Abstract
Due to the unique affinity of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to the angiotensin-converting enzyme 2 (ACE2) receptor in patients, the foremost recent evidence indicated that ACE1 and ACE2 polymorphisms could affect the susceptibility of individuals to SARS-CoV-2 infection and also the disease outcome. Here, we aimed to assess the possible association between two polymorphisms and the severity of disease in patients. In the present study, 146 patients with COVID-19 who were admitted to the Mazandaran University of Medical Sciences hospitals between March 2020 and July 2020 were enrolled in this case–control study. The patients were divided into four groups based on clinical symptoms and severity of the diseases (mild, moderate, severe, and critical). After DNA extraction, the ACE gene I/D polymorphism (rs4646994) and ACE2 gene polymorphism (rs2285666) were genotyped using Gap-PCR and PCR–RFLP techniques, respectively. Then, five samples from each obtained genotype were confirmed by Sanger sequencing technique. Data were analyzed with SAS software version 9.1 using appropriate statistical procedures. The ACE gene I/D polymorphism (rs4646994) genotypes were classified into three types: I/I, I/D, and D/D. Our finding indicated that the prevalence of ACE1 D/D genotype was significantly higher in severe and critical COVID-19 patients (P = 0.0016). Additionally, the analysis revealed a remarkable association between rs4646994 SNP and the HB and ESRI levels in patients (P < 0.05). Although the ACE2 rs2285666 SNP was not related to the severity of disease, this variant was significantly associated with ALT, ESRI, and P. These results provide preliminary evidence of a genetic association between the ACE-D/D genotype and the D allele of ACE1 genotype and the disease severity. Therefore, our findings might be useful for identifying the susceptible population groups for COVID-19 therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to the SARS-CoV-2 virus, a novel virus from the Coronaviridae family, the world has been struggling with the epidemic of the COVID-19 (World Health Organization 2020). COVID-19 has been impacting an outsized number of individuals worldwide. Evidence indicated the COVID-19 case mortality rate had varied among nations. So far, around 7.2 million cases have been reported in Iran in keeping with data reported by WHO with 140,000 confirmed deaths.
The clinical spectrum of the COVID-19 virus appears very diverse, starting from mild flu to severe pneumonia and even acute respiratory distress syndrome (ARDS) (Xia et al. 2020). Other complications include septic shock, metabolic acidosis, coagulopathy, acute cardiac and arrhythmic damage, secondary infections and multiple organ damage, neurological and ocular manifestations, and renal and hepatic dysfunction (Chang et al. 2020; Pambuccian 2020). It has been hypothesized that viral infection has a severe inflammatory response and leads to severe lung damage that may require ICU admission, mechanical ventilation, and increase the risk of multiple organ failure and death (Jose and Manuel 2020).
The renin–angiotensin–aldosterone (RAAS) system appears to play a crucial role in the COVID-19 pathogenesis (Ingraham et al. 2020). Angiotensin-converting enzyme (ACE or ACE1) catalyzes the synthesis of angiotensin-II (Ang-II) from Ang-I, and ACE2 hydrolyzes Ang-II to Ang-1-7. Ang-II binds to the AT1 receptor, causing vasoconstriction, fibrosis, inflammation, thrombosis, and other responses. Ang-1-7, on the other hand, binds to the AT2 receptor, dilating blood vessels and reducing fibrosis, inflammation, and thrombosis. Thus, ACE and ACE2 play a key and opposite role in the balance that leads to the risk of hypertension and cardiovascular disease. In the lung, ACE2 produces a protective response by reducing edema, permeability, and lung damage (Kuba et al. 2005). Previous studies revealed that the high ACE activity increases the risk of pulmonary and cardiovascular disease by increasing Ang-II/AT1R axis activity.
