Abstract
The expression of inulinolytic genes in Aspergillus niger is co-regulated and induced by inulin and sucrose. We have identified a positive acting transcription factor InuR, which is required for the induced expression of inulinolytic genes. InuR is a member of the fungal specific class of transcription factors of the Zn(II)2Cys6 type. Involvement of InuR in inulin and sucrose metabolism was suspected because of the clustering of inuR gene with sucB, which encodes an intracellular invertase with transfructosylation activity and a putative sugar transporter encoding gene (An15g00310). Deletion of the inuR gene resulted in a strain displaying a severe reduction in growth on inulin and sucrose medium. Northern analysis revealed that expression of inulinolytic and sucrolytic genes, e.g., inuE, inuA, sucA, as well as the putative sugar transporter gene (An15g00310) is dependent on InuR. Genome-wide expression analysis revealed, three additional putative sugar transporters encoding genes (An15g04060, An15g03940 and An17g01710), which were strongly induced by sucrose in an InuR dependent way. In silico analysis of the promoter sequences of strongly InuR regulated genes suggests that InuR might bind as dimer to two CGG triplets, which are separated by eight nucleotides.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Some plants store part of their carbon as sucrose and as polymers of fructose (fructans). Plant fructans, often referred to as inulins, have predominantly β-2,1 linkages between the fructose moieties and terminate with a sucrosyl residue. Inulin is present in a range of plant species. In some species such as chicory, garlic, Jerusalem artichoke and Dahlia tubers about 15–20% of the plant fresh weight consists of inulin (Gupta and Kaur 1997; Kaur and Gupta 2002). Apart from the use of inulin for the production of fructose rich syrups, inulin and fructo-oligosaccharides (FOS) have attracted considerable research attention because of the large number of health benefits obtained from inulin and FOS in the diet of humans (see for a review Kaur and Gupta 2002).
Aspergillus niger is a saprophytic fungus, mainly present in the soil, feeding preferably on organic matter such as plant cell wall polysaccharides (cellulose, hemi-cellulose and pectin), and on plant storage polysaccharides (starch and inulin). A. niger is able to produce various inulin degrading enzymes including an exo-inulinase, an endo-inulinase and an invertase (Moriyama et al. 2003; Ohta et al. 1998; Akimoto et al. 1999; Berges et al. 1993; Boddy et al. 1993; L’Hocine et al. 2000). The inulinolytic enzymes are simultaneously induced in the presence of inulin or sucrose, indicating that the expression of the genes encoding these enzymes is co-regulated and controlled by a common transcription factor (Yuan et al. 2006).
Transcriptional activation of catabolic enzyme networks is often accomplished through transcription factors of the Zn(II)2Cys6 type. This type of transcription factor is unique to fungi. Gal4 is the transcriptional activator for the expression of genes involved in galactose utilization, and is the best characterized Zn(II)2Cys6 transcription factor. In general, Zn(II)2Cys6 transcription factors possess a well conserved N-terminal localized DNA binding motif (CX2CX6CX5–16CX2CX6–8), followed by a less well defined conserved domain known as a fungal specific transcription factor domain (Pfam04082). The C-terminal part of the Zn(II)2Cys6 transcription factor normally contains the activation domain. In the genome sequence of Saccharomyces cerevisiae, 54 putative Zn(II)2Cys6 transcription factors have been identified (Akache et al. 2001). They are involved in the transcriptional control of a wide variety of cellular processes (Todd and Andrianopoulos 1997; Akache et al. 2001) including genes involved in amino acid metabolism (ArgRp, Crabeel et al. 1995; Leu3p, Kohlhaw 2003; Lys14p, El Alami et al. 2000), sugar metabolism (Mal63p, Ni and Needleman 1990), pyrimidine biosynthesis (Ppr1p, Flynn and Reece 1999). In the genome of filamentous fungi even more Zn(II)2Cys6 transcription factors are present. In the recently released genome sequence of A. niger CBS 513.88, 296 ORFs were identified that contain the Zn(II)2Cys6 motif and similar numbers are predicted from the genomes of A. fumigatus and A. nidulans (see for comparison Pel et al. 2007). The role of only a few of the Zn(II)2Cys6 transcription factors in filamentous fungi have been studied in detail. Filamentous fungal Zn(II)2Cys6 proteins are involved in the regulation of various processes which include secondary metabolite production (e.g. AflR, Woloshuk et al. 1994), pigmentation (e.g. Cmr1, Tsuji et al. 2000), nitrogen metabolism (NirA, Burger et al. 1991), proline utilization (PrnA, Cazelle et al. 1998; Leucine biosynthesis (Leu3p, Kirkpatrick and Schimmel 1995), alcohol utilization (AlcR, Kulmburg et al. 1992) and sexual development (RosA, Vienken et al. 2005). Well studied examples of fungal Zn(II)2Cys6 transcription factors required for polysaccharide catabolism include AmyR, a transcriptional activator of starch degrading enzymes (Petersen et al. 1999; Gomi et al. 2000; Tani et al. 2001a), and XlnR, a transcriptional activator for xylanolytic enzymes (van Peij et al. 1998a, 1998b; Gielkens et al. 1999; Hasper et al 2004).
It is well established that the genes encoding many proteins required for secondary metabolite production are clustered in the genome (Woloshuk et al. 1994; Walton 2000). In addition to several biosynthetic enzymes that synthesize the secondary metabolites, such clusters also contain a Zn(II)2Cys6 transcription factor which is required for the transcriptional activation of the genes within the cluster. Gene clustering is not limited to secondary metabolite pathways, but also found for the proline utilization genes (PrnA, Gomez et al. 2002) and for some of the amylolytic genes under the control of the regulator AmyR. Adjacent to the gene for the AmyR transcription factor are genes encoding alpha-glucosidase (AgdA) and an alpha-amylase (AmyA) (Gomi et al. 2000). Also in A. parasiticus, a sugar utilization gene cluster has been identified (Yu et al. 2000). However, it is clear that clustering of genes with related functions is not a general rule, since the genes adjacent to the XlnR transcription factor do not seem to be involved in xylan degradation.
