Abstract
Bud dormancy in deciduous fruit trees is an important adaptive mechanism for their survival in cold climates. The WRKY genes participate in several developmental and physiological processes, including dormancy. However, the dormancy mechanisms of WRKY genes have not been studied in detail. We conducted a genome-wide analysis and identified 58 WRKY genes in peach. These putative genes were located on all eight chromosomes. In bioinformatics analyses, we compared the sequences of WRKY genes from peach, rice, and Arabidopsis. In a cluster analysis, the gene sequences formed three groups, of which group II was further divided into five subgroups. Gene structure was highly conserved within each group, especially in groups IId and III. Gene expression analyses by qRT-PCR showed that WRKY genes showed different expression patterns in peach buds during dormancy. The mean expression levels of six WRKY genes (Prupe.6G286000, Prupe.1G393000, Prupe.1G114800, Prupe.1G071400, Prupe.2G185100, and Prupe.2G307400) increased during endodormancy and decreased during ecodormancy, indicating that these six WRKY genes may play a role in dormancy in a perennial fruit tree. This information will be useful for selecting fruit trees with desirable dormancy characteristics or for manipulating dormancy in genetic engineering programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
To endure harsh environmental conditions in winter, perennial deciduous fruit trees have developed adaptation mechanisms such as dormancy and overwintering (bud dormancy). In recent years, with global warming, perennial deciduous fruit trees have shown irregular phenologies because of inadequate winter chilling. These irregularities adversely affect fruit production (Luedeling et al. 2011). Thus, bud dormancy plays a critical role in the development of perennial plants, and research on bud dormancy is useful for the development of perennial deciduous fruit trees. There are several types of dormancy in plants; ecodormancy, paradormancy, and endodormancy (Lang 1987). Endormant buds cannot initiate growth under favorable conditions without prior chilling (Crabbe and Barnola 1996; Faust et al. 1997). Because endodormancy is irreversible, dormancy is one of the key factors limiting fruit production. The WRKY transcription factors (TFs) are involved in seed dormancy in Arabidopsis, but it is unknown whether they are involved in bud dormancy in perennial plants. Therefore, the identification of WRKY and dormancy-related WRKY genes in peach not only provides a molecular foundation for studies on bud dormancy, but will also be useful for exploring the functions of WRKY gene products.
Dormancy is an important state in which meristem activity ceases and the meristem is insensitive to growth-promoting signals (Rohde and Bhalerao 2007). The plant cannot resume growth until dormancy is released (Rohde and Bhalerao 2007). In bud dormancy, FLOWERING LOCUS T (FT) is a versatile regulator in the environmental control of meristem transitions including the cessation of growth and the establishment of dormancy (Henrik et al. 2006). In Populus, FT was rapidly induced by the short day-length signal (Henrik et al. 2006). In grapevine, VvFT was detected in leaves and buds under a short day-length photoperiod (Vergara et al. 2015), indicating that FT played a key role in regulating the cessation of growth and establishment of endodormancy. A natural mutant of peach (Prunus persica) that could not express six MICK-type MADS genes at the EVG locus was unable to cease growth and form buds. This observation suggested that MADS box genes may be major candidates for controlling growth cession (Bielenberg et al. 2004, 2008). Subsequently, PpDAM1, PpDAM2, and PpDAM4 were shown to be closely related to terminal bud formation (Li et al. 2009) and the transcript abundances of PpDAM5 and PpDAM6 were inversely with the sprouting rate of terminal buds in peach (Jiménez et al. 2009). PpDAMs were shown to play roles not only in inducing endodormancy, but also in releasing endodormancy (Yamane et al. 2011). Recently, SOC1 (encoding a MADS-domain TF) was shown to affect the duration of dormancy in kiwifruit (Voogd et al. 2015). In seed dormancy, DELAY OF GERMINATION1 (DOG1) is a major regulator of dormancy. In Arabidopsis, the levels of the DOG1 protein, which were largely independent of abscisic acid (ABA), functioned as a timer for seed dormancy release in freshly harvested seeds (Nakabayashi et al. 2012). SNL1 (SIN3-LIKE1) and SNL2 were also shown to be related to seed dormancy via their role in mediating the ABA-ethylene antagonism in Arabidopsis (Wang et al. 2013). AtWRKY41 was shown to control both primary seed dormancy and thermo inhibition via directly regulating ABI3 expression in mature imbibed seeds (Ding et al. 2014). As we known, both types of dormancy are characterized by a temporary insensitivity to growth-promoting signals and may have similar molecular mechanisms (Fu et al. 2014). In Persian walnut, Vahdati et al. (2012) also confirmed a relationship between the two types of dormancy breaking mechanism. WRKY TFs involved in seed dormancy have been identified, and whether they have a relationship with bud dormancy is unknown. Thus, it is necessary to verify the hypothesis.
