Abstract
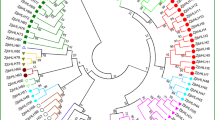
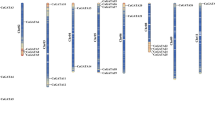
The ZF-HD gene family plays an important role in plant developmental processes and stress responses. However, the function of the ZF-HD genes in Chinese cabbage remains largely unknown. Chinese cabbage (Brassica rapa ssp. pekinensis) is a member of one of the most important leaf vegetables grown worldwide. The entire Chinese cabbage genome sequence has been determined, and more than forty thousand proteins have been identified to date. In this study, 31 ZF-HD genes were identified in Chinese cabbage. We show here that the BraZF-HD genes could be categorized into ZHD and MIF subfamilies. Among them, ZHD genes are plant-specific, nearly all intronless, and related to MINI ZINC FINGER genes that possess only the zinc finger. Phylogenetic analysis suggested that ZHDs have expanded considerably during angiosperm evolution. In addition, the ZHD group has 24 members, which is twice as much as the Arabidopsis ZHD group, indicating that the Chinese cabbage ZHD genes have been retained more frequently than other group genes. Real-time PCR analysis showed that most of BraZF-HD genes are preferentially expressed in flower. Furthermore, most of these genes are significantly induced under photoperiod or vernalization conditions, as well as abiotic stresses. Thereby implying that they may play important roles in these processes. This study provides insight into the evolution of ZF-HD genes in Chinese cabbage genome and may aid efforts to further characterize the function of these predicted ZF-HD genes in flowering and resistance.








Similar content being viewed by others
References
Akin Z, Nazarali A (2005) Hox genes and their candidate downstream targets in the developing central nervous system. Cell Mol Neurobiol 25(3–4):697–741
Albert VA, Soltis DE et al (2005) Floral gene resources from basal angiosperms for comparative genomics research. BMC Plant Biol 5(1):5
Ariel FD, Manavella PA et al (2007) The true story of the HD-Zip family. Trends Plant Sci 12(9):419–426
Bai Y, Meng Y et al (2011) Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98(2):128–136
Bailey TL, Boden M et al (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Barth O, Vogt S et al (2009) Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol Biol 69(1–2):213–226
Birchler JA, Veitia RA (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19(2):395–402
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Bürglin TR (1994) A comprehensive classification of homeobox genes. Guidebook to the homeobox genes, pp 25–71
Cheng F, Mandáková T et al (2013) Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25(5):1541–1554
Englbrecht CC, Schoof H et al (2004) Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom 5(1):39
Finn RD, Clements J et al (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37
Freeling M (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60:433–453
Friedman R, Hughes AL (2001) Gene duplication and the structure of eukaryotic genomes. Genome Res 11(3):373–381
Gehring WJ, Affolter M et al (1994) Homeodomain proteins. Annu Rev Biochem 63(1):487–526
Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr Opin Plant Biol 4(4):301–308
Halbach T, Scheer N et al (2000) Transcriptional activation by the PHD finger is inhibited through an adjacent leucine zipper that binds 14-3-3 proteins. Nucleic Acids Res 28(18):3542–3550
Hu W, Ma H (2006) Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. Plant J 45(3):399–422
Hu W, Wang Y et al (2003) Isolation, sequence analysis, and expression studies of florally expressed cDNAs in Arabidopsis. Plant Mol Biol 53(4):545–563
Hu W, DePamphilis CW et al (2008) Phylogenetic analysis of the plant specific zinc finger homeobox and mini zinc finger gene families. J Integr Plant Biol 50(8):1031–1045
Hunter CS, Rhodes SJ (2005) LIM-homeodomain genes in mammalian development and human disease. Mol Biol Rep 32(2):67–77
Ito M, Sato Y et al (2002) Involvement of homeobox genes in early body plan of monocot. Int Rev Cytol 218:1-36e
Jain M, Tyagi AK et al (2008) Genome wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J 275(11):2845–2861
Kim YB, Li X et al (2013) MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 18(7):8682–8695
Klug A, Schwabe J (1995) Protein motifs 5. Zinc fingers. FASEB J 9(8):597–604
Knight CA, Vogel H et al (2006) Expression profiling and local adaptation of Boechera holboellii populations for water use efficiency across a naturally occurring water stress gradient. Mol Ecol 15(5):1229–1237
Kosarev P, Mayer K et al (2002) Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol 3(4):0016.0011–0016.0012
Krishna SS, Majumdar I et al (2003) Structural classification of zinc fingers SURVEY AND SUMMARY. Nucleic Acids Res 31(2):532–550
Lee TH, Tang H, Wang X, Paterson AH (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41:D1152–D1158
Li J, Jia D et al (2001) HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 13(10):2269–2281
Nakashima K, Yamaguchi-Shinozaki K (2006) Regulons involved in osmotic stress responsive and cold stress responsive gene expression in plants. Physiol Plant 126(1):62–71
Punta M, Coggill PC et al (2011) The Pfam protein families database. Nucleic Acids Res 49:D290–D301
Schoof H, Zaccaria P et al (2002) MIPS Arabidopsis thaliana Database (MAtDB): an integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res 30(1):91–93
Schranz ME, Lysak MA et al (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11(11):535–542
Singh KB, Foley RC et al (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5(5):430–436
Song X, Li Y et al (2013) Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom 14(1):573
Takatsuji H (1999) Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol 39(6):1073–1078
Tamura K, Peterson D et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tan QK-G, Irish VF (2006) The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol 140(3):1095–1108
Tang H, Lyons E (2012) Unleashing the genome of Brassica rapa. Front Plant Sci 3:172
Thomas BC, Pedersen B et al (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16(7):934–946
Tong C, Wang X et al (2013) Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom 14(1):689
Town CD, Cheung F et al (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18(6):1348–1359
Tran LSP, Nakashima K et al (2007) Co-expression of the stress inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J 49(1):46–63
Vision TJ, Brown DG et al (2000) The origins of genomic duplications in Arabidopsis. Science 290(5499):2114–2117
Wang X, Wang H et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43(10):1035–1039
Williams RW (1998) Plant homeobox genes: many functions stem from a common motif. BioEssays 20(4):280–282
Windhövel A, Hein I et al (2001) Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol Biol 45(2):201–214
Wolberger C (1996) Homeodomain interactions. Curr Opin Struct Biol 6(1):62–68
Yanagisawa S (2004) Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol 45(4):386–391
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Key Program, No. 31330067), National Program on Key Basic Research Projects (The 973 Program: 2012CB113900), and National High Technology Research and Development Program of China (863 Program, No. 2012AA100101).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
W. Wang and P. Wu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, W., Wu, P., Li, Y. et al. Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol Genet Genomics 291, 1451–1464 (2016). https://doi.org/10.1007/s00438-015-1136-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1136-1