In a previous study, we showed that the expression of the genes encoding inulin degrading enzymes is co-regulated and specifically induced in the presence of inulin and sucrose (Yuan et al. 2006). The availability of the genome sequence of A. niger allowed the rapid identification of Zn(II)2Cys6 transcription factors adjacent to or in close proximity to the genes encoding (putative) enzymes involved in inulin catabolism. We have identified a Zn(II)-cluster transcription factor (InuR) which is adjacent to a gene encoding a sugar transporter and a recently identified intracellular invertase (SucB) with transfructosylation activity (Goosen et al. 2007).
In this paper, we show that deletion of the inuR gene results in strongly impaired growth on inulin and sucrose, and also abolishes the induction of the genes encoding the extracellular enzymes involved in inulin and sucrose degradation. The gene encoding the putative sugar transporter located next to inuR is co-regulated with the genes encoding the inulinolytic enzymes. These results show that InuR acts as a positive acting transcriptional activator for the induced expression of genes involved in the breakdown of inulin and sucrose and the uptake of inulin breakdown products.
Materials and methods
Strains, culture conditions and transformation
Aspergillus niger strain N402 used in this study was derived from the wild-type strain A. niger van Tieghem (CBS 120.49, ATCC 9029) (Bos et al. 1988). The A. niger strain used by DSM to sequence the genome is NRRL 3122. Strain AB4.1 is a pyrG negative derivative of N402 (van Hartingsveldt et al. 1987) and was used to construct disruption strains. A. niger strains were grown in Aspergillus minimal medium (MM) (Bennet and Lasure 1991), or Aspergillus complete medium (CM) consisting of MM medium with the addition of 0.5% (w/v) yeast extract and 0.1% (w/v) casamino acids. Growth medium was supplemented with 10 mM uridine (Serva) when required. For shake flask cultures, A. niger strains were grown in MM supplemented with 1% (w/v) carbon source and 0.1% (w/v) casamino acids. The conidiospores were inoculated at a concentration of 2 × 106 spores per ml. Glucose and sucrose (BDH chemicals), maltodextrin (Avebe), starch (Windmill Starch, Avebe), xylose, fructose and maltose (Sigma-Aldrich), raffinose (Sigma chemicals) and inulin (Sensus Frutafit, Cosun) were used as carbon sources.
For transfer experiments, A. niger strains were pre-grown in MM supplemented with 2% (w/v) xylose and 0.1% (w/v) casamino acids for 18 h at 30°C on a rotary shaker at 300 rpm. Then mycelium was harvested by suction over a nylon membrane and washed with MM without carbon source. Aliquots of 1.6 g wet weight of mycelium were transferred to 300-ml Erlenmeyer flasks containing MM supplemented with 1% (w/v) carbon source and incubated at 30°C for the time indicated in the text. The mycelium was harvested over Myracloth (Calbiochem) and frozen in liquid nitrogen, followed by storage at −80°C prior to the isolation of total RNA. Conidiospores were obtained by harvesting spores from CM plates after 4–6 days of growth at 30°C, using a 0.9% NaCl solution. Transformation of A. niger AB4.1 was done as described by Punt and van den Hondel (1992) using lysing enzymes (L1412, Sigma) for protoplastion. The bacterial strain used for transformation and amplification of recombinant DNA was Escherichia coli XL1-Blue (Stratagene). Transformation of XL1-Blue was performed according to the heat shock protocol as described by Inoue et al. (1990).
Construction of the inuR::pyrG deletion strain
The A. niger inuR gene was deleted by the replacement of the complete open reading frame of the inuR gene with the Aspergillus oryzae pyrG gene. The plasmid used to disrupt inuR was constructed as follows. The DNA fragments flanking the inuR ORF were amplified by PCR using N402 genomic DNA as template. Fragments with a length of 0.8 kb of 5′ flanking DNA and 1.0 kb of 3′ flanking DNA were amplified using primers inuRP1 and inuRP2, inuRP3 and inuRP4 (Supplementary Table 1), respectively. Each primer was adapted with restriction sites as indicated for further cloning. The amplified PCR fragments were digested with NotI and XhoI or XhoI and KpnI, respectively, and cloned into pBlue-Script II to obtain plasmid pInuRF5 and pInuRF3. Subsequently, the 0.8 kb NotI-XhoI fragment containing 5′ flanking of inuR from pInuRF5, was ligated into NotI/XhoI digested pInuRF3 to give pInuRF53. The A. oryzae pyrG gene was isolated as a 2.8 kb SalI–BamHI fragment, obtained from plasmid pAO4-13 (de Ruiter-Jacobs et al. 1989) and ligated into XhoI/BamHI digested pInuRF53 to obtain pΔinuR (pXY3.1). Before transformation, the plasmid pΔinuR was linearized with NotI and transformed into A. niger pyrG - strain AB4.1. Uridine prototrophic transformants were selected by incubating protoplasts on agar plates containing MM without uridine. Transformants were screened by PCR using primers PAO9 and inuRP5 or PAO10 and inuRP6 (Supplementary Table 1). Only transformants with a targeted deletion of the inuR gene should result in the amplification of a 1.2 or 1.4 kb PCR fragment, respectively. PCR positive transformants were verified by Southern blot analysis as described by Sambrook et al. (1989).
Chromosomal DNA from positive PCR transformants was isolated as described by Kolar et al. (1988). Ten μl of genomic DNA was digested for 3 h with 10 U of XhoI or EcoRI, respectively. The 1.0 kb of the 3′-flanking DNA fragment of inuR was used as probe for the detection of inuR disruptants. A probe was generated by digestion of the pInuRF3 plasmid containing the inuR 3′-flanking fragment with XhoI and BamHI. Fragments were purified from gel and [α-32P]dCTP labeled probes were synthesized using Rediprime II DNA labeling System (Amersham Pharmacia Biotech) according to the instructions of the manufacturer.