The WRKY family is one of the ten largest families of TFs. This family, which is predominant in plants, is considered to be plant specific. Members of the WRKY family play crucial roles in regulating plant growth and development. The name, WRKY, is derived from the conserved domain of a WRKYGQK hexapeptide sequence at the N-terminus. These TFs also have a novel zinc-finger-like motif at the C-terminus, and form a four-stranded β-sheet and a zinc-binding pocket in which zinc coordinates with Cys/His residues to form the WRKY domain (Rushton et al. 2010). Although research on WRKY TFs has rapidly expanded from model plants to crop species, our knowledge of WRKY TFs in fruit trees, including peach, is limited. Since the first isolation of a WRKY protein (SPF1) from sweet potato in 1994, many other WRKY TFs have been identified from various plants including parsley Arabidopsis, wild oat, tobacco, and cucumber (Ishiguro and Nakamura 1994; Rushton et al. 1995, 1996). Previous studies have shown that WRKY TFs bind to certain promoters containing a W box (TTGACC/T), a cognate cis-acting element. Early research on WRKY TFs suggested that their main roles were in responses to pathogens (Eulgem and Somssich 2007). For example, PopP2 and AvrRps4 were shown to interact with WRKY domain-containing proteins (e.g. NB-LRR proteins) in Arabidopsis (Sarris et al. 2015). Recently, however, WRKY TFs have been shown to function in diverse processes such as germination, dormancy, and responses to abiotic stresses (Rushton et al. 2010). Many studies have demonstrated that members of the WRKY family play complex and sometimes contradictory regulatory roles in biotic stress responses. For example, OsWRKY45-1 and OsWRKY45-2 were shown to play opposite roles in regulating resistance to Xanthomonas oryzae, but identical roles in regulating resistance to Magnaporthe grisea (Masaki et al. 2007; Tao et al. 2009). Several WRKY TFs have been implicated in seed development, such as Arabidopsis AtWRKY10 and Solanum chacoense ScWRKY1 (Sun et al. 2003; Lagacé and Matton 2004). In wild oat, ABF1 and ABF2 were shown to bind to W boxes in the promoters of genes encoding α-amylases, which are crucial for starch hydrolysis during germination in cereals. Thus, ABF1 and ABF2 were shown to affect germination, and indirectly, post-germination. AtWRKY6 was implicated in the regulation of leaf senescence and was strongly up-regulated during senescence (Robatzek and Somssich 2001; Silke and Somssich 2002). In subsequent studies, AtWRKY53, AtWRKY70, and OsWRKY23 were also shown to regulate senescence. Other studies have shown that WRKY TFs participate in multiple processes. For example, HvWRKY38 and HvWRKY1 were shown to provide a mechanistic link among biotic stress responses, germination, and abiotic stress responses. OsWRKY53 acted as a negative feedback modulator of MPK3 and MPK6 (Hu et al. 2015). Cai et al. (2015) demonstrated that CaWRKY6 activates CaWRKY40, which functions as a positive regulator of Ralstonia solanacearum resistance and heat tolerance. Few dormancy-related WRKY genes have been identified so far. Therefore, it is important to identify which WRKY genes, if any, are related to dormancy in perennial species. The full genome sequence of peach is now available in public databases. Therefore, the aim of this study was to identify the WRKY genes, and specifically the dormancy-related WRKY genes, in the peach genome.
We searched the recently released peach genome and identified 58 candidate WRKY genes, which were distributed on all eight chromosomes. The genes were classified into three main groups according to their predicted WRKY domains and zinc finger structures. Gene structure analyses showed that the structures were highly conserved within each group. Finally, we analyzed the expression profiles of the WRKYs in bud dormancy, and identified six WRKY genes that may play important roles in dormancy.
Materials and methods
Plant materials
Peach samples were obtained from the Horticulture Experimental Station of Shandong Agricultural University, Tai’an, China. The plant materials (‘Prunus persica L. cv Zhong You Tao 4’) were grown under standard agricultural practices for 5 years. Bud samples for this study were collected from at least 30 independent trees. For analyses of gene expression at different stages of bud dormancy, peach bud samples were collected before leaf abscission, during dormancy, and during the dormancy-release period, on 16 and 31 October, 15 November, 1, 15 and 31 December, 15 and 25 January, and 15 February. At each time point, flower buds (500 mg) with scales were collected from first-year branches of different vigorous individual trees, and then the samples were immediately frozen in liquid nitrogen. The samples were stored at −80 °C until use.
