Abstract
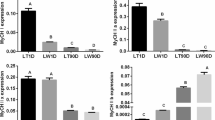
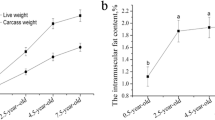
Circular RNA (circRNA) is a class of non-coding RNA (ncRNA) that plays an important regulatory role in various biological processes of the organisms and has a major function in muscle growth and development. However, its molecular mechanisms of how it regulates pork quality remain unclear at present. In this study, we compared the longissimus dorsi (LD) muscle expression profiles of Queshan Black (QS) and Large White (LW) pigs to explore the role of circRNAs in meat quality using transcriptome sequencing. A total of 62 differentially expressed circRNAs (DECs), including 46 up- and 16 down-regulated, 39 differentially expressed miRNAs (DEmiRNAs), including 21 up- and 18 down-regulated and 404 differentially expressed mRNAs (DEMs), including 174 up- and 230 down-regulated were identified, and most circRNAs were composed of exons. Our results indicated that the DEC parent genes and DEMs were enriched in the positive regulation of fast-twitch skeletal muscle fiber contraction, relaxation of skeletal muscle, regulation of myoblast proliferation, AMPK signaling pathway, Wnt and Jak-STAT signaling pathway. Furthermore, circSETBP1/ssc-miR-149/PIK3CD and circGUCY2C/ssc-miR-425-3p/CFL1 were selected by constructing the competitive endogenous RNA (ceRNA) regulatory network, circSETBP1, circGUCY2C, PIK3CD and CFL1 had low expression level in QS, while ssc-miR-149 and ssc-miR-425-3p had higher expression level than LW, our analysis revealed that circSETBP1, circGUCY2C, ssc-miR-149, ssc-miR-425-3p, PIK3CD and CFL1 were associated with lipid regulation, cell proliferation and differentiation, so the two ceRNAs regulatory networks may play an important role in regulating intramuscular fat (IMF) deposition, thereby affecting pork quality. In conclusion, we described the gene regulation by the circRNA–miRNA–mRNA ceRNA networks by comparing QS and LW pigs LD muscle transcriptome, and the two new circRNA-associated ceRNA regulatory networks that could help to elucidate the formation mechanism of pork quality. The results provide a theoretical basis for further understanding the genetic mechanism of meat quality formation.

source statistics and d density distribution on the chromosomes of circRNAs




Similar content being viewed by others
References
Arcinas C, Tan W, Fang W, Desai TP, Teh DCS, Degirmenci U, Xu D, Foo R, Sun L (2019) Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat Metab 1:16. https://doi.org/10.1038/s42255-019-0078-z
Aronoff SL, Berkowitz K, Shreiner B, Want L (2004) Glucose metabolism and regulation: beyond insulin and glucagon. Diabetes Spectr 17:183–190. https://doi.org/10.2337/diaspect.17.3.183
Cen J, Liang Y, Huang Y, Pan Y, Shu G, Zheng Z, Liao X, Zhou M, Chen D, Fang Y, Chen W, Luo J, Zhang J (2021) Circular RNA circSDHC serves as a sponge for miR-127-3p to promote the proliferation and metastasis of renal cell carcinoma via the CDKN3/E2F1 axis. Mol Cancer 20:1. https://doi.org/10.1186/s12943-021-01314-w
Chen Q, Liu M, Luo Y, Yu H, Zhang J, Li D, He Q (2020a) Maternal obesity alters circRNA expression and the potential role of mmu_circRNA_0000660 via sponging miR_693 in offspring liver at weaning age. Gene 731:1. https://doi.org/10.1016/j.gene.2020.144354
Chen Q, Liu M, Luo Y, Yu H, Zhang J, Li D, He Q (2020b) Downregulation of SETBP1 promoted non-small cell lung cancer progression by inducing cellular EMT and disordered immune status. Am J Transl Res 12:16
Cui HX, Liu RR, Zhao GP, Zheng MQ, Chen JL, Wen J (2012) Identification of differentially expressed genes and pathways for intramuscular fat deposition in pectoralis major tissues of fast-and slow-growing chickens. BMC Genomics 13:1. https://doi.org/10.1186/1471-2164-13-213