The human ACE gene is located on chromosome 17 with 21 kb long, and contains 26 exons and 25 introns (Riordan 2003). The genetic variants in the two ACE1 and ACE2 genes are associated with various diseases such as the risk of hypertension, heart disease, kidney failure, and lung disease. Reports revealed associations of COVID-19 severity with genetic variants in RAAS-related genes including ACE1, ACE2. From a systematic review, ACE1 rs4646994 and ACE2 rs2285666 were noted as common polymorphisms of RAAS-related genes. Also, meta-analyses studies uncovered that both ACE1 rs4646994 and ACE2 rs2285666 were significantly associated with the severity of COVID-19 (Saengsiwaritt et al. 2022). The functional polymorphism observed in this gene is insertion/deletion (rs4646994 I/D SNP) associated with increased activity and concentration of the enzyme. The two alleles D and I of the ACE gene are different in size due to the absence of a 287 bp sequence in intron 16 of this gene and three genotypes DD, DI, and II are created. Individuals with the D/D genotype indicated the highest blood ACE levels, and this rise in expression increases the risk of cardiovascular and respiratory diseases among people with the homozygous elimination genotype. This polymorphism is associated with acute respiratory distress syndrome (ARDS) as well as the progression of pneumonia (SARS) in SARS (Marshall et al. 2002; Matsuda et al. 2012).
The human ACE2 gene is located on the X chromosome and has been linked to several single nucleotide polymorphisms (SNPs) that have been reviewed as important risk factors for hypertension and heart failure, including G to A change in nucleotide + 4 intron 3 (SNP rs2285666). In fact, the location of the ACE2 gene on the X chromosome is considered a disadvantage for male carriers who have alleles associated with low ACE2 expression, and this may explain the higher prevalence of severe COVID-19 in men. SARS-CoV reduces cardiac ACE2 expression and this may explain inflammation and myocardial damage and adverse cardiac outcomes in patients with SARS (Oudit et al. 2009). rs2285666 genetic variant is one of tagged-SNP that could alter mRNA alternate splicing and affect ACE2 receptor gene expression. Also, the SNP is found to be associated with lower plasma ACE2 level (Chen and Yu 2018). It has also been reported that this polymorphism shows a strong linkage disequilibrium with the other SNPs (rs1978124 intron 1 and rs714205 intron 16) in the ACE2 receptor gene (Çelik et al. 2021).
To date, it is clear that ACE/ACE2 imbalance plays an eminent role in the pathogenesis of COVID-19. Therefore, different types of genetic variants in these genes may increase the risk of disease in susceptible individuals by altering gene expression and protein function. Some researchers have also reported that regional differences in allele abundance may explain differences in incidence and mortality rate of the disease. Although some studies proposed a role for rs4646994 and rs2285666 SNPs in COVID-19 severity, the evidence has not been conclusive among different nations as some findings have not been replicated. Therefore, our study aimed to identify different allelic forms in ACE and ACE2 genes associated with COVID-19 among Iranian patients to better understand the pathogenic mechanism of SARS-CoV-2 virus and how the immune system responds to the virus to explain the wide range of disease severity in individuals.
Materials and methods
Subjects
In the present study, 146 patients with COVID-19 infection admitted to Mazandaran University of Medical Sciences hospitals with the diagnosis of COVID-19 during late March, April and May 2020 (first peak), June and July 2020 (second peak) were examined. After obtaining consent from the study participants, all information related to age, sex, history of chronic diseases, early symptoms, CT scan, laboratory tests, medications used, and the course of the disease were recorded in a questionnaire.