Definition of bud dormancy
To evaluate bud dormancy, we used 120 first-year branches incubated in 5 % (w/v) sucrose solution each time. The branches were collected on 16 and 31 October, 15 November, 1, 15 and 31 December, 15 and 25 January, and 15 February, and were incubated in a growth chamber. Trials were conducted in a completely randomized design with three replicates, each with 40 cuttings. The branches were kept under a 16-h light/8-h dark photoperiod with artificial fluorescent light (200 μmol m−2 s−1) with day/night temperatures of 25/18 °C and 70 % relative humidity. The basal ends of the shoots were cut weekly, and the sucrose solution was replaced daily. Sprouting was recorded after 25 days and 50 % bud sprouting marked the beginning of dormancy release. The results are expressed as percentage of budbreak for the three replicates.
Identification of WRKY genes in peach
To identify the members of the WRKY gene family in peach, we conducted BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches using the proteome sequences as a database. Annotated peach WRKY proteins were used as query sequences to perform BLAST searches against the proteome and genome files downloaded from the peach genome database (https://www.rosaceae.org). To verify the authenticity of candidate sequences, the hidden Markov model (HMM) profile of the WRKY domain (PF03106) was used as a query to identify WRKYs using the program HMMER3.0 (http://hmmer.janelia.org). Finally, the sequences were compared with cDNA sequences of WRKY genes in PlantTFDB (http://planttfdb.cbi.pku.edu.cn) (Zhang et al. 2011) and the integrity of the WRKY domain was confirmed by SMART with default cut-off parameters (http://smart.embl-heidelberg.de/). After manually removing incorrect and overlapping predicted genes, 58 protein sequences were identified.
Mapping WRKY genes on peach chromosomes
The locations of the WRKY genes on the chromosomes were obtained from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html). The WRKY genes were mapped to the chromosomes using Circos software (http://circos.ca/tutorials/lessons/).
Phylogenetic analysis of WRKY genes in peach, Arabidopsis, and rice
The WRKY family sequences for Arabidopsis were retrieved from TAIR (http://www.arabidopsis.org/) and those for rice were retrieved from NCBI (http://www.ncbi.nlm.nih.gov). Peach sequences were identified by local BLASTP searches and HMM profiling as described above. The WRKY sequences were aligned using the ClustalX 2.1 (Larkin et al. 2007) with the default settings. Phylogenetic and molecular trees based on the protein sequences predicted from WRKY gene sequences were constructed using the neighbor-joining algorithm with the program MEGA6.0 (http://www.megasoftware.net/mega6/faq.html), with parameters set according to the JTT model. The reliability of the obtained trees was tested by conducting 1000 bootstrap sampling steps.
Gene structure construction
The coding sequences (CDS) and genome sequences of WRKY genes in peach were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppersica_er). The gene structures were predicted using GSDS online (http://gsds.cbi.pku.edu.cn/).
qRT-PCR analysis of WRKY gene expression during bud dormancy
Total RNA was extracted from buds with scales (500 mg) using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. For the qRT-PCR, cDNA was synthesized using the PrimeScript RT reagent kit with gDNA Eraser (Takara, Dalian, China). The qRT-PCR was performed with SYBR Premix Ex Taq (Takara) following the manufacturer’s instructions. The sequences of gene primers (β-actin primer pair as an internal control)used for qRT-PCR (Table S1) were designed with BD software and synthesized by BGI (http://www.genomics.cn). The expression of the reference gene is not changed during all the development stages. We selected 36 WRKY genes for qRT-PCR analysis (Table S2). The thermal cycling conditions were as follows: 10 min at 95 °C for pre-denaturation, followed by 40 cycles of 15 s at 95 °C for denaturation and 60 s at 60 °C for annealing and extension. The specificity of the qRT-PCR was confirmed by the presence of a single peak in the melting curve and by size estimation of the amplified qRT-PCR product. To quantify cDNAs with amplification efficiencies, the comparative cycle threshold (CT) method (2−ΔΔCT) method was used. To further observe the changes of expression profiles between endodormancy and ecodormancy, the mean expression of each genes during endodormancy and ecodormancy was used. As shown in Fig. 5, endodormancy represents the mean expression of 16 and 31 October, 15 November, 1, 15 and 31 December and ecodormancy represents the mean expression of 15 and 25 January, and 15 February. Each reaction was repeated three times. Results are the average of three independent biological replicates.
Results
Identification of WRKY genes in peach
Members of the WRKY-gene family have been identified in many species, but not in peach until now. We used two approaches to identify members of the WRKY family in peach. First, all annotated proteins of peach were used as query sequences to perform BLASTP searches in the NCBI database. Then, the hidden Markov model (HMM) profile of the WRKY domain (PF03106) was used as a query to identify WRKY genes using the program HMMER3.0. After manually removing redundant sequences, the remaining genes were further analyzed to confirm the integrity of the WRKY domain using SMART with the default cut-off parameters. Finally, 58 non-redundant putative WRKY genes were identified in peach. The length, putative molecular weight, and theoretical isoelectric points of the WRKY TFs were analyzed in this study varied widely (Table 1). The length of the predicted WRKY TFs ranged from 170 to 751 amino acids, their putative molecular weights ranged from 19.6 to 82.2 kDa, and their theoretical isoelectric points ranged from 4.7 to 10.6.