Complementation of inuR mutant
The A. niger inuR gene was amplified by PCR using primers inuRP7 and inuRP8 (Supplementary Table 1) and N402 genomic DNA as a template. The primers corresponded to 1 kb upstream and 0.5 kb downstream of the A. niger inuR gene, respectively. PCR was performed using Phusion™ High-Fidelity DNA Polymerase (Finnzymes) according to the manufacturer’s instruction. The amplified 4.1 kb PCR fragment was cloned into pGEMT-easy vector (Promega) to obtain the inuR complementation plasmid pXY5.1. pXY5.1 was then co-transformed with pAN7.1 harboring hygromycin gene as selection marker (Punt et al. 1987) into A. niger ΔinuR strain XY3.1 (see in the text). Hygromycin resistant transformants were selected by incubating protoplasts on agar plates containing MM with 200 mg/ml of hygromycin. Southern blot analysis was performed on selected transformants as described by Sambrook et al. (1989).
Construction of the ΔinuR ΔamyR double deletion strain
To make a pyrG-strain of the ΔinuR strain (XY3.1) this strain was incubated on MM plates containing 1 mg/ml of 5′FOA and 10 mM uracil. 5′FOA resistant, uracil requiring mutants were transformed with pAB4.1 harboring the A. niger pyrG or with pPyrE harboring the A. niger pyrE gene on a 4.3 kb SstII subclone, respectively. One of the selected 5′FOA resistant mutants, XY4.1, was complemented with the pyrG gene, indicating that the uracil auxotrophy was caused by a mutation in the pyrG gene. The strain XY4.1 was then used to construct a ΔinuR ΔamyR double disruption strain by transforming plasmid pΔamyR which also contains the pyrG selection marker (Yuan et al., submitted). Uridine prototrophic transformants were selected by incubating protoplasts on agar plates containing MM without uridine. Transformants that showed normal growth on glucose but not on starch were selected. One possible ΔinuRΔamyR double mutant (XY5.1) was verified by Southern analysis and used for further analysis.
Northern blot analysis
Total RNA isolation, Northern analysis and synthesis of DNA probes was performed as described in Yuan et al. (2006). The primers used to generate probe An15g00310 are shown in Supplementary Table 1.
Microtiter plate growth assay
Growth of A. niger strains were determined using a HTS7000 BioAssay Reader (Perkin Elmer Life Sciences). Spores (1 × 104) were inoculated in each well of a 96-well microtiter plate (Nalge Nunc International, USA) and incubated at 32°C for 56 h. Each well contained 200 μl of MM containing 1% (w/v) of one of the various carbon sources each supplemented with 0.01% (w/v) casamino acids to stimulate spore germination. Six replicates of each condition were made. Growth was monitored by measuring turbidity (OD595) at 2-h intervals.
Microarray experiments and data analysis
RNA extracted from the A. niger ΔinuR strain and wild-type strain (N402) grown on different carbon sources were used for microarray experiments using custom made “dsmM_ANIGERa_coll” Affymetrix GeneChip® microarrays kindly provided by DSM Food Specialties (Delft, The Netherlands). For microarray experiments, mycelia were isolated from grown cultures by transfer experiment (see above) and each growth condition was performed in duplicate as independent biological experiments.
Total RNA was isolated from mycelia using TRIzol reagent (Invitrogen) and RNA quality was verified by analyzing aliquots with glyoxal/DMSO gel electrophoresis and Agilent Bioanalyzer “Lab-on-chip” system (Agilent Technologies, USA). Processing, labeling and hybridization of cRNA to A. niger Affymetrix GeneChips were performed according to the corresponding Affymetrix protocols for “Eukaryotic Target Preparation” and “Eukaryotic Target Hybridization”. For probe array washing and staining, the protocol “Antibody Amplification for Eukaryotic Targets” was followed. Hybridized probe array slides were scanned with Agilent technologies G2500A Gene Array Scanner at 3 μm resolution and a wavelength of 570 nm. Affymetrix Microarray Suite software MAS5.0 was used to calculate the signal and P values and to set the algorithm’s absolute call flag, which indicates the reliability of the data points according to P (present), M (marginal) and A (absent). The data on each chip were globally scaled to an arbitrary target gene intensity of 500.
The pre-scaled chip data from each hybridization experiment were normalized using Genespring 7.0 software (Silicon Genetics). Normalization with default parameters in Genespring software (Per Chip: Normalize to 50th percentile, Per Gene: Normalize to median) was used. For this study, we focused on sucrose induced genes. Therefore, pre-filtering of data was performed to select for genes whose detection calls are present in both sucrose duplicate samples of wild-type strain N402. The selected data set was further performed for one-way ANOVA analysis. Fold changes in expression levels between two different conditions were then computed for genes with P < 0.05 based on One-way ANOVA analysis and changes by more than twofold were considered significant and are reported here. Microarray data were deposited into ArrayExpress with an accession number 19095 at http://www.ebi.ac.uk/at-miamexpress.
In silico analysis of regulatory elements of co-regulated genes
The 1.0 kb promoter sequences of the seven sucrose induced InuR dependant genes: An12g08280 (InuE), An11g03200 (InuA), An08g11070 (SucA), An15g04060, An15g00310, An15g03940, and An17g01710 were obtained from the CBS513.88 strain. The 1.0 kb promoter regions were analyzed on both strands using DNA Analyzer module of Phylosopher 6.5.1 (Genedata A.G., Basel, Switzerland) using the Gibbs Sampling Strategy for Multiple Alignment (Lawrence et al. 1993).
Results
Identification of potential transcriptional regulators involved in inulin catabolism
A. niger is able to produce various inulinolytic enzymes involved in the modification or degradation of inulin and sucrose. The transcriptional regulation of the genes encoding the inulinolytic enzymes of A. niger, including an exo-inulinase (InuE; An12g08280), an endo-inulinase (InuA; An11g03200), and an invertase (SucA; An08g11070), has been studied previously (Moriyama et al. 2003; Yuan et al. 2006). In addition to the extracellur enzymes, two additional potential intracellular inulinolytic enzymes (SucB; An15g00320 and SucC; An06g02420) were predicted from the A. niger genome sequence (Yuan et al. 2006). These proteins contain all the conserved domains of the GH32 family to which inulinolytic enzymes belong. Finally, a potential ORF was found in the genome which showed homology to the group of exo-inulinases. Because the predicted ORF lacked several of the conserved domains and contained several frame shift mutations, this gene (inuQ, An11g03210) was considered to be a pseudogene (Yuan et al. 2006).