The severity of the disease is defined into four groups by the WHO including mild, moderate, severe, and critical. The manifestation of “Mild” stage included usually fever less than 38°, sore throat with or without dry cough, chills, headache, loss of appetite, loss of taste or smell, nausea, vomiting, anorexia, diarrhea, weakness, and fatigue. At this stage, vital signs (pulse, pressure, and number of breaths) are stable and oxygen saturation level (SPO2) is greater than or equal to 93%. In the “Moderate” stage, gastrointestinal and neurological symptoms (severe headache) are observed. Criteria for entering this stage are including (1) shortness of breath, feeling of pain, and pressure in the chest with or without fever equal to or greater than 38° (2) SPO2 between 90% and 93%. In the “severe” stage, there is a range of more severe clinical symptoms. Criteria for entering this stage are including (1) rapid progression of respiratory symptoms (2) tachypnea (RR > 30) and shortness of breath (3) SPO2 less than 90 and (4) increased involvement of more than 50% of the lungs in CT-scan. In critical stage, the patient needs special care. The criterion for entering this stage is the presence of at least one of the following symptoms: (1) patient with symptoms of respiratory failure despite non-invasive oxygen therapy (2) patient with shock symptoms (3) patient in need or under mechanical ventilation and (4) patient with multiple organ failure.
DNA analysis
DNA extraction
Blood samples were collected in EDTA-containing tubes and DNA was isolated using the commercial DNA extraction kit of Aria Gene Farzam Madiar (AGFM) Company according to the protocol of the DNA extraction manufacturer. Then, the quantity and quality of the extracted DNA were determined using two methods of spectrophotometry and agarose gel electrophoresis.
Amplification of gene fragments
Specific primers were designed using Primer premier software to amplify fragments of the ACE and ACE2 genes (Table 1), and using the NCBI website, a sequence of primers designed with the entire human genome will be blasted and featured. The desired fragments for both two genes are then amplified using Amplicon Mastermix in Biorad thermocycler and agarose gel electrophoresis is used to determine the quantity and quality of the amplified fragments.
Genotyping
The I/D polymorphism (rs4646994) located in intron 16 of the ACE gene was determined by Gap-polymerase chain reaction (PCR) followed by agarose gel electrophoresis to visualize the I and D alleles (Alvarez et al. 1998).
For the ACE2 rs2285666 A/G SNP the PCR products were digested with the AluI restriction enzyme and followed by electrophoresis on 2% agarose gels. To perform polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP), 5 μL of the PCR product of ACE2 gene was digested with 0.3 μL of restriction enzymes at 37 °C for 14 h. Digested products were segregated by agarose gel electrophoresis, then visualized by Gel Doc instrument.
Sequencing of different genotypes
We used the direct sequencing technique to confirm the obtained results and also to determine the exact length of the obtained parts. We examine five samples of each genotype (wild and mutant forms) using ABI Genetic Analyzer 3130XL. After obtaining the results, evaluation of reads was done by bioinformatics software such as BioEdit.
Statistical analysis
In this experimental and laboratory study, due to the limitations in sample sizes and selection of samples with complete information, 146 patients were evaluated with complete information. Genotypes and clinical data collected in Excel software were entered and then analyzed using SAS statistical software version 9.1 by appropriate statistical tests such as chi-square and logistic regression. The chi-square test (χ2) was used to verify the equilibrium of the genotypic frequencies with the Hardy–Weinberg equilibrium (HWE). The effects of observed genotypes on the clinical values were evaluated with the GLM procedure. Continuous and categorical variables were expressed as mean ± standard deviation and as percentages, respectively. Significant level in all tests is considered 0.05.
Results
ACE I/D polymorphism
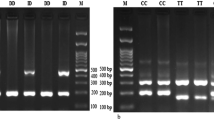
After isolating DNA from blood samples, the ACE I/D polymorphism was genotyped by Gap-PCR. PCR products of ~ 490 and ~ 200 bp characterize as I and D alleles, respectively (Fig. 1).
ACE2 rs2285666 SNP
For ACE2 rs2285666 polymorphism, PCR–RFLP was applied. After digestion of the PCR products with restriction enzyme AluI, fragments of 499 and 277 bp identify A allele (AA genotype) and a 776 bp band identifies G (GG genotype) (Fig. 2). For confirmation of genotyping, five samples were randomly evaluated by Sanger sequencing. The sequencing results confirmed the results of PCR–RFLP technique (Fig. 2).