Phylogenetic analysis and classification of WRKYs in peach, Arabidopsis, and rice
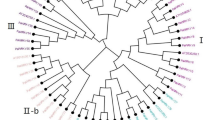
To evaluate the phylogenetic relationships of the WRKY genes in peach and classify them within the established subfamilies, we analyzed 225 amino acid sequences containing the WRKY domain. These sequences consisted of 58 sequences from peach, 73 sequences from Arabidopsis, and 94 sequences from rice. The sequences of Arabidopsis WRKY proteins were downloaded from TAIR, those of peach were downloaded from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppersica_er), and those of rice were downloaded from NCBI (http://www.ncbi.nlm.nih.gov). An unrooted phylogenetic tree (Fig. 1) was constructed using the neighbor-joining method with MEGA6.0 (http://www.megasoftware.net/history.php) (Tamura et al. 2013). In the phylogenetic tree, the WRKY genes in peach were divided into three main groups: PpWRKY I, II, and III, according to their predicted WRKY domains and zinc finger structures. There were 10 WRKY TFs in group I, 40 PpWRKYs in group II, and 8 PpWRKYs in group III. Members of group I contained two conserved domains and one C2H2 zinc finger motif, members of group II contained one conserved domain, and members of group III harbored the other conserved domain. The difference between groups II and III was the type of zinc finger motif; group II members had the same zinc finger motif as that in group I, while group III members contained the C2HC zinc finger motif. Group II contained five subgroups (PpWRKY IIa, b, c, d, and e, containing 3, 8, 15, 7, and 7 WRKY TFs, respectively). As shown in the phylogenetic tree, all the WRKY TFs from different species were clustered into three groups, and the WRKY TFs of different species in the same group were more similar than those from the same species in different groups. A previous study showed that WRKY TFs from various species harbor different variants of the WRKY domain (WRKYGQK), such as WRKYGKK, WRKYGSK, and WRKYGEK (Zhang and Wang 2005). In our study, we detected the WRKYGKK variant in Prupe.1G407500 and Prupe.6G169700 in group IIc. This was the only variant in peach (Yang et al. 2015). Interestingly, members of group IIb contained another highly conserved tetrapeptide sequence, the LDLT sequence (Supplementary 3).
Phylogenetic tree of peach, rice, and Arabidopsis WRKY proteins. The 58 peach, 73 Arabidopsis and 94 rice protein sequences were aligned by ClustalW and the phylogenetic tree was constructed using MEGA6.0 by the neighbor-joining method with 1000 bootstrap replicates. WRKY proteins clustered into three main groups
Distribution of WRKY genes on peach chromosomes
Figure 2 shows the distribution of the 58 WRKY genes on peach chromosomes. As shown in the figure, the WRKY genes were unevenly distributed throughout all eight peach chromosomes, and the number on each chromosome was not related to its length. Chromosome 6 had the most WRKY genes (13 genes, or 22.4 % of the total) followed by Chr1 (12 genes), while Chr7 had the least (one WRKY gene). Nine WRKY genes were located on Chr2, 10 on Chr3, 6 on Chr4, 5 on Chr5, and 2 on Chr8. The nomenclature of the PpWRKYs was established from the exact position of the WRKY genes on peach chromosomes 1 to 8, from top to bottom, and from their classifications.
Previous reports have indicated that gene duplication and positive selection have significantly contributed to the expansion of gene families and the diversification of protein functions (Wei et al. 2012). To understand the role of gene duplication in the expansion of the WRKY gene family, we analyzed the tandem and segmental duplications of this gene family. The results revealed 30 colinear WRKY genes and nine tandem WRKY genes. The collinear genes are consecutive genes along a genomic region that by a duplication event have paralogs in the same consecutive order in another genomic region. Interestingly, all the colinear WRKY genes within the syntenic regions belonged to the same group. Only two sets of triplicate WRKY genes were identified (Prupe.1G223200/Prupe.5G148700/Prupe.3G308200 and Prupe.5G187800/Prupe.1G269200/Prupe.3G270800). It is interesting to find that the two sets of triplicate WRKY genes are located on triplicated regions in the peach genome (Verde et al. 2013). Surprisingly, all chromosomes except Chr7 had WRKY genes located in the colinear duplicated regions.