The expression of the genes encoding the extracellular inulinolytic enzymes is co-regulated and induced on inulin and sucrose, suggesting that these genes are under control of a single transcription factor (Yuan et al. 2006). Some transcriptional activators involved in sugar catabolism are clustered in the genome with their target genes (Gomi et al. 2000; Yu et al. 2000). To identify possible candidate transcription factors involved in the regulation of inulinolytic genes, we searched the A. niger genome sequence for transcription factors located close to the genes encoding the inulinolytic enzymes. We found that inuA (An11g03200) and inuQ (An11g03210) are located next to each other and that also a Zn(II)2Cys6 transcription factor encoding gene (An11g03220) was located adjacent to inuQ. The orientation of the three ORFs is in the same direction indicating that they do not share a common promoter region (Fig. 1).
Schematic representation of the clustering of putative transcription factors with inulinolytic genes in the genome of A. niger. An11g03220 and An08g11040 and inuR encode putative transcription factors. The genes related to inulin degradation include: inuA (endo-inulinase), inuQ (pseudogene), sucA (invertase), sucB (homologous to sucA) and An15g00310 (putative sugar transporter). An08g00160 encodes a hypothetical protein without known function. The arrows indicate the transcriptional orientations of the genes
In the proximity of sucA (An08g11070), a second transcription factor encoding gene was found. This transcription factor contains a Cys2His2 zinc finger DNA binding motif. Between this gene, An08g11040 and sucA an additional ORF (An08g11060) is present. This 273 amino-acid protein encodes a hypothetical protein without any conserved motifs. A BlastP search revealed that this protein has no close orthologs in other fungal genomes. The transcription factor, encoded by An08g11040, shows strong sequence similarity to the S. cerevisiae Zpr1p (e-value 1e-100), an essential transcription factor that contributes to normal cell proliferation (Gangwani et al. 1998).
Clustered with sucB (An15g00320) a third putative transcription factor encoding gene (An15g00300) was found which also belonged to the Zn(II)2Cys6 transcription factor family. Between sucB and An15g00300, an additional ORF (An15g00310) is located (Fig. 1). The protein encoded by this gene has all the characteristics of a sugar transporter protein. In the direct proximity of the two remaining genes encoding inulinolytic enzymes (InuE and SucC) no potential transcription factors were found. The three transcription factors identified via clustering were considered to be good candidates to be involved in the transcriptional regulation of the inulinolytic system of A. niger. In order to study their possible role in inulin utilization, deletion strains of these transcription factors were constructed. Deletion of An11g03220 did not have any measurable effect on growth on inulin or sucrose in comparison with the wild-type strain, which indicates that this gene is not required for inulin utilization. Growth of the deletion strain on xylose, glucose, fructose, starch and maltose was also identical to the growth of the wild-type strain (data not shown). A deletion strain of An08g11040 was never obtained although over 500 transformants were screened. A possible explanation might be that An08g11040 is an essential gene. As indicated above, An08g11040 is most homologous to the essential Zpr1p transcription factor of S. cerevisiae. Since the homology with Zpr1p does not indicate involvement of this transcription factor in inulin utilization, no further attempts were made to obtain the deletion strain. As shown in detail below, deletion of An15g00300 resulted in a strain which showed a severe growth defect on inulin and on sucrose. For the remainder of the paper, we will refer to An15g00300 as inuR.
InuR homologs were also identified in the genomes of other Aspergilli. The alignment of the InuR proteins is given in Fig. 2. The strong conservation of the transcription factor among Aspergilli, suggest that the regulation of inulinolytic gene expression is mediated in the various Aspergilli by the InuR proteins present in the different fungi.
Alignment of InuRp and its orthologs in Aspergilli. The amino acid sequences of the Zn2Cys6 binuclear DNA-binding motif (Pfam00172) and the Fungal specific transcription factor domain (Pfam04082) are underlined. The conserved six cystein residues are indicated by stars. AoInuRp, AgInuRp, AfInuRp and AdInuRp are InuR protein orthologs from A. oryzae, A. niger, A. fumigatus and A. nidulans, respectively
InuR encodes a Zn(II)2Cys6 transcription factor that is required for growth on inulin
Based on the analysis of the CBS513.88 genomic sequence, the predicted open reading frame of the inuR encoding gene is 2,466 bp long and interrupted by 4 introns with sizes of 153, 80, 58 and 48 bp, respectively. The predicted InuR protein sequence is comprised of 709 amino acid residues which would result in a protein with a calculated molecular mass of 78.3 kDa. Analysis of the predicted InuR protein indicated that InuR contains two conserved domains. One is the Zn(II)2Cys6 (CX2CX6CX5CX2CX6C) binuclear cluster which represents the DNA-binding domain at the NH2-terminal end (residues 35–61). The domain is very well conserved (Pfam00172) and binds two Zn atoms which coordinate folding of the domain. A BlastP search revealed orthologous transcription factors in the genome of A. nidulans, A. oryzae and A. fumigatus (see above, Fig. 2). The most similar transcription factors to InuR which have been functionally characterized, apart from the proposed orthologs in A. oryzae, A. fumigatus and A. nidulans, are the AmyR transcription factors from the different Aspergillus species. Subsequent phylogenetic analysis indicated that the subgroup of InuR transcription factors is most closely related to the group of AmyR transcription factors (data not shown). Besides the Zn(II)2Cys6 motif, a second conserved domain, known as the middle homology region (MHR) or Pfam04082 domain, is present in Zn(II)2Cys6 transcription factors. The region is thought to assist the Zn(II)2Cys6 cluster in DNA target discrimination (Schjerling and Holmberg 1996). In the A. niger InuR protein, the MHR/Pfam04082 domain compromises aa 353–425, and the domain is also present in the InuR proteins of the other Aspergilli.
Construction and growth properties of the inuR disruptant in A. niger N402
An inuR disruptant mutant was constructed as described in the “Materials and methods”. Putative disruption strains were identified by PCR screening, and further verified by Southern blot analysis. Strain XY3.1 was selected to further characterize the InuR function and we will refer to this strain as the ΔinuR strain.
