivars.
Asian rice is largely classified into two groups: indica and japonica. In both subspecies, two seed-shattering loci of qSH3 and sh4 are commonly involved in reducing seed shattering, while those at qSH1 and qCSS3 or qCSS2 and qCSS7 are specific to their cultivar group. Interestingly, circum-aus rice cultivars carry a functional (non-domesticated) allele at qSH3 (Ishikawa et al. 2022). This finding is of particular interest since we do not know whether the seed-shattering loci detected using Nipponbare and IR36 are also involved in reduced seed-shattering behaviour for circum-aus rice cultivars. Once the causal mutations at the qCSS2, qCSS3, and qCSS7 loci are identified, the survey of mutations at these loci for indica, japonica, and circum-aus cultivars will provide an important scope for understanding the genetic basis of reduced seed shattering in rice. Our findings imply that a distinct history of selections may underlie the process of rice domestication. Seed-shattering loci with specific roles can be important for controlling seed shattering in cross-breeding programmes among rice cultivars with diverse origins, and are also informative in clarifying the processes of rice domestication.
Quadratic increase in BTS followed by integration of domesticated alleles at seed-shattering loci
We previously reported that a single mutation at one of the known seed-shattering loci was not sufficient to inhibit abscission layer formation in wild rice genetic background (Ishii et al. 2013, Htun et al. 2014; Inoue et al. 2015). Accordingly, further investigation of the individual effects of qCSS2 or qCSS7 domesticated alleles alone on the loss of seed shattering is necessary to confirm their effect in wild rice. Notably, these regions have not been previously studied in the genetic analyses of seed shattering in rice, and it is likely that their effects are smaller than those of major loci, thus, an effect from either single mutation may not be observed in wild rice. In IL(qSH3-IR, sh4-IR), a slight inhibition of the abscission layer was observed around the central vascular bundles (Fig. 6b). Additionally, the IL(qSH3-IR, sh4-IR, qCSS2-IR) or IL(qSH3-IR, sh4-IR, qCSS7-IR), which have an IR36 chromosomal segment at both of qSH3 and sh4 and either of qCSS2 or qCSS7 in the wild genetic background, resulted in increased inhibition of the abscission layer from the central vascular bundles (Fig. 6b). This increase in abscission layer inhibition was associated with a slight increase in the BTS values (Fig. 6a). A large increase in the BTS value was observed for plants with quadruple introgressions; IL(qSH3-IR, sh4-IR, qCSS2-IR, qCSS7-IR). Compared to IL(qSH3-IR, sh4-IR, qCSS2-IR) or IL(qSH3-IR, sh4-IR, qCSS7-IR), a further increase in the length of inhibited region in abscission layer in IL(qSH3-IR, sh4-IR, qCSS2-IR, qCSS7-IR) in vertical sections was similar to that caused by the additional effects of qCSS2 or qCSS7 with qSH3 and sh4 (Fig. 6b). The notably high BTS value of IL(qSH3-IR, sh4-IR, qCSS2-IR, qCSS7-IR) can be explained by the inhibited area of the abscission layer, as the area of abscission layer inhibition can be calculated using the radius squared (Ishikawa et al. 2022). These results suggest that the quadratic increase in the BTS values with the accumulation of the domesticated alleles at seed-shattering loci may be the genetic basis of the reduced seed-shattering behaviour of cultivated rice. This increase in the degree of seed shattering may be associated with the development of harvesting tools during domestication. In the early stages of domestication, primitive tools were ideal for harvesting, whereas more advanced tools were employed in the later eras. Evaluating the effect of common and subspecies-specific seed-shattering loci that are associated with reduced seed shattering will be particularly informative in understanding the process of rice domestication in terms of phenotypic changes to rice and thus the development of human civilisation.
Data availability
Raw fastq reads for High-bulk, Low-bulk, IL(qSH3-Npb, sh4-Npb), and IR36 were deposited in the Sequence Read Archive (SRA) under DRA accession number DRA016170.