The present study included 146 COVID-19 patients of which 102 were in the mild and moderate and 44 are in the severe and critical group. 76 subjects were women, and 70 were men enrolled in the study. Additionally, our analysis has indicated that gender, location, and blood groups were insignificant between two severity groups (P > 0.05) (Table 2).
Table 3 shows the distribution of Allelic and genotypic frequencies for ACE I/D (rs4646994) and ACE2 rs2285666 SNPs in the COVID-19 patients. The analysis revealed a significant association between groups according to genotype (P = 0.0016) for ACE I/D gene polymorphism. It was found that the distribution of genotype of ACE2 rs2285666 gene polymorphisms was similar in two groups according to the severity of disease. There was no statistically significant association between groups and genotypic frequencies (P = 0.8472).
Association analysis was also performed between observed genotypes and clinical values using GLM procedure (Table 4). The analysis revealed a remarkable association between rs4646994 SNP and the HB and ESRI levels in patients (P < 0.05). Although the ACE2 rs2285666 SNP was not related to the disease severity, the SNP was significantly associated with ALT, ESRI, and P (Table 4).
Discussion
Since the outbreak of COVID-19, several researchers have noted the importance of ACE and ACE2 gene polymorphisms in COVID-19 outcomes. Finding the genetic risk factors for assessing the individual’s sensitivity to COVID-19 could be important. Although several studies reported various factors including male sex, age, obesity, diabetes and ethnicity are reported to be associated with an increased risk of COVID-19 infection (Hastie et al. 2020), no significant difference in age, gender, location, and blood groups was observed in our findings.
The COVID outbreak has evoked a wave of interest focused on potential SNPs correlated with the severity of the disease. So far, several polymorphisms (Di Maria et al. 2020) with potential to affect SARS-CoV-2 susceptibility were noted by researchers, but undoubtedly confirmed and widely replicated genetic determinants of SARS-CoV-2 infection and COVID-19 severity remain unknown and vary among different populations. The genome-wide association (GWAS) method was applied to the detection of genetic susceptibility to COVID-19, suggesting promising candidate genes such as CCR9 and CXCR6, and SLC6A20, which interacts with ACE2 (Group 2020). Considering the role of ACE gene in the renin–angiotensin (RA) system and consequently, in the pathogenesis of COVID-19, there have been many trials verifying the hypothetical effect of genetic variants in ACE1 and ACE2 genes.
In this regard, the ACE gene I/D (insertion or deletion) SNP (rs4340, rs4646994), is probably the most studied polymorphism. In the present study, we found a significant association between ACE gene I/D SNP and COVID-19 severity which is consistent with former reports (Bellone and Calvisi 2020; Delanghe et al. 2020a; Saadat 2020; Yamamoto et al. 2020). Conclusion based on which allele (D or I) is as deleterious, is controversial among different studies. Our finding indicated that ACE1 D/D genotype was significantly higher in severe and critical COVID-19 patients (P = 0.0016) which is in agreement with the results described by studies performed on Asian populations (Pourbagheri-Sigaroodi et al. 2020; Yamamoto et al. 2020; Verma et al. 2021), reflecting the ethnicity-specific allelic effect. A linear regression analysis indicated an association between D/D genotype and COVID-19 mortality in 25 European nations (Bellone and Calvisi 2020). Other researchers have noted opposite results and have recognized I allele as a predictive marker of COVID-19 severity (Delanghe et al. 2020a, b; Hubacek et al. 2021). Additionally, a few studies have reported no association between this SNP and the disease severity (Çelik et al. 2021). However, the probable reasons for this difference among the abovementioned studies are due to ethnicity, sample size, and the protocol performed.