Doumit ME, Bates RO (2000) Regulation of pork water holding capacity, color and tenderness by protein phosphorylation. Pork Qual 63:6
Fernandez X, Monin G, Talmant A, Mourot J, Lebret B (1999a) Influence of intramuscular fat content on the quality of pig meat—1. Composition of the lipid fraction and sensory characteristics of m. longissimus lumborum. Meat Sci 53:7. https://doi.org/10.1016/s0309-1740(99)00037-6
Fernandez X, Monin G, Talmant A, Mourot J, Lebret B (1999b) Influence of intramuscular fat content on the quality of pig meat—2. Consumer acceptability of m. longissimus lumborum. Meat Sci 53:6. https://doi.org/10.1016/s0309-1740(99)00038-8
Folgueira C, Torres-Leal FL, Beiroa D, Pena-Leon V, Da Silva LN, Milbank E, Senra A, Al-Massadi O, Lopez M, Dieguez C, Seoane LM, Nogueiras R (2020) Oral pharmacological activation of hypothalamic guanylate cyclase 2C receptor stimulates brown fat thermogenesis to reduce body weight. Neuroendocrinology 110:13. https://doi.org/10.1159/000505972
Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucl Acids Res 40:16. https://doi.org/10.1093/nar/gkr688
Gao Y, Zhang J, Zhao F (2018) Circular RNA identification based on multiple seed matching. Brief Bioinform 19:8. https://doi.org/10.1093/bib/bbx014
Hao F, Itoh T, Morita E, Shirahama-Noda K, Yoshimori T, Noda T (2016) The PtdIns3-phosphatase MTMR3 interacts with mTORC1 and suppresses its activity. FEBS Lett 590:13. https://doi.org/10.1002/1873-3468.12048
Hocquette JF, Gondret F, Baeza E, Medale F, Jurie C, Pethick DW (2010) Intramuscular fat content in meat-producing animals: development, genetic and nutritional control, and identification of putative markers. Anim Int J Anim Biosci 4:17. https://doi.org/10.1017/S1751731109991091
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:2. https://doi.org/10.1038/280339a0
Huang K, Chen M, Zhong D, Luo X, Feng T, Song M, Chen Y, Wei X, Shi D, Liu Q, Li H (2021) Circular RNA profiling reveals an abundant circEch1 that promotes myogenesis and differentiation of bovine skeletal muscle. J Agric Food Chem 69:10. https://doi.org/10.1021/acs.jafc.0c06400
Jiang R, Li H, Yang J, Shen X, Song C, Yang Z, Wang X, Huang Y, Lan X, Lei C, Chen H (2020) circRNA profiling reveals an abundant circFUT10 that promotes adipocyte proliferation and inhibits adipocyte differentiation via sponging let-7. Mol Ther Nucl Acids 20:3. https://doi.org/10.1016/j.omtn.2020.03.011
Jiao Y, Huang B, Chen Y, Hong G, Xu J, Hu C, Wang C (2018) Integrated analyses reveal overexpressed Notch1 promoting porcine satellite cells’ proliferation through regulating the cell cycle. Int J Mol Sci 19:1. https://doi.org/10.3390/ijms19010271
Joo ST, Kim GD, Hwang YH, Ryu YC (2013) Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci 95:828–836. https://doi.org/10.1016/j.meatsci.2013.04.044
Kamal AH, Kim WK, Cho K, Park A, Min JK, Han BS, Park SG, Lee SC, Bae KH (2013) Investigation of adipocyte proteome during the differentiation of brown preadipocytes. J Proteomics 94:10. https://doi.org/10.1016/j.jprot.2013.10.005
Lanferdini E, Andretta I, Fonseca LS, Moreira RHR, Cantarelli VS, Ferreira RA, Saraiva A, Abreu MLT (2018) Piglet birth weight, subsequent performance, carcass traits and pork quality: a meta-analytical study. Livest Sci 214:5. https://doi.org/10.1016/j.livsci.2018.05.019
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20:1829–1842. https://doi.org/10.1261/rna.047126.114
Latorre MA, García-Belenguer E, Ariño L (2008) The effects of sex and slaughter weight on growth performance and carcass traits of pigs intended for dry-cured ham from Teruel (Spain). J Anim Sci 86:10. https://doi.org/10.2527/jas.2007-0764
Li A, Zhang J, Zhou Z (2014) PLEK a tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform 15:1. https://doi.org/10.1186/1471-2105-15-311