Change history
15 June 2023
Error in the reference has been updated
23 June 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00438-023-02047-9
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178. https://doi.org/10.1038/nbt.2095
Ballini E, Berruyer R, Morel JB, Lebrun MH, Nottéghem JL, Tharreau D (2007) Modern elite rice varieties of the ‘Green Revolution’ have retained a large introgression from wild rice around the Pi33 rice blast resistance locus. New Phytol 175:340–350
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H (2021) Twelve years of SAMtools and BCFtools. GigaScience 10:008. https://doi.org/10.1093/gigascience/giab008
Debernardi JM, Lin HQ, Chuck G, Faris JD, Dubcovsky J (2017) microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144:1966–1975
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321. https://doi.org/10.1016/j.cell.2006.12.006
Htun TM, Inoue C, Chhourn O, Ishii T, Ishikawa R (2014) Effect of quantitative trait loci for seed shattering on abscission layer formation in Asian wild rice Oryza rufipogon. Breed Sci 64:199–205. https://doi.org/10.1270/jsbbs.64.199
Inoue C, Htun TM, Inoue K, Ikeda K, Ishii T, Ishikawa R (2015) Inhibition of abscission layer formation by an interaction of two seed-shattering loci, sh4 and qSH3, in rice. Genes Genet Syst 90:1–9. https://doi.org/10.1266/ggs.90.1
Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, Ishikawa R, Ashikari M (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45:462–465. https://doi.org/10.1038/ng.2567
Ishikawa R, Nishimura A, Htun TM, Nishioka R, Oka Y, Tsujimura Y, Inoue C, Ishii T (2017) Estimation of loci involved in non-shattering of seeds in early rice domestication. Genetica 145:201–207. https://doi.org/10.1007/s10709-017-9958-x
Ishikawa R, Castillo CC, Fuller DQ (2020) Genetic evaluation of domestication-related traits in rice: implications for the archaeobotany of rice origins. Archaeol Anthropol Sci 12:197. https://doi.org/10.1007/s12520-020-01112-3
Ishikawa R, Castillo CC, Htun TM, Numaguchi K, Inoue K, Oka Y, Ogasawara M, Sugiyama S, Takama N, Orn C, Inoue C, Nonomura KI, Allaby R, Fuller DQ, Ishii T (2022) A stepwise route to domesticate rice by controlling seed shattering and panicle shape. Proc Natl Acad Sci USA 119:e2121692119. https://doi.org/10.1073/pnas.2121692119
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:25–34. https://doi.org/10.1023/A:1005810616885
Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312:1392–1396. https://doi.org/10.1126/science.1126410
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
Li C, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939. https://doi.org/10.1126/science.1123604
Li YL, Weng JC, Hsiao CC, Chou MT, Tseng CW, Hung JH (2015) PEAT: an intelligent and efficient paired-end sequencing adapter trimming algorithm. BMC Bioinform 16:S2. https://doi.org/10.1186/1471-2105-16-S1-S2
Li F, Numa H, Hara N, Sentoku N, Ishii T, Fukuta Y, Nishimura N, Kato H (2019) Identification of a locus for seed shattering in rice (Oryza sativa L.) by combining bulked segregant analysis with whole-genome sequencing. Mol Breed 39:36. https://doi.org/10.1007/s11032-019-0941-3
Lin Z, Li X, Shannon LM, Yeh CT, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, Doebley J, Schnable PS, Tuinstra MR, Tesso TT, White F, Yu J (2012) Parallel domestication of the Shattering1 genes in cereals. Nat Genet 44:720–724. https://doi.org/10.1038/ng.2281
Onishi K, Horiuchi Y, Ishigoh-Oka N, Takagi K, Ichikawa N, Maruoka M, Sano Y (2007) A QTL cluster for plant architecture and its ecological significance in Asian wild rice. Breed Sci 57:7–16. https://doi.org/10.1270/jsbbs.57.7
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99:9043–9090
Sugihara Y, Young L, Yaegashi H, Natsume S, Shea DJ, Takagi H, Booker H, Innan H, Terauchi R, Abe A (2022) High performance pipeline for MutMap and QTL-seq. PeerJ 10:e13170
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183. https://doi.org/10.1111/tpj.12105
Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493. https://doi.org/10.1007/s00122-003-1270-8
Tsujimura Y, Inoue C, Htun TM, Oka Y, Ishii T, Ishikawa R (2017) Investigation of genetic loci controlling non-shattering behaviour in an Indica rice cultivar ‘IR36'. J Crop Res 62:19–23. https://doi.org/10.18964/jcr.62.0_19
Tsujimura Y, Sugiyama S, Otsuka K, Htun TM, Numaguchi K, Castillo C, Akagi T, Ishii T, Ishikawa R (2019) Detection of a novel locus involved in non-seed-shattering behaviour of Japonica rice cultivar, Oryza sativa “Nipponbare”. Theor Appl Genet 132:2615–2623. https://doi.org/10.1007/s00122-019-03376-3
Van der Auwera GA, O’Connor BD (2020) Genomics in the cloud: using Docker, GATK, and WDL in Terra, 1st edn. O’Reilly Media, Sebastopol
Zhang LB, Zhu O, Wu ZO, Ross-Ibarra J, Gaut BS, Ge S, Sang T (2009) Selection on grain shattering genes and rates of rice domestication. New Phytol 184:708–720. https://doi.org/10.1111/j.1469-8137.2009.02984.x
Acknowledgements
The wild rice accession, O. rufipogon W630, was provided by the National Institute of Genetics supported by the National Bioresource Project, MEXT, Japan. This study was supported in part by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) (Grant numbers: 15KK0280, 18K05594, and 23H02183) to R.I. and by the JSPS Bilateral Open Partnership Joint Research Project (Grant numbers: JPJSBP120189948 and JPJSBP120219922) to R.I., and the Nikki Saneyoshi and Kinoshita Foundations to R.I.
Funding
Open access funding provided by Kobe University.
Author information
Authors and Affiliations
Contributions
RI conceived and designed the study. SS, SM, YT, YY. TMH, CI, and RI performed the experiments. SS, SM, TI and RI analysed the data. SS, YT, YY, and KN contributed to bioinformatic analysis. SS, SM, TI and RI prepared the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all the authors, the corresponding author states that there are no conflicts of interest.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sugiyama, S., Sakuta, M., Tsujimura, Y. et al. Detection of novel loci involved in non-seed-shattering behaviour of an indica rice cultivar, Oryza sativa IR36. Mol Genet Genomics 298, 943–953 (2023). https://doi.org/10.1007/s00438-023-02027-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-023-02027-z