Abstract
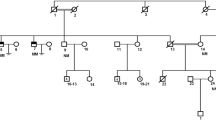
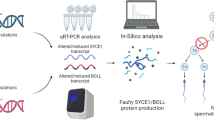
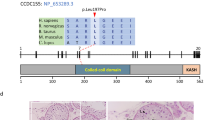
Infertility is a multifactorial disorder that affects approximately 12% of couples of childbearing ages worldwide. Few studies have been conducted to understand the genetic causes of infertility in depth. The synaptonemal complex (SC), which is essential for the progression of meiosis, is a conserved tripartite structure that binds homologous chromosomes together and is thus required for fertility. This study investigated genetic causes of infertility in a Pakistani consanguineous family containing two patients suffering from non-obstructive azoospermia (NOA). We performed whole-exome sequencing, followed by Sanger sequencing, and identified a novel pathogenic variant (c.7G > A [p.D3N]) in the SC coding gene C14orf39, which was recessively co-segregated with NOA. In silico analysis revealed that charges on wild-type residues were lost, which may result in loss of interactions with other molecules and residues, and a reduction in protein stability occurred, which was caused by the p.D3N mutation. The novel variant generated the mutant protein C14ORF39D3N, and homozygous mutations in C14orf39 resulted in NOA. The transcriptome profile of C14ORF39 shows that it is specifically expressed in early brain development, which suggests that research in this area is required to study other functions of C14ORF39 in addition to its role in the germline. This research highlights the conserved role of C14orf39/SIX6OS1 in assembly of the SC and its indispensable role in facilitating genetic diagnosis in patients with infertility, which may enable the development of future treatments.






Similar content being viewed by others
Data availability
The variation data reported in this paper have been deposited in the Genome Variation Map (Song et al. 2018) in BIG Data Center (BIGDC 2017), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number GVM000307 that can be publicly accessible at (http://bigd.big.ac.cn/gvm/getProjectDetail?project=PRJCA008169).
References
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249. https://doi.org/10.1038/nmeth0410-248 (PMID:20354512;PMCID:PMC2855889)
BIGDC (2017) Database resources of the BIG data center in 2018. Nucleic Acids Res 46:D14
Bolcun-Filas E et al (2007) SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol 176:741–747
Bolcun-Filas E et al (2009) Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet 5:e1000393
Bolor H et al (2009) Mutations of the SYCP3 gene in women with recurrent pregnancy loss. Am J Hum Genet 84:14–20
Caburet S et al (2014) Mutant cohesin in premature ovarian failure. N Engl J Med 370:943–949
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. https://doi.org/10.1093/nar/gki375 (PMID:15980478;PMCID: PMC1160136)
Cardoso-Moreira M, Halbert J, Valloton D et al (2019) Gene expression across mammalian organ development. Nature 571:505–509. https://doi.org/10.1038/s41586-019-1338-5
Cheng J, Randall A, Baldi P (2006) Prediction of protein stability changes for single-site mutations using support vector machines. Proteins 62(4):1125–1132
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7(10):e46688. https://doi.org/10.1371/journal.pone.0046688
Du K, Sharma M, Lukacs GL (2005) The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol 12(1):17–25. https://doi.org/10.1038/nsmb882 (Epub 2004 Dec 26 PMID: 15619635)
Fan S, Jiao Y, Khan R, Jiang X, Javed AR, Ali A, Shi Q (2021) Homozygous mutations in C14orf39/SIX6OS1 cause non-obstructive azoospermia and premature ovarian insufficiency in humans. Am J Hum Genet 108(2):P324-336
Ferdouse B, Reshmi C, Cheung VG, Sherman SL, Feingold E (2016) Genome-wide association study of meiotic recombination phenotypes. GenesGenomesGenetics 6(12):3995–4007. https://doi.org/10.1534/g3.116.035766
Gao J, Colaiacovo MP (2018) Zipping and unzipping: protein modifications regulating synaptonemal complex dynamics. Trends Genet TIG 34:232–245
Geisinger A, Benavente R (2016) Mutations in genes coding for synaptonemal complex proteins and their impact on human fertility. Cytogenet Genome Res 150:77–85
Geisinger A, Benavente R (2016) Mutations in genes coding for synaptonemal complex proteins and their impact on human fertility. Cytogenet Genome Res 150(77–85):525
Gomez HL et al (2016) C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat Commun. https://doi.org/10.1038/ncomms13298
Hamer G, Wang H, Bolcun-Filas E, Cooke HJ, Benavente R, Hoog C (2008) Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J Cell Sci 121:2445–2507
Handel MA, Schimenti JC (2010) Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11:124–136
Hartsuiker E, Vaessen E, Carr AM, Kohli J (2001) Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J 20:6660–6671
Hernandez-Lopez D et al (2020) Familial primary ovarian insufficiency associated with an SYCE1 point mutation: defective meiosis elucidated in humanized mice. Mol Hum Reprod 26:485–497
Klein F et al (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98:91–103
Kong A et al (2014) Common and low-frequency variants associated with genome wide recombination rate. Nat Genet 46:11–16
Krausz C, Cioppi F, Riera-Escamilla A (2018) Testing for genetic contributions to infertility: potential clinical impact. Expert Rev Mol Diagn 18:331–346