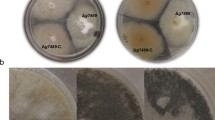
The effect of the disruption of the inuR gene on the utilization of various carbon sources was analyzed on agar plates. Growth and conidiation of the ΔinuR strain was strongly reduced on plates containing inulin in comparison to the wild-type strain (Fig. 3). As shown, growth of the ΔinuR strain on inulin was not completely abolished. After 7 days of growth, a thin colony with a reduced degree of conidiation was formed. Some effect on growth and conidiation was also observed for the ΔinuR strain growing on plates containing sucrose and to a lesser extent on raffinose. The differences in growth on sucrose and raffinose were most clear after 3 days of growth (Fig. 3). Both the colony size and the degree of conidiation of the ΔinuR strain and the wild-type strain after 7 days of growth were very similar (data not shown). The ΔinuR strain showed a similar growth phenotype to the wild-type on MM plates containing fructose, glucose, maltose, starch, xylose, sorbitol or glycerol as sole carbon source (Fig. 3 and data not shown).
Growth of the ΔinuR strain on different carbon sources was also determined by analyzing growth in a microtiter plate well containing MM with various carbon sources. Equal numbers of spores of the wild-type strain (N402) and ΔinuR strain were inoculated and plates were incubated at 32°C. For each condition at least four replicates were made and the average value of the replicates and its standard deviation value were determined as shown in Fig. 4. The ΔinuR showed a clear growth defect on inulin and sucrose. A small, but reproducible growth difference in growth on raffinose on agar plates was also observed in the microtiter plate analysis. Initially, the growth curves of the wild-type and the ΔinuR strain on raffinose were identical, but after 24–32 h growth of the ΔinuR strain seem to be retarded (Fig. 4). The ΔinuR strain grew equally well as the wild-type strain on MM with maltodextrin, maltose, glucose, xylose or fructose as sole carbon source (Fig. 4 and data not shown). In conclusion, the growth analysis of the ΔinuR strain indicates that the Zn(II)2Cys6 transcription factor is required for the efficient utilization of inulin and and to a lesser extent raffinose, and it may therefore act as a positive transcriptional activator of the genes encoding the inulinolytic enzymes. To further substantiate that the defective growth of the ΔinuR strain on inulin was caused by the disruption of the inuR gene, a plasmid containing the inuR gene with 1 kb upstream and 0.5 kb downstream region was constructed and co-transformed into the ΔinuR strain with pAN7.1 (Punt et al. 1987). Hygromycin resistant transformants were analyzed for growth on inulin and subjected to Southern blot analysis. Transformants that showed restored growth on inulin all harbor the inuR gene, while transformants showing defective growth on inulin did not contain the inuR gene (data not shown). The restoration of the wild-type growth on inulin upon retransformation of the ΔinuR strain with a functional copy of the inuR gene indicated that the inability to grow on inulin was indeed a consequence of inuR disruption.
Microtiter plate assay of growth of inuR disruptant on different carbon sources. A. niger wild-type (N402; open circles) strain and inuR disruptant (ΔinuR; solid circles) were inoculated in microtiter plate wells and each well was inoculated with 1 × 104 spores in 200 μl of MM containing 1% carbon source and 0.1% casamino acids. The growth was monitored by measuring culture turbidity (OD595) and read every 2 h. Each condition was performed in six replicates. The standard deviation value is indicated
InuR is required for the induction of inulinolytic genes
To further establish the role of InuR as a positive acting transcriptional regulator for inulinolytic gene expression, Northern blot analysis was performed to determine the mRNA level of the various inulinolytic enzyme encoding genes both in the ΔinuR strain and the WT strain. We previously showed that the expression of the genes encoding the exo-inulinase (inuE), endo-inulinase (inuA), invertase (sucA), was detected only in the presence of inulin and sucrose and not on other carbon sources tested such as xylose, glucose, maltose or starch. Expression of the sucB gene was low on all the carbon sources tested and slightly induced in the presence of inulin and sucrose (Yuan et al. 2006).
In order to clarify whether the expression of these inulinolytic genes was controlled by InuR, both the wild-type strain and the ΔinuR strain were pre-grown for 18 h in MM containing xylose as a carbon source. After the pre-growth, equal amounts of mycelium was transferred to fresh MM containing 1% (w/v) inulin or 1% (w/v) sucrose as a carbon source. At different time points after the transfer RNA was extracted from mycelia and subjected to Northern blot analysis (Fig. 5). The transfer to inulin and sucrose resulted in an induction of the inulinolytic enzymes in the wild-type strain. The induction of genes is much faster on sucrose than on inulin and is explained by a much more rapid degradation and uptake of the carbon source as observed previously (Yuan et al. 2006). In the ΔinuR strain, no induction of expression was found for the inuE and inuA genes after transfer to inulin and a very low level of sucA mRNA was detected after the transfer to inulin. The transfer to sucrose did not result in a detectable induction of any of the three inulinolytic genes in the ΔinuR strain. A slight induction of expression of sucB in the presence of inulin and sucrose seems also dependent on InuR (Fig. 5). The expression of the gene encoding a putative sugar transporter, An15g00310 in relation to inulin and sucrose was also examined. As discussed above, An15g00310 is clustered and located between the inuR and the sucB genes (Fig. 1). Expression of An15g00310 is strongly induced in response to the presence of inulin and sucrose and dependant on the InuR transcription factor (Fig. 5). No or very low expression levels of An15g00310 were detected when grown on maltose, glucose, fructose or xylose or after a transfer to maltose indicating that An15g00310 expression is specific for growth on inulin and sucrose (data not shown). The inuR gene was constantly expressed at a relatively low level on the various carbon sources tested (inulin, sucrose, maltose, fructose or glucose, data not shown), suggesting that the activation of the InuR transcription factor is regulated post-transcriptionally.
Genome wide transcriptional profiling of the InuR regulon in A. niger
The experiments described above suggest that InuR is a transcriptional activator of inulinolytic genes and required for the efficient utilization of inulin and sucrose. In addition to regulating the inulinolytic genes, it is possible that InuR also regulates other genes in the genome, through direct or indirect processes in the pathway of inulin or sucrose utilization. To identify additional genes with a possible role in inulin or sucrose metabolism, we performed genome wide expression analysis using A. niger Affymetrix GeneChip arrays. The A. niger wild-type strain N402 and the ΔinuR strain were pre-grown for 18 h in 2% (w/v) xylose and transferred to fresh medium containing 1% (w/v) sucrose as carbon source and grown for 2 h on sucrose. As a control, the mycelium from the N402 strain was also transferred to fresh 1% (w/v) xylose medium. For each culture condition a duplicate experiment was carried out. RNA was isolated from each of the samples and subjected to microarray analysis (see “Materials and methods”).