It has been stated that several ACE2 variants are correlated to various diseases such as hypertension, whose incidence depends on the balance in the renin–angiotensin–aldosterone pathway (Lu et al. 2012; Hidalgo-Tenorio et al. 2017; Pan et al. 2018). Some of these variants influenced the response to ACE inhibitors (Fan et al. 2007; Chen et al. 2016). These variants may be particularly undesirable in men who have only one copy of the X-linked ACE2 gene. The rs2285666 polymorphism of the ACE2 gene is located at the splicing site, which can affect the processing of total ACE2 RNA into mRNA and, ultimately, the amount of protein. In this regard, a study reported higher levels of circulating ACE2 in male than in female (Salah and Mehta 2021). Furthermore, rs2285666 (A allele) is related with a lower probability of cardiovascular death in female (Vangjeli et al. 2011; Malard et al. 2013; Pan et al. 2018). Our finding shows no association between rs2285666 SNP and the severity of the disease which is in agreement with previous studies (Gómez et al. 2020).
Another study verified the serum ACE2 levels in A-carriers of rs2285666 was significantly higher compared to G-homozygotes reflecting the effect of rs2285666 on serum ACE2 levels (Wu et al. 2017). Therefore, it is hypothesized that these functional variants of ACE2 can affect the severity of pathogenicity. Moreover, pathogenic variants of ACE2 are very rare on a population scale, and survival is not possible without the receptors for this gene. In addition, according to the databases associated with human genome variants, there is no missense change in the encoded sequence of the ACE2 gene. At a frequency of minor alleles less than 0.01, only four missense SNPs have been investigated, and rs2285666 is the variant that can affect splicing (Gómez et al. 2020).
Recently, rs190509934 genetic variant was identified through a genome-wide association study as a low-frequency variant (minor allele frequency 0.2–2%) near ACE2 gene that downregulates ACE2 expression and affect the risk of SARS-CoV-2 infection, providing evidence that ACE2 expression levels influence COVID-19 risk (Horowitz et al. 2022). We exploited GWAS meta-analysis data from the COVID-19 Host Genetics Initiative (Initiative 2020) for two genetic variants were associated with severe COVID-19 at the genome-wide significance level (P ≤ 5 × 10− 8). The results indicated that both rs190509934 and rs2285666 SNPs at the ACE2 gene showing suggestive association with the severity of disease (Supplementary Table 1).
Furthermore, to evaluate the association between SNPs and gene expression level to better understanding the mechanism of disease susceptibility, the expression quantitative trait locus (eQTL) analysis in different tissues was performed using the GTEx database (http://www.gtexportal.org/home/). The eQTL analysis from GTEx indicated that rs2285666 variant (chrX_15592225_C_T_b38) was associated with ACE2 gene expression in the different tissues, including nerve tibial (NERVET), brain nucleus accumbens (BRNNCC), brain hypothalamus (BRNHPT), brain frontal cortex (BRNCTXB), brain cortex (BRNCTXA), and brain caudate (BRNCDT). The relative expression of ACE2 gene was significantly lower in subjects with the CC genotype of rs2285666 compared with those carrying the CT or TT genotype (Fig. 3).
As presented, the association of the rs4646994 and rs2285666 polymorphisms with COVID-19 severity is inconsistent among former studies around the world, and possible reason is due to ethnic variations between nations as the above-mentioned variants indicate some population-based differences (Srivastava et al. 2020; Sarangarajan et al. 2021). I/D ACE polymorphism revealed significant geographical and ethical variability like an increase at the I allele frequency in Asian and African population compared to that in Caucasians (Hubacek et al. 2021). In our study, we worked on patient subjects, individuals who suffering from COVID-19 disease, and assessed association between obtained genotypes and COVID-19 severity. Regardless of variety at methodology, sample size, ethnicity among former researches, we have obtained similar results. Therefore, we may encounter contradictory findings but this should not undervalue the importance of ACE and ACE2 genes variants in COVID-19 severity.
Additionally, our analysis revealed a remarkable association between rs4646994 SNP and the HB and ESRI levels in patients (P < 0.05). Although the ACE2 rs2285666 SNP was not related to the severity of the disease, the SNP was significantly associated with ALT, ESRI, and P. Erythrocyte sedimentation rate (ESR) is used as a marker for indicating inflammation, and mostly shows changes in a variety of plasma proteins (Tan et al. 2020). In the present study, ESR showed a significant association with the rs4646994 and rs2285666 SNPs, which could play an eminent role in assessing the severity of COVID-19. Although there are few studies available evaluating ESR as a single predictor of the prognosis and mortality in COVID-19 patients (Kaya et al. 2021), However, these results remain controversial because some other studies have reported no significant difference in the serum levels of ESR between the two severe and no-severe groups (Zeng et al. 2020).