Mayer S, Rüdiger S, Ang HC, Joerger AC, Fersht AR (2007) Correlation of levels of folded recombinant p53 in escherichia coli with thermodynamic stability in vitro. J Mol Biol 372(1):268–276. https://doi.org/10.1016/j.jmb.2007.06.044 (Epub 2007 Jun 22 PMID: 17631895)
Pashaei M et al (2020) The second mutation of SYCE1 gene associated with autosomal recessive nonobstructive azoospermia. J Assist Reprod Genet 37:451–458
Qin Y, Zhang F, Chen ZJ (2019) BRCA2 in ovarian development and function. N Engl J Med 380:1086
Sanchez-Saez F et al (2020) Meiotic chromosome synapsis depends on multivalent SYCE1-SIX6OS1 interactions that are disrupted in cases of human infertility. Sci Adv. https://doi.org/10.1126/sciadv.abb1660
Schilit SLP et al (2020) SYCP2 translocation-mediated dysregulation and frameshift variants cause human male infertility. Am J Hum Genet 106:41–57
Schramm S et al (2011) A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet 7:e1002088
Shao Y et al (2019) GenTree, an integrated resource for analyzing the evolution and function of primate-specific coding genes. Genome Res 29(4):682–696. https://doi.org/10.1101/gr.238733.118
Sim N-L, Kumar P, Jing Hu, Henikoff S, Schneider G, Ng PC (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40(W1):W452–W457. https://doi.org/10.1093/nar/gks539
Singh SM, Kongari N, Cabello-Villegas J, Mallela KM (2010) Missense mutations in dystrophin that trigger muscular dystrophy decrease protein stability and lead to cross-beta aggregates. Proc Natl Acad Sci USA 107(34):15069–15074. https://doi.org/10.1073/pnas.1008818107 (Epub 2010 Aug 9. PMID: 20696926; PMCID: PMC2930578)
Song S, Tian D, Li C, Tang B, Dong L, Xiao J, Bao Y, Zhao W, He H, Zhang Z (2018) Genome variation map: a data repository of genome variations in BIG data center. Nucl Acids Res 46:D944–D999
Vander Borght M, Wyns C (2018) Fertility and infertility: definition and epidemiology. Clin Biochem 62:2–10
Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G (2010) Protein structure analysis of mutations causing inheritable diseases An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics 11:548. https://doi.org/10.1186/1471-2105-11-548 (PMID: 21059217; PMCID: PMC2992548)
Vries FA et al (2005) Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19:1376–1389
Wang J, Zhang W, Jiang H, Wu BL (2014) Primary ovarian insufficiency collaboration. Mutations in HFM1 in recessive primary ovarian insufficiency. N Engl J Med 370:972–974
Weinberg-Shukron A et al (2018) Essential role of BRCA2 in ovarian development and function. N Engl J Med 379:1042–1049
World Health Organization (2010) WHO laboratory manual for the examination and processing of human semen, 5th edn. WHO Press, Geneva
Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ (2006) 518 Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol 173(520):497–507
Yang F et al (2015) TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in the mouse. EMBO Mol Med 7:1198–1210
Yao C et al (2020) Bi-allelic SHOC1 loss-of-function mutations cause meiotic arrest and non-obstructive azoospermia. J Med Genet 58:679
Yatsenko AN et al (2015) X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med 372:2097–2107
Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C (2000) The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 5:73–83
Zhou J, Gao J, Zhang H, Zhao D, Li A, Iqbal F, Shi Q, Zhang Y (2020) PedMiner: a tool for linkage analysis-based identification of disease-associated variants using family based whole-exome sequencing data. Brief Bioinform. https://doi.org/10.1093/bib/bbaa077
Acknowledgements
The present study was financially supported by the National Key Research and Developmental Program of China (2018YFC1004700 and 2016YFC1000600), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), the National Natural Science Foundation of China (31890780, 31630050 and 31871514), the Major Program of Development Foundation of Hefei Centre for Physical Science and Technology (2018ZYFX005), and the Fundamental Research Funds for the Central Universities (YD2070002006).
Author information
Authors and Affiliations
Contributions
HA, AU, and SD wrote the manuscript, MZ, and AH collected the patient’s samples and performed semen analysis. HA and AU performed the bioinformatics analysis. HA, AU, and SD, FU, IK analyzed the data; Qinghua Shi modified the manuscript and supervised the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by Shuhua Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
438_2022_1876_MOESM1_ESM.pdf
Supplementary file1 (PDF 457 KB) Figure S1. The logarithm of the odds scores; Genome-wide logarithm of the odds scores using WES-derived genotypes for the family. LOD, the logarithm of the odds
438_2022_1876_MOESM3_ESM.pdf
Supplementary file3 (PDF 138 KB) Figure S3. Developmental stages and Transcriptomic profile of C14ORF39 in other organisms
Rights and permissions
About this article
Cite this article
Ali, H., Unar, A., Zubair, M. et al. In silico analysis of a novel pathogenic variant c.7G > A in C14orf39 gene identified by WES in a Pakistani family with azoospermia. Mol Genet Genomics 297, 719–730 (2022). https://doi.org/10.1007/s00438-022-01876-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01876-4