To identify genes that were induced by sucrose, we first compared the expression profile of A. niger wild-type strain N402 grown in sucrose to that of A. niger wild-type strain N402 grown in xylose 2 h after transfer. The expression of 97 genes was more than twofold increased by sucrose (One-way ANOVA analysis P < 0.05) (Supplementary Table 2). To identify genes that were induced on sucrose in an InuR dependant way, the expression profile of the A. niger wild-type strain N402 grown in sucrose was compared to that of the ΔinuR strain after the 2 h transfer to sucrose. A total of 40 genes were identified where expression was significantly increased on sucrose in the N402 strain, but not in the ΔinuR strain (One-way ANOVA analysis P < 0.05 and >2-fold change) (Supplementary Table 3.) Finally, by combining these 2 data sets, 26 genes were identified for which expression was induced by sucrose in an InuR dependent manner (Table 1).
The predicted protein sequences of these 26 genes were assigned to various functional categories (Ruepp et al. 2004; Mewes et al. 1997) and shown in Table 1. Interestingly, genes with the highest fold changes (>10-fold change) almost all fall into the category of carbohydrate transport and metabolism. Among them are the genes encoding the extracellular inulinolytic enzymes, sucA, inuE and inuA, as well as the above mentioned putative sugar transporter gene (An15g00310), supporting the previous Northern analysis. An additional gene that showed a very high fold-change and belongs to the category of carbohydrate transport and metabolism is An15g04060. An15g04060 encodes a putative sugar transporter which is over 200-fold induced by sucrose in a wild-type strain N402 when compared to xylose and the induction was greatly reduced by 127-fold when inuR gene was deleted. BlastP search revealed that the transporter displayed the highest identity to a fructose transporter protein (Frz1) from Zygosaccharomyces bailii (Pina et al. 2004). An additional putative sugar transporter gene (An15g03940) was induced on sucrose in an InuR dependant way. In contrast to the other two transporters (An15g00310 and An15g04060) this transporter is relatively highly expressed on both xylose and in the ΔinuR mutant, resulting in relative small fold-changes. An additional gene (An17g01710) was strongly induced on sucrose (about 19-fold), in an InuR dependant way. The protein encoded by An17g01710 is annotated as a multidrug resistance (MDR) protein because of its highest identity to other MDR proteins of the Major Facilitator Superfamily of transporters. Since the substrates of MDR proteins are hard to predict on the basis of the amino acid sequence, the annotation should be interpreted with care and does not exclude the possibility that this protein has a role in sugar transport. Apart from the An15g04060 transporter which might function as a fructose transporter based on the strong homology towards Frz1p (Pina et al. 2004), a useful prediction of substrates of the other putative transporters can not be made.
In the list of induced genes (Table 1), a considerable number are related to transcription, translation and amino acid metabolism. The increase in expression of those genes might indicate an overall increase of metabolic activity on sucrose compared to xylose and a subsequent higher expression of the protein transcription/translation machinery.
The genome wide analysis indicated that three extracellular inulinolytic genes and four putative sugar transporter genes were strongly induced on sucrose in an InuR dependant way. To identify possible elements in the promoter regions of these genes that could mediate InuR binding an in silico analysis of 1-kb upstream promoter sequences of these seven genes was performed. As shown in Fig. 6, optimal alignment of the promoter sequences revealed a consensus sequence (CGGN8CGG) which is present at least once in the promoter region of all genes. At least one of the triplets consistently is preceded by a T, resulting in a conserved TCGG motif in every proposed element. Within the second triplet, the conservation of the GG is consistent, but conservation of the C is not. This putative binding site of the InuR protein (CGGN8CGG) is identical to the binding site of the AmyR transcription factor which binds and induces the expression of amylolytic genes (Petersen et al. 1999). To examine whether AmyR is also involved in the regulation of inulinolytic genes, the ΔamyR mutant was constructed and analysed for growth on various carbon sources. As shown in Fig. 3, deletion of the amyR transcription factor did not result in a growth defect on inulin, sucrose or other carbon sources. The ΔamyR strain did show a strong growth reduction on maltose (data not shown). To examine a possible overlapping role of the InuR and AmyR transcription factors for inulin and/or sucrose utilization, the ΔamyR/ΔinuR double mutant was constructed. Growth analysis of the double mutant on inulin showed a minor further reduction in growth and sporulation on inulin and sucrose (Fig. 3) indicating AmyR and InuR have independent functions.
Discussion
We previously showed that the genes encoding extracellular inulinolytic enzymes in A. niger are co-regulated at the transcription level and identified sucrose, but not fructose or glucose as an efficient inducer of inulinolytic genes. It was proposed that there might be a common transcription factor to activate the expression of inulinolytic genes in response to the presence of inducing carbon sources (Yuan et al. 2006), similar to that which has been found in other carbohydrate degrading enzymes networks such as the amylolytic genes (AmyR) or the xylanolytic genes (XlnR). In this study we reported the successful identification of an inulinolytic transcriptional activator, InuR. Previous studies to isolate the AmyR and XlnR transcription factors were based on genetic screens to identify mutants with impaired expression of amylolytic and xylanolytic enzymes (Petersen et al. 1999; Gomi et al. 2000; van Peij et al. 1998a). Here, we used a different approach which made use of the full genome sequence of A. niger. The approach is based on the observation that in some cases, e.g., in the case of the AmyR transcription factor, the target genes of the transcription factor are adjacent to the transcription factor itself. In the case of the inulinolytic genes, three possible inulinolytic gene clusters were identified, each of which contains a gene encoding a putative transcription factor (Fig. 1). Disruption mutants were successfully obtained for two of the three transcription factors and subsequent phenotypic analysis indicated that only InuR (An15g00300) was involved in the inulin utilization pathway. The ΔinuR mutant showed severe reduced growth on agar plates and in submerged cultures containing sucrose or inulin as sole carbon source. Furthermore, we showed that the induction of the inulinolytic genes by inulin and sucrose is dependant on the presence of InuR. In addition to the transcription factor itself the inulinolytic gene cluster identified in A. niger consists of a sugar transporter and sucB which encodes an intracellular invertase with transfructosylating activity (Goosen et al. 2007).