Conclusion
The results of our study suggest that ACE D/D homozygote patients may be at an increased risk of symptomatic COVID-19. Thus, ACE I/D SNP may be a biomarker to predict the COVID-19 severity and may influence on its treatment strategies to offer a population-based therapeutic development. The ACE2 rs2285666 variant was not associated with severity of the disease. A significant association between rs4646994 SNP and the HB and ESRI levels in patients was observed (P < 0.05). ACE2 rs2285666 SNP was also significantly associated with ALT, ESRI, and P.
Limitation
Our study has several limitations, mainly the reduced sample size of the patients. In addition, we could not assess individuals exposed to the virus and show resistant to infection; thus, they have not any disease symptoms. These individuals are important for the detection of genetic variants related to disease susceptibility.
References
Alvarez V, López-Larrea C, Coto E (1998) Mutational analysis of the CCR5 and CXCR4 genes (HIV-1 co-receptors) in resistance to HIV-1 infection and AIDS development among intravenous drug users. Hum Genet 102:483–486
Bellone M, Calvisi SL (2020) ACE polymorphisms and COVID-19-related mortality in Europe. J Mol Med 98:1505–1509
Çelik SK, Genç GÇ, Piskin N, Acikgoz B, Altınsoy B, İşsiz BK, Dursun A (2021) Polymorphisms of ACE I/D and ACE2 receptor gene (RS2106809, RS2285666) are not related to the clinical course of COVID-19: a case study. J Med Virol. 93(10):5947–5952. PMID: 34170561; PMCID: PMC8426884. https://doi.org/10.1002/jmv.27160
Chang D, Lin M, Wei L, Xie L, Zhu G, Cruz CSD, Sharma L (2020) Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA 323:1092–1093
Chen Y, Yu H (2018) A3216 Relationship between genetic variants of ACE2 gene and circulating levels of ACE2 and its metabolites. J Hypertens 36:e2
Chen Y, Liu D, Zhang P, Zhong J, Zhang C, Wu S, Zhang Y, Liu G, He M, Jin L (2016) Impact of ACE2 gene polymorphism on antihypertensive efficacy of ACE inhibitors. J Hum Hypertens 30:766–771
Delanghe JR, Speeckaert MM, De Buyzere ML (2020a) COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin Chem Lab Med 58:1125–1126
Delanghe JR, Speeckaert MM, De Buyzere ML (2020b) The host’s angiotensin-converting enzyme polymorphism may explain epidemiological findings in COVID-19 infections. Clin Chim Acta Int J Clin Chem 505:192
Di Maria E, Latini A, Borgiani P, Novelli G (2020) Genetic variants of the human host influencing the coronavirus-associated phenotypes (SARS, MERS and COVID-19): rapid systematic review and field synopsis. Hum Genomics 14:1–19
Fan X, Wang Y, Sun K, Zhang W, Yang X, Wang S, Zhen Y, Wang J, Li W, Han Y (2007) Study group for pharmacogenomic based antihypertensive drugs selection, effects and side effects. Rural area Chinese. Polymorphisms of ACE2 gene are associated with essential hypertension and antihypertensive effects of captopril in women. Clin Pharmacol Ther 82:187–196
Gómez J, Albaiceta GM, García-Clemente M, López-Larrea C, Amado-Rodríguez L, Lopez-Alonso I, Hermida T, Enriquez AI, Herrero P, Melón S (2020) Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene 762:145102
Group SC-G (2020) Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med 383:1522–1534
Hastie CE, Mackay DF, Ho F, Celis-Morales CA, Katikireddi SV, Niedzwiedz CL, Jani BD, Welsh P, Mair FS, Gray SR (2020) Vitamin D concentrations and COVID-19 infection in UK Biobank. Diabetes Metab Syndr 14:561–565