Structure of WRKY genes
Considering that gene structure is a typical imprint of evolution within a gene family, we analyzed the WRKY genes in peach using tools at the GSDS website (Fig. 3). Interestingly, all of the WRKY genes in peach had one or more (up to five) introns, so each PpWRKY sequence was divided into many segments by introns. The genes in each group showed similar structures and similar intron phases. The numbers and phases of introns were more conserved in groups II and III than in group I. Members of group I contained phase 0, phase 1, and phase 2 introns. Compared with Prupe.3G202000, Prupe.4G232600 had two phase 0 introns before a phase 1 and two phase 2 introns. We inferred that there was no significant impact on mRNA level, but further research is required to determine whether the two phase 0 introns played a role in processing of the primary transcripts.
Phylogenetic analysis and structures of WRKY genes in peach. Phylogenetic tree was constructed by MEGA6.0 with the neighbor-joining method based on alignments of complete predicted protein sequences of WRKY genes. In gene structure diagram, green boxes and lines represent exons and introns, respectively
Measurement of bud dormancy status and evaluation of stage-specific expression of WRKY genes during dormancy by qRT-PCR
To measure the transcript profiles of WRKY genes during dormancy in peach, the dormancy status of buds was defined for shoots of 5-year-old ‘Prunus persica L. cv Zhong You Tao 4’ peach trees collected on nine dates. As shown in Fig. 4, the first buds sprouted on 15 December, there was a marked increase in sprouting from early January, and then sprouting reached almost 100 %. Thus, the buds sampled from 15 October to 1 January represent endodormant buds, and those sampled from 15 January to 15 February represent ecodormant buds.
WRKY genes are thought to be involved in regulating dormancy. To identify dormancy-related WRKY genes, the transcript profiles of WRKY genes at different stages of bud dormancy were analyzed by qRT-PCR. The 36 WRKY genes were classified into four gene expression groups using MeV software (Eisen et al. 1998) according to the chronological stages of bud dormancy (Fig. 5). Considering the inherent errors in the experimental set-up, we chose a three-fold change in expression as the definition of a dormancy-related WRKY gene. From the mean expression level, the expressions of Prupe.6G286000, Prupe.1G393000, Prupe.1G114800, Prupe.1G071400, Prupe.2G185100 and Prupe.2G307400 were up-regulated in endodormancy and down-regulated in ecodormancy, whereas during endodormancy the expressions were down-regulated concomitantly with endodormancy release. The other genes including Prupe.1G280700, Prupe.3G202000 Prupe.4G232600, Prupe.6G036300 had no obviously changes during dormancy.
Discussion
With the development of genome sequencing projects, genome-wide analysis of the WRKY gene family has been reported for various species, such as Arabidopsis (Wu et al. 2005), soybean (Ülker and Somssich 2004), Carica papaya (Pan and Jiang 2014), cotton (Dou et al. 2014), Gossypium raimondii (Cai et al. 2014), grape (Wang et al. 2014), Aegilops tauschii (Ma et al. 2015), Cucumis sativus (Jian et al. 2011), Brachypodium distachyon (Feng et al. 2014), Solanum lycopersicum (Chen et al. 2015), Gossypium (Ding et al. 2015), and Camellia sinensis (Wu et al. 2015). However, the WRKY genes in Prunus persica had not been characterized until now. Therefore, the identification of WRKY genes in the peach genome and analyses of their expression patterns are important topics.
Previous studies have shown that the bHLH family is the largest TF family in peach, while the WRKY gene family is the eighth-largest. To investigate the evolution of WRKY genes in plants, we compared 21 diverse plant species including those from the Chlorophyta to Embryophyta subkingdoms and determined how many WRKY genes were present in each species (Fig. 6). The number of WRKY genes in the various species ranged from 2 to 233. Embryophyte species had more WRKY genes than Chlorophyte species, suggesting that WRKY genes have played a vital role during evolution. Within the Rosaceae, Malus domestica had more WRKY genes than did peach, possibly because of the two genome replication events that occurred in M. domestica during evolution.
Duplication of WRKY genes in peach
In peach, all seven paleosets of paralogs can be detected in fragmentary triplicated blocks. Peach has not undergone a recent whole genome duplication (Verde et al. 2013). The WRKY transcription factors have a long history and ancient origin in eukaryotes, which originally had one WRKY gene (Zhang and Wang 2005). The results of our analyses indicate that the gene duplication in peach and the distribution of the 58 WRKY genes on peach chromosomes are non-uniform. Gene duplication is the main driving force in evolution, and takes several forms; tandem duplication, segmental duplication, and whole genome duplication (Xu et al. 2012). The homologous genes (12 pairs of WRKY genes plus two sets of triplets) in peach were identified in the same homologous blocks, and the two sets of triplets were consistent with the fragmentary triplicated blocks. Compared with peach, Arabidopsis has 25.9 % more WRKY genes, and rice has 62.1 % more. The smaller number of WRKY genes in peach may be because unlike Arabidopsis and rice, peach has not undergone a recent whole-genome duplication after the differentiation of eudicots and monocots. There are tandem duplicates in the peach WRKY gene family; for some pairs, the two genes may have different functions. Alternatively, two functional genes may be required when a large transcript abundance is necessary for specific responses at specific times.
Colinear orthologs of WRKY genes among peach, rice, Arabidopsis
Comparisons of genomic data between well-characterized and less-studied taxa can allow us to infer details of genome structure, function, and evolution of less-studied species based on knowledge gained from model species. Therefore, comparative genomic analysis is a relatively rapid and effective method for evaluating less-studied taxa (Lyons et al. 2008). The functions of the WRKY gene family have been widely studied in model plants such as Arabidopsis and rice. In theory, we can estimate the potential functions of WRKYs in peach by comparisons with their well-characterized homologs in Arabidopsis and rice. In the phylogenetic tree, many homologous genes from Arabidopsis, rice, peach with common conserved motifs clustered into the same clade.
In a previous study, ABF1 and ABF2, two WRKY TFs from wild oat were found to be GA-inducible and ABA-repressible, like their homologs in rice and barley (Rushton et al. 1995). In rice, OsWRKY51 and OsWRKY71 were shown to function synergistically by forming a heterotetramer to control the production of α-amylase. OsWRKY51/OsWRKY71 not only antagonized GAMYB but also prevented it forming a complex with other proteins, thus repressing ABA-induced dormancy (Xie et al. 2005, 2006; Zhang et al. 2004). In our BLAST searches, we identified the colinear orthologs of OsWRKY51/OsWRKY71 in peach (Prupe.1G071400 and Prupe.3G098100). These genes showed almost identical expression profiles; therefore, we speculated that they may have the same functions in dormancy. Further studies are required to determine whether the peach orthologs form a tetramer, like their orthologs in rice.
Expression profiles among paralogs in peach WRKY gene family
Some genes and their paralogs play redundant roles in planta, such as AtWRKY18, AtWRKY40, AtWRKY60 (Xu et al. 2006), AtWRKY54, and AtWRKY70 (Besseau et al. 2012; Li et al. 2013). However, some paralogs, such as AtWRKY4 and AtWRKY3, have different functions. Previous studies have shown that the expression level of AtWRKY4 but not AtWRKY3 increased in response to B. cinerea infection. In our study, we analyzed the expression profiles of paralog pairs or triplets (Prupe.1G393000 and Prupe.1G393100; Prupe.6G168200 and Prupe.6G169700; Prupe.2G264900 and Prupe.2G265000; Prupe.6G294900, Prupe.6G295000, and Prupe.6G295100) in the peach WRKY gene family. Prupe.6G168200 and Prupe.6G169700 showed the same expression profiles, while the others had non-identical profiles and showed different expression levels during dormancy. We inferred that differences in expression between and among paralogs may be related to sequences outside the conserved motif. Our results suggested that some paralogs in peach are redundant, while others have diverse functional roles.
WRKY gene family may be involved in dormancy
WRKY transcription factors have been shown participate in many plant processes such as biotic stress, abiotic stress, seed development, seed dormancy and germination, senescence, and development (Craig and Ling 2014). According to QTLs in peach, G1, G4, G6/8 and G7 were detected associated with controlling seed dormancy (Blaker et al. 2013). Romeu et al. (2014) also identified that QTLs related to bud dormancy in peach mainly mapped to LG1which closes to the evergrowing locus. Based on these, we performed analysis which compares the genomic sites for the QTLs with reference to the location of the relevant WRKY genes on the peach genome. Interestingly, we found that some WRKY genes including Prupe.1G071400 mapped to LG1, Prupe.7G262600 mapped to G7 and Prupe.4G232600 mapped to G4. All these demonstrated the WRKY genes may be involved in dormancy. A previous study showed that 15 WRKY genes in grape had identical expression patterns under cold treatment (Wang et al. 2014). Another study showed that Prupe.1G071400 might play roles in the early responses to abiotic stress, in acquiring resistance, and in controlling dormancy. In our study, the mean expression of Prupe.1G071400 was at relatively high level in endodormancy and at lower level in ecodormancy, further indicating that this gene may participate in regulating dormancy. Similarly, other WRKY genes (Prupe.6G286000, Prupe.1G393000, Prupe.1G114800, Prupe.2G185100 and Prupe.2G307400) were detected specifically in endodormant buds compared with ecodormant buds, suggesting that they may be related to dormancy. Along with the changes in the process of endodormancy, most of the studied genes including Prupe.1G393000, Prupe.1G 071400 reduced their expression at December 15 previous to dormancy release which further illustrates WRKY genes may be involved in endodormancy. Considering that internal factors inhibit the growth of endodormant buds even in favorable conditions (Lang et al. 1987), we inferred that WRKY genes may act as internal factors controlling endodormancy. However, transgenic studies are required to evaluate the functions of WRKYs.
In conclusion, the peach genome contains 58 WRKY genes that are unevenly distributed on all eight chromosomes. The phylogenetic, gene structure and chromosomal location analyses have provided complete information for this gene family in peach. Segmental duplication has played a vital role in the expansion of the WRKY gene family in peach. The expression profiles of WRKY genes during dormancy demonstrated that some of them may be involved in dormancy. In summary, the results of this study provide the foundation for further studies on the roles of WRKY genes in bud dormancy and will be useful for further research on the evolutionary history of WRKY genes in eukaryotes.
References
Besseau S, Li J, Palva ET (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63:2667–2679
Bielenberg DG, Wang Y, Fan S et al (2004) A deletion affecting several gene candidates is present in the evergrowing peach mutant. J Hered 95:436–444
Blaker KM, Chaparro JX, Beckman TG (2013) Identification of QTLs controlling seed dormancy in peach (Prunus persica). Tree Genet Genomes 9(3):659–668
Cai C, Niu E, Du H, Zhao L, Feng Y, Guo W (2014) Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. Crop J 2:87–101
Cai H, Yang S, Yan Y, Xiao Z, Cheng J, Wu J et al (2015) CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J Exp Bot 66:3163–3174
Chen L, Yang Y, Liu C, Zheng Y, Xu M, Wu N et al (2015) Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem Biophys Res Commun 464:962–968
Crabbe J, Barnola P (1996) A new conceptual approach to bud dormancy in woody plants. In: Lang GA (ed) Plant dormancy: physiology, biochemistry and molecular biology. CAB International, Wallingford, pp 83–113
Craig S, Ling Y (2014) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol 167:295–306
Ding ZJ, Yan JY, Li GX, Wu ZC, Zhang SQ, Zheng SJ (2014) WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J 79:810–823
Ding M, Chen J, Jiang Y, Lin L, Cao YF, Wang M et al (2015) Genome-wide investigation and transcriptome analysis of the WRKY gene family in gossypium. Mol Genet Genomics 290:151–171
Dou L, Zhang X, Pang C, Song M, Wei H, Fan S et al (2014) Genome-wide analysis of the WRKY gene family in cotton. Mol Genet Genomics 289:1103–1121
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95:14863–14868
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371
Faust M, Erez A, Rowland LJ, Wang SY, Norman HA (1997) Bud dormancy in perennial fruit trees: physiological basis for dormancy induction, maintenance, and release. HortScience 32:623–629
Feng W, Hong Z, Peng L, Min J, Mao W, Ong C et al (2014) Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res 21:327–339
Fu XL et al (2014) Roles of endoplasmic reticulum stress and unfolded protein response associated genes in seed stratification and bud endodormancy during chilling accumulation in Prunus persica. PLoS ONE 9(7):e101808
Henrik B, Tao H, Laurence CC, Brunner AM, Stefan J, Strauss SH et al (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312:1040–1043
Hu L, Ye M, Li R, Zhang T, Zhou G, Wang Q, Lou Y (2015) The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of map kinase activity. Plant Physiol. doi:10.1104/pp.15.01090
Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes sp8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Genet Genomics 244:563–571
Jian L, Jiang W, Ying Z, Yu H, Mao Z, Gu X et al (2011) Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom 12:12368–12375
Jiménez S, Lawton-Rauh AL, Reighard GL, Abbott AG, Bielenberg DG (2009) Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol 9:81
Lagacé M, Matton DP (2004) Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219:185–189
Lang GA (1987) Dormancy: a new universal terminology. HortScience 22:817–820
Lang GA, Early JD, Martin GC, Darnell RL (1987) Endo-, para-, and ecodormancy: physiological terminology and classification for dormancy research. Hortscience 22(3):371–377
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Letunic I, Bork P (2007) Interactive tree of life (itol): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128
Letunic I, Bork P (2011) Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478
Li Z, Reighard GL, Abbott AG, Bielenberg DG (2009) Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot 60:3521–3530
Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L et al (2013) Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol 200:457–472
Luedeling E, Girvetz EH, Semenov MA, Brown PH (2011) Climate change affects winter chill for temperate fruit and nut trees. PLoS ONE 6:1575–1581
Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, Freeling M (2008) Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol 148:1772–1781
Ma J, Zhang D, Shao Y, Liu P, Jiang L, Li C (2015) Genome-wide analysis of the WRKY transcription factors in Aegilops tauschii. Cytogenetic Genome Research 144:243–253
Masaki S, Shoji S, Akira N, Chang-Jie J, Kazuko O, Seiichi T et al (2007) Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19:2064–2076
Nakabayashi K, Bartsch M, Xiang Y, Miatton E, Pellengahr S, Yano R et al (2012) The time required for dormancy release in Arabidopsis is determined by delay of germination1 protein levels in freshly harvested seeds. Plant Cell 24:2826–2838
Pan LJ, Jiang L (2014) Identification and expression of the WRKY transcription factors of Carica papaya in response to abiotic and biotic stresses. Mol Biol Rep 41:1215–1225
Robatzek S, Somssich IE (2001) A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant Journal 28:123–133
Rohde A, Bhalerao RP (2007) Plant dormancy in the perennial context. Trends Plant Sci 12:217–223
Romeu JF, Monforte AJ, Sánchez G, Granell A, García-Brunton J et al (2014) Quantitative trait loci affecting reproductive phenology in peach. BMC Plant Biol 14(3):1–16
Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-amy2 genes. Plant Mol Biol 29:691–702
Rushton PJ, Torres JM, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley pr1 genes. EMBO J 15:5690–5700
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Sarris PF, Duxbury Z, Huh SU, Ma Y, Segonzac C, Sklenar J et al (2015) A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161:1089–1100
Silke R, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Develop 16:1139–1149
Sun CX, Sara P, Helena O, Mats B, Staffan A, Christer J (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15:2076–2092
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tao Z, Liu HB, Qiu DY, Zhou Y, Li XH, Xu CG et al (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151:936–948
Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Vahdati K, Aslamarz AA, Rahemi M et al (2012) Mechanism of seed dormancy and its relationship to bud dormancy in Persian walnut. Environ Exp Bot 75(4):74–82
Verde I, Abbott AG, Scalabrin S et al (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45:487–494
Vergara R, Noriega X, Parada F, Dantas D, Pérez FJ (2015) Relationship between endodormancy, FLOWERING LOCUS T and cell cycle genes in Vitis vinifera. Planta http://link.springer.com/article/10.1007%2Fs00425-015-2415-0
Voogd C, Wang T, Varkonyi-Gasic E (2015) Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J Exp Bot 66:4699–4710
Wang Z, Hong C, Yongzhen S, Xiaoying L, Fengying C, Annaick C et al (2013) Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid-ethylene antagonism mediated by histone deacetylation. Plant Cell 25:149–166
Wang L, Zhu W, Fang L, Sun X, Su L, Liang Z, Xin H (2014) Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol 14:160–162
Wei K, Chen Y, Chen J, Wu L, Xie D (2012) Evolution and adaptation of hemagglutinin gene of human H5N1 influenza virus. Virus Genes 44:450–458
Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12:9–26
Wu Z-J, Li X-H, Liu Z-W, Li H, Wang Y-X, Zhuang J (2015) Transcriptome-wide identification of camellia sinensis WRKY transcription factors in response to temperature stress. Mol Genet Genomics 291(1):255–269
Xie Z, Zhang ZL, Zou XL, Huang J, Ruas P, Thompson D et al (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137:176–189
Xie Z, Zhang Z, Zou XG, Komatsu S, Shen Q (2006) Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J 46:231–242
Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors. Plant cell 18:1310–1326
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon–intron structure. Proc Natl Acad Sci 109:1187–1192
Yamane H, Tao R, Ooka T, Jotatsu H, Sasaki R, Yonemori K et al (2011) Comparative analyses of dormancy-associated MADS-box genes, PpDAM5 and PpDAM6, in low- and high-chill peaches (Prunus persica). J Jpn Soc Hortic Sci 80:276–283
Yang X, Deng C, Zhang Y, Cheng YF, Huo QY, Xue LB (2015) The WRKY transcription factor genes in eggplant (Solanum melongenal.) and turkey berry (Solanum torvumsw.). Int J Mol Sci 16:7608–7626
Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5:1–12
Zhang ZL, Xie Z, Zou X, Casaretto J, Ho THD, Shen QJ (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134:1500–1513
Zhang H, Jin JP, Tang L, Zhao Y, Gu XC, Gao G, Luo JC (2011) PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39:D1114–D1117
Acknowledgments
This study was funded by the National Natural Science Foundation of China (Grant number 31372050) and the Modern Agricultural Industry Technology System of Fruit Industry in Shandong Province-Equipment of Cultivation and Facilities (Grant number SDAIT-03-022-05).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This study was funded by the National Natural Science Foundation of China (Grant number 31372050) and the Modern Agricultural Industry Technology System of Fruit Industry in Shandong Province-Equipment of Cultivation and Facilities (Grant number SDAIT-03-022-05).
Conflict of interest
All authors declare they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This article does not contain any studies with human participants.
Additional information
Communicated by R. Velasco.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, M., Tan, Q., Sun, M. et al. Genome-wide identification of WRKY family genes in peach and analysis of WRKY expression during bud dormancy. Mol Genet Genomics 291, 1319–1332 (2016). https://doi.org/10.1007/s00438-016-1171-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1171-6