To examine whether the position and clustering of the InuR orthologs was conserved among aspergilli, we compared the genes adjacent to the InuR homologous transcription factors in A. nidulans, A. oryzae and A. fumigatus. The clustering of the three genes, inuR (An15g00300), An15g00310 (putative sugar transporter) and sucB (An15g00320) in A. niger was conserved in A. nidulans (AN3835; e-value 5e-144, AN3836; e-value 7e-158 and AN3837; e-value 5e-142), and A. oryzae (AO090701000036; e-value 0.0, AO090701000037; e-value 0.0 and AO090701000038; e-value 0.0, respectively). In A. fumigatus, the InuR orthologue (Afu6g05010; e-value 0.0) is located next to the SucB orthologue (Afu6g05000; e-value 0.0), but no homolog to the putative sugar transporter (An15g00310) was present in the gene cluster in A. fumigatus. The initial gene model of the inuR gene in the A. niger annotation was lacking the ZnII(2)Cys6 binuclear cluster domain. Repositioning the intron/exon positions in N-terminal region resulted in the prediction of a 709 amino acid long protein containing a complete Zn(2)Cys6 binuclear cluster domain. BlastP searches with the A. niger InuR protein revealed orthologous genes in the three Aspergillus species examined (A. nidulans, A. oryzae and A. fumigatus). The closest homologs in A. oryzae and A. nidulans did not contain a Zn(II)2Cys6 domain and the A. fumigatus homolog only a partial Zn(II)2Cys6 cluster. The gene models of the other Aspergilli were manually inspected and by improving the gene model a Zn(II)2Cys6 domain was found to be present in the proteins. Interestingly, it turned out that the gene model of A. niger was completely conserved in the other Aspergili. Thus, the gene model of the A. niger inuR gene could be used as a template for the prediction of the inuR gene models in the other Aspergilli. Both the numbers of introns as well as the intron/exon positions were conserved among the four Aspergilli. This result implies that gene model prediction in Aspergilli can be improved by comparison of the gene models of orthologous genes.
The inuR gene is situated in the genome next to a putative sugar transporter gene (An15g00310) and sucB. We recently showed that the sucB gene encodes an intracellular invertase with transglycosylating activity (Goosen et al. 2007). As depicted in Fig. 1, the putative sugar transporter (An15g00310) in the cluster shares the same promoter region with the sucB gene. The Northern analysis and microarray analysis showed that the two genes are not coregulated indicating that the common promoter region does not act bidirectionally as has been observed, e.g., in the case of the niiA and niaD genes (Punt et al. 1995). Expression of the gene An15g00310 was highly induced by sucrose or inulin and the induction was dependent on InuR (Fig. 5), while the sucB gene was lowly expressed on various carbon sources and hardly induced on sucrose and inulin. The function of An15g00310 remains to be elucidated but it is tempting to suggest that it involved in the transport of sucrose, or its hydrolysis products fructose or glucose.
The function of SucB in relation to inulin or sucrose metabolism is also currently unknown. We showed that deletion of the sucB did not result in a phenotype related to growth on inulin or sucrose (Goosen et al. 2007). The constitutive low level of expression of the sucB gene might suggest that SucB acts as a scouting enzyme to “sense” the presence of a useful carbon source by generating an inducer molecule which can further activate transcription of additional inulinolytic genes. A possible explanation for the lack of phenotype of the sucB deletion mutant is the presence of an addition intracellular invertase, encoded by the sucC gene. Both the enzymatic activity of the SucC protein as well as the effect of deletion of sucC and simultaneously deletion of sucB and sucC will be the subject of future research.
The genome wide transcription analysis using Affymetrix microarrays identified a limited number of genes that were strongly induced by sucrose in an InuR dependent way. These genes encode the extracellular inulinolytic enzymes and four genes encoding putative transporters. The specificity of the sugar transporters should be determined biochemically as has been done previously for A. niger sugar transporters (vanKuyk et al. 2004) since phylogenetic comparison to suggest possible substrates is not reliable (Patricia vanKuyk, personal communication). The limited number of genes that are induced by sucrose in an InuR dependant way suggests a relatively simple regulatory mechanism regarding sucrose catabolism in A. niger. Unpublished additional microarray data showed that the same three extracellular enzymes were induced on inulin in an inuR dependant way. The expression of the putative sugar transporters on inulin differed from the pattern observed on sucrose. Two of the four transporters (An17g01710 and An15g04060) were not induced on inulin after 2 and 8 h of growth on inulin. The transporter encoded by the An15g03940 gene was four to fivefold induced on inulin compared to xylose after 8 h of the transfer. Expression of this sugar transporter is also high on xylose (Table 1) and maltose, Yuan et al., unpublished results), indicating that the expression of this sugar transporter is not very tightly regulated and suggest a broad specificity of the sugar transporter. The fourth transporter (An15g00310) was more highly expressed on inulin 8 h after the transfer than on maltose and xylose (four to fivefold induction), and the induction was InuR dependant. The difference between the observed expression patterns of the extracellular enzymes and the putative sugar transporters suggest that under the different growth conditions the fungus expresses the same set of extracellular enzymes, but that the fungus fine-tunes the expression of the sugar transporters in order to allow efficient uptake. Such a fine-tuning mechanism could explain why such a large number of sugar transporters is present in the genome of A. niger as 461 proteins have been identified that belong to the Major Facilititator Superfamily of transporters (Pel et al. 2007). Such a mechanism allows the fungus to express the sugar transporters with the optimal properties in relation to substrate diversity, substrate concentration, environmental pH, and other environmental factors and nutrient conditions.