Hidalgo-Tenorio C, Gil-Anguita C, Ramírez-Taboada J, Esquivias J, López-Ruz MA, Balgahata OM, Javier-Martinez R, Pasquau J (2017) Risk factors for infection by oncogenic human papillomaviruses in HIV-positive MSM patients in the ART era (2010–2016). Medicine 96:e8109
Horowitz JE, Kosmicki JA, Damask A, Sharma D, Roberts GH, Justice AE, Banerjee N, Coignet MV, Yadav A, Leader JB (2022) Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat Genet 54:382–392
Hubacek JA, Dusek L, Majek O, Adamek V, Cervinkova T, Dlouha D, Adamkova V (2021) ACE I/D polymorphism in Czech first-wave SARS-CoV-2-positive survivors. Clin Chim Acta 519:206–209
Ingraham NE, Barakat AG, Reilkoff R, Bezdicek T, Schacker T, Chipman JG, Tignanelli CJ, Puskarich MA (2020) Understanding the renin–angiotensin–aldosterone–SARS-CoV axis: a comprehensive review. Eur Respir J 56:2000912
Initiative C-HG (2020) The COVID-19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet 28:715–718
Jose RJ, Manuel A (2020) COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med 8:e46–e47
Kaya T, Nalbant A, Kılıçcıoğlu GK, Çayır KT, Yaylacı S, Varım C (2021) The prognostic significance of erythrocyte sedimentation rate in COVID-19. Rev Assoc Med Bras 67:1305–1310
Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W (2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 11:875–879
Lu N, Yang Y, Wang Y, Liu Y, Fu G, Chen D, Dai H, Fan X, Hui R, Zheng Y (2012) ACE2 gene polymorphism and essential hypertension: an updated meta-analysis involving 11,051 subjects. Mol Biol Rep 39:6581–6589
Malard L, Kakinami L, O’Loughlin J, Roy-Gagnon M-H, Labbe A, Pilote L, Hamet P, Tremblay J, Paradis G (2013) The association between the angiotensin-converting enzyme-2 gene and blood pressure in a cohort study of adolescents. BMC Med Genet 14:1–7
Marshall RP, Webb S, Bellingan GJ, Montgomery HE, Chaudhari B, McAnulty RJ, Humphries SE, Hill MR, Laurent GJ (2002) Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am J Respir Crit Care Med 166:646–650
Matsuda A, Kishi T, Jacob A, Aziz M, Wang P (2012) Association between insertion/deletion polymorphism in angiotensin-converting enzyme gene and acute lung injury/acute respiratory distress syndrome: a meta-analysis. BMC Med Genet 13:1–8
Oudit G, Kassiri Z, Jiang C, Liu P, Poutanen S, Penninger J, Butany J (2009) SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Investig 39:618–625
Pambuccian SE (2020) The COVID-19 pandemic: implications for the cytology laboratory. J Am Soc Cytopathol 9:202–211
Pan Y, Wang T, Li Y, Guan T, Lai Y, Shen Y, Zeyaweiding A, Maimaiti T, Li F, Zhao H (2018) Association of ACE2 polymorphisms with susceptibility to essential hypertension and dyslipidemia in Xinjiang, China. Lipids Health Dis 17:1–9
Pourbagheri-Sigaroodi A, Bashash D, Fateh F, Abolghasemi H (2020) Laboratory findings in COVID-19 diagnosis and prognosis. Clin Chim Acta 510:475–482
Riordan JF (2003) Angiotensin-I-converting enzyme and its relatives. Genome Biol 4:1–5
Saadat M (2020) No significant correlation between ACE Ins/Del genetic polymorphism and COVID-19 infection. Clin Chem Lab Med 58:1127–1128
Saengsiwaritt W, Jittikoon J, Chaikledkaew U, Udomsinprasert W (2022) Genetic polymorphisms of ACE1, ACE2, and TMPRSS2 associated with COVID-19 severity: a systematic review with meta-analysis. Rev Med Virol 32(4):e2323. PMID: 34997794. https://doi.org/10.1002/rmv.2323
Salah HM, Mehta JL (2021) Hypothesis: sex-related differences in ACE2 activity may contribute to higher mortality in men versus women with COVID-19. J Cardiovasc Pharmacol Ther 26:114–118
Sarangarajan R, Winn R, Kiebish MA, Bountra C, Granger E, Narain NR (2021) Ethnic prevalence of angiotensin-converting enzyme deletion (D) polymorphism and COVID-19 risk: rationale for use of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers. J Racial Ethn Health Disparities 8:973–980
Srivastava A, Pandey RK, Singh PP, Kumar P, Rasalkar AA, Tamang R, van Driem G, Shrivastava P, Chaubey G (2020) Most frequent South Asian haplotypes of ACE2 share identity by descent with East Eurasian populations. PLoS One 15:e0238255
Tan C, Huang Y, Shi F, Tan K, Ma Q, Chen Y, Jiang X, Li X (2020) C-reactive protein correlates with computed tomographic findings and predicts severe COVID-19 early. J Med Virol 92:856–862
Vangjeli C, Dicker P, Tregouet D-A, Shields DC, Evans A, Stanton AV (2011) A polymorphism in ACE2 is associated with a lower risk for fatal cardiovascular events in females: the MORGAM project. J Renin Angiotensin Aldosterone Syst 12:504–509
Verma S, Abbas M, Verma S, Khan FH, Raza ST, Siddiqi Z, Ahmad I, Mahdi F (2021) Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect Genet Evol 91:104801
World Health Organization (2020) WHO Director-general’s remarks at the media briefing on 2019-nCoV. 11 February 2020. https://covid19.who.int/
Wu YH, Li JY, Wang C, Zhang LM, Qiao H (2017) The ACE 2 G8790A polymorphism: involvement in type 2 diabetes mellitus combined with cerebral stroke. J Clin Lab Anal 31:e22033
Xia W, Shao J, Guo Y, Peng X, Li Z, Hu D (2020) Clinical and CT features in pediatric patients with COVID-19 infection: different points from adults. Pediatr Pulmonol 55:1169–1174
Yamamoto N, Ariumi Y, Nishida N, Yamamoto R, Bauer G, Gojobori T, Shimotohno K, Mizokami M (2020) SARS-CoV-2 infections and COVID-19 mortalities strongly correlate with ACE1 I/D genotype. Gene 758:144944
Zeng F, Huang Y, Guo Y, Yin M, Chen X, Xiao L, Deng G (2020) Association of inflammatory markers with the severity of COVID-19: A meta-analysis. Int J Infect Dis. 96:467–474. PMID: 32425643; PMCID: PMC7233226. https://doi.org/10.1016/j.ijid.2020.05.055
Acknowledgements
This work was supported by a grant from Mazandaran University of Medical Sciences (IR-9183). We would also like to show our gratitude to the Dr. Saeed Hassani for provide full assistance in analysis and interpretation of data.
Author information
Authors and Affiliations
Contributions
All the authors contributed to this work by performing the genetic and statistical analysis. MN wrote the manuscripts. All the authors read and approved the submission of this paper.
Corresponding author
Ethics declarations
Conflict of interest
The study was approved by Ethics Committees of Mazandaran University of Medical Sciences (IR.MAZUMS.REC.1400.9183), and written informed consent was obtained from all subjects. The authors declare that there is no conflict of interest.
Additional information
Communicated by Shuhua Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Najafi, M., Mahdavi, M.R. Association investigations between ACE1 and ACE2 polymorphisms and severity of COVID-19 disease. Mol Genet Genomics 298, 27–36 (2023). https://doi.org/10.1007/s00438-022-01953-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01953-8