Li X, Qiao R, Ye J, Wang M, Zhang C, Lv G, Wang K, Li X, Han X (2019) Integrated miRNA and mRNA transcriptomes of spleen profiles between Yorkshire and Queshan black pigs. Gene 688:11. https://doi.org/10.1016/j.gene.2018.11.077
Li J, Tu J, Gao H, Tang L (2021a) MicroRNA-425-3p inhibits myocardial inflammation and cardiomyocyte apoptosis in mice with viral myocarditis through targeting TGF-beta1. Immun Inflamm Dis 9:11. https://doi.org/10.1002/iid3.392
Li X, Zhang D, Ren C, Bai Y, Ijaz M, Hou C, Chen L (2021b) Effects of protein posttranslational modifications on meat quality: a review. Compr Rev Food Sci Food Saf 20:43. https://doi.org/10.1111/1541-4337.12668
Liu Y, Liu H, Li Y, Mao R, Yang H, Zhang Y, Zhang Y, Guo P, Zhan D, Zhang T (2020) Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 10:15. https://doi.org/10.7150/thno.42417
Lu M, Xu L, Wang M, Guo T, Luo F, Su N, Yi S, Chen T (2018) miR149 promotes the myocardial differentiation of mouse bone marrow stem cells by targeting Dab2. Mol Med Rep 17:8. https://doi.org/10.3892/mmr.2018.8903
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. https://doi.org/10.1038/nature11928
Mohamed JS, Hajira A, Pardo PS, Boriek AM (2014) MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT-1/PGC-1alpha network in skeletal muscle. Diabetes 63:1546–1559. https://doi.org/10.2337/db13-1364
Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13:12. https://doi.org/10.1038/nrg3162
Peng W, Li T, Pi S, Huang L, Liu Y (2020) Suppression of circular RNA circDHCR24 alleviates aortic smooth muscle cell proliferation and migration by targeting miR-149-5p/MMP9 axis. Biochem Biophys Res Commun 529:7. https://doi.org/10.1016/j.bbrc.2020.06.067
Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, Antolini L, Mologni L, Donadoni C, Papaemmanuil E, Schnittger S, Kim DW, Boultwood J, Rossi F, Gaipa G, De Martini GP, di Celle PF, Jang HG, Fantin V, Bignell GR, Magistroni V, Haferlach T, Pogliani EM, Campbell PJ, Chase AJ, Tapper WJ, Cross NC, Gambacorti-Passerini C (2013) Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet 45:7. https://doi.org/10.1038/ng.2495
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H (2015) Circular RNA: a new star of noncoding RNAs. Cancer Lett 365:8. https://doi.org/10.1016/j.canlet.2015.06.003
Reiss N, Kanety H, Schlessinger J (1986) Five enzymes of the glycolytic pathway serve as substrates for purified epidermal-growth-factor-receptor kinase. Biochem J 239:7. https://doi.org/10.1042/bj2390691
Rodriguez A, Gomez-Ambrosi J, Catalan V, Ezquerro S, Mendez-Gimenez L, Becerril S, Ibanez P, Vila N, Margall MA, Moncada R, Valenti V, Silva C, Salvador J, Fruhbeck G (2016) Guanylin and uroguanylin stimulate lipolysis in human visceral adipocytes. Int J Obes (lond) 40:11. https://doi.org/10.1038/ijo.2016.66
Sale EM, White MF, Kahn CR (1987) Phosphorylation of glycolytic and gluconeogenic enzymes by the insulin receptor kinase. J Cell Biochem 33:12. https://doi.org/10.1002/jcb.240330103
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell 146:6. https://doi.org/10.1016/j.cell.2011.07.014
Shen Y, Mao H, Huang M, Chen L, Chen J, Cai Z, Wang Y, Xu N (2016) Long noncoding RNA and mRNA expression profiles in the thyroid gland of two phenotypically extreme pig breeds using Ribo-Zero RNA sequencing. Genes (basel) 7:1. https://doi.org/10.3390/genes7070034
Shen X, Tang J, Ru W, Zhang X, Huang Y, Lei C, Cao H, Lan X, Chen H (2020) CircINSR regulates fetal bovine muscle and fat development. Front Cell Dev Biol 8:1. https://doi.org/10.3389/fcell.2020.615638
Siede D, Rapti K, Gorska AA, Katus HA, Altmuller J, Boeckel JN, Meder B, Maack C, Volkers M, Muller OJ, Backs J, Dieterich C (2017) Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol 109:9. https://doi.org/10.1016/j.yjmcc.2017.06.015
Song K, Wang S, Mani M, Mani A (2014) Wnt signaling, de novo lipogenesis, adipogenesis and ectopic fat. Oncotarget 5:4. https://doi.org/10.18632/oncotarget.2769
Tang L, Xiong W, Zhang L, Wang D, Wang Y, Wu Y, Wei F, Mo Y, Hou X, Shi L, Xiong F, Zhang S, Gong Z, Liao Q, Xiang B, Zhang W, Zhou M, Li X, Li G, Guo C, Zeng Z (2021) CircSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene 40:15. https://doi.org/10.1038/s41388-020-01531-5
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:5. https://doi.org/10.1038/nbt.1621
Wang MY, Zhang C, Xue YH, Ye JW, Han XL, Qiao RM, Li XL, Liu X, Wang XW, Wu SJ, Wang MM, Li XJ (2019) Comparative analysis of body size indexes, slaughter performance, meat quality and blood indicators between Yuxi Black pigs and Queshan Black pigs (Chinese). China Anim Husb Vet Med 46:10. https://doi.org/10.16431/j.cnki.1671-7236.2019.07.007
Wang L, Liang W, Wang S, Wang Z, Bai H, Jiang Y, Bi Y, Chen G, Chang G (2020) Circular RNA expression profiling reveals that circ-PLXNA1 functions in duck adipocyte differentiation. PLoS ONE 15:1. https://doi.org/10.1371/journal.pone.0236069
Wen M, Shen Y, Shi S, Tang T (2012) miREvo an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform 13:1. https://doi.org/10.1186/1471-2105-13-140
Xing K, Wang K, Ao H, Chen S, Tan Z, Wang Y, Xitong Z, Yang T, Zhang F, Liu Y, Ni H, Sheng X, Qi X, Wang X, Guo Y, Wang C (2019) Comparative adipose transcriptome analysis digs out genes related to fat deposition in two pig breeds. Sci Rep 9:12925. https://doi.org/10.1038/s41598-019-49548-5
Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, Yuan X, Yin W, Xu J, Chen K, He C, Wei L (2020) CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer 19:1. https://doi.org/10.1186/s12943-020-01246-x
Yan XM, Zhang Z, Meng Y, Li HB, Gao L, Luo D, Jiang H, Gao Y, Yuan B, Zhang JB (2020) Genome-wide identification and analysis of circular RNAs differentially expressed in the longissimus dorsi between Kazakh cattle and Xinjiang brown cattle. PeerJ 8:1. https://doi.org/10.7717/peerj.8646
Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q, Wang Y, Wang X (2019) METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPbeta pathway via an m(6)A-YTHDF2-dependent manner. FASEB J off Publ Fed Am Soc Exp Biol 33:16. https://doi.org/10.1096/fj.201802644R
Zhang M, Han Y, Zhai Y, Ma X, An X, Zhang S, Li Z (2020) Integrative analysis of circRNAs, miRNAs, and mRNAs profiles to reveal ceRNAs networks in chicken intramuscular and abdominal adipogenesis. BMC Genomics 21:1. https://doi.org/10.1186/s12864-020-07000-3
Acknowledgements
This work was supported by Henan Key Research & Development Program (192102110067 and 202102110242), Pig Industry Technology System Innovation Team Project of Henan Province (S2012-06-G03), Grand Science and Technology Special Project in Tibet (212102110003), and the National Natural Science Foundation of China (32002142).
Author information
Authors and Affiliations
Contributions
Author contributions XH proposed conceptualization, methodology, wrote reviews and editing, administrated project; KQ performed verification and visualization, wrote original draft preparation; XH, YL and CL investigated; XH, XL and XL provided resources; KW and RQ performed supervision. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All of the experiments involving animals were carried out in accordance with the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of the People’s Republic of China (Approval Number 2006–398).
Additional information
Communicated by Joan Cerdá.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Qi, K., Liu, Y., Li, C. et al. Construction of circRNA-related ceRNA networks in longissimus dorsi muscle of Queshan Black and Large White pigs. Mol Genet Genomics 297, 101–112 (2022). https://doi.org/10.1007/s00438-021-01836-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01836-4