The InuR transcription factor is a member of the Zn(II)2-Cys6 DNA transcription factor family which is a fungal specific type of transcription factor. In Aspergilli, this family of transcription factors has expanded significantly when genome data is compared to that of the yeast S. cerevisiae or Neuropora crassa. In S. cerevisiae and N. crassa 54 and 89 genes, respectively (Akache et al. 2001; Borkovich et al. 2004), have been identified that contain the Zn(II)2-Cys6 DNA binding domain, the genomes of Aspergilli contain 216 (A. nidulans), 188 (A. fumigatus) or 296 (A. niger) Zn(II)2-Cys6 DNA transcription factors. The actual number of proteins containing a Zn(II)2-Cys6 DNA binding domain might still increase in Aspergilli and other fungi. We showed that the predicted gene model for the inuR gene in several Aspergilli was incorrect, resulting in a protein lacking the Zn(II)2-Cys6 DNA binding domain. Closer examination of the A. nidulans and A. niger genome annotations revealed that the difficulty of the automatic annotation in predicting the correct gene model for this class of transcription factors is not limited to inuR gene, but is a more general problem. Manual improvement of the gene models identified 326 Zn(II)2-Cys6 DNA binding domain containing proteins in A. nidulans and 480 Zn(II)2-Cys6 DNA binding domain containing proteins in A. niger (Ram and Punt, unpublished data). Thus, it is likely that other fungal genomes contain more as yet unidentified Zn(II)2-Cys6 DNA binding domain containing proteins, which could be revealed with improved gene models or manual annotation.
Transcription factors containing the Zn(II)2-Cys6 DNA binding domain usually bind the promoter region of their target genes via a conserved DNA binding site. Often, but not always, the binding site consists of conserved terminal trinucleotides, usually in symmetrical configuration (direct repeat, everted repeat or an inverted repeat) and spaced by an internal variable sequence of a defined length. Examples are the Gal4p and Lac9p transcription factors that bind to CGGN11CCG, Ppr1p and UaY that bind to CGGN6CGG, and Put3 that binds to CGGN10CCG (Todd and Andrianopoulos 1997). The binding sites for the AmyR transcription factor have been studied in detail. Several promoter regions of amylolytic genes contain a CGGN8(A/C)GG site in their promoter region to which AmyR can bind and activate transcription (Tani et al. 2001b; Ito et al. 2004).
Comparison of the 1-kb upstream promoters of the strongly InuR dependent sucrose induced genes revealed that a common consensus sequence (CGGN8CGG) is present in the promoters of all genes. Strikingly, in every putative element at least one repeat consists of a TCGG sequence (Fig. 6). This observation might be explained by a speculative, but testable, model of InuR binding and activation. We propose that InuR will bind as a dimer to the CGGN8CGG motif, and that the first InuR molecule will bind with high affinity to the TCGG DNA sequence. This binding may recruit a second InuR molecule to the site that will bind to the second motif, possibly with a weaker affinity for the DNA because of the sequence variation. Such a model for cooperative binding of a dimeric transcription factor has been experimentally substantiated for the AmyR transcription factor (Ito et al. 2004). The proposed InuR binding site (CGGN8CGG) might be conserved among filamentous fungi, as the shared promoter of the inuC and inuD genes in Penicillium sp. strain TN-88 contains a similar site (Moriyama et al. 2006).
The proposed InuR binding site is very similar to the AmyR binding site which is present in the promoter of many amylolytic genes in A. niger (Yuan et al., unpublished results) and raises immediately the question of how the specificity of the InuR and AmyR transcription factors is accomplished. Comparison of the Zn(II)2Cys6 domain of several Zn(II)2Cys6 transcription factors revealed that the Zn(II)2Cys6 domain of InuR and AmyR is not more related to each other than they are related to unrelated Zn(II)2Cys6 transcription factors (XlnR and FacB). Interestingly, the MHD of InuR and AmyR showed a high degree of identity and suggest that the MHD domain may assist in binding to the DNA target sequence. After establishing that the conserved CGGN8CGG motif in the promoter of inulinolytic genes is involved in the binding and activation of expression of the target genes, the AmyR and the InuR transcription factors are interesting subjects for further research aimed to understand how Zn(II)2-Cys6 transcription factors recognize their target genes and what determines the specificity of the binding. Analysis of the ΔamyRΔinuR double mutant indicated that both transcription factors have independent functions. The slightly decreased growth and sporulation of the ΔamyRΔinuR double mutant on sucrose compared to growth of the ΔinuR mutant on sucrose might be explained by the activity of α-glucosidases that are under control of the AmyR transcription factor. It is well established that sucrose can be hydrolyzed either by β-fructosidase, that splits sucrose by attacking the terminal, non-reducing α1, β2 linked fructose moiety, but also by an α-glucosidase catalyzing the hydrolysis of sucrose at the terminal non-reducing α1–4 linked glucose residue (Geber et al. 1992; Kelly and Kwon-Chung 1992; Alberto et al. 2004). A. niger contains several α-glucosidase encoding gene of which two of them are under control of the AmyR transcription factor. Disruption of the amyR transcription factor further decreases the level of agdA and agdB transcripts (Yuan et al., unpublished results), which might result in lower α-glucosidase activity and a subsequent decrease in the efficiency by which sucrose can be hydrolyzed.
In conclusion, we have identified InuR as the transcriptional activator required for utilization of inulin and sucrose. The results in this paper suggest that A. niger metabolizes inulin and sucrose by inducing the expression of a limited number of extracellular enzymes and sugar transporters that are under transcriptional control of a single transcription factor, InuR. The mechanism by which the InuR transcription factor is activated in response to the presence of inulin and sucrose and the mechanism that determines the specificity of the binding to the InuR target genes are the subjects of future research.
References
Akache B, Wu K, Turcotte B (2001) Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res 29:2181–2190
Akimoto H, Kushima T, Nakamura T, Ohta K (1999) Transcriptional analysis of two endoinulinase genes inuA and inuB in Aspergillus niger and nucleotide sequences of their promoter regions. J Biosci Bioeng 88:599–604
Alberto F, Bignon C, Sulzenbacher G, Henrissat B, Czjzek M (2004) The three-dimensional crystal structure of invertase (β-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases. J Biol Chem 279:18903–18910
Bennet JW, Lasure LL (1991) Growth media. In: Bennet JW, Lasure LL (eds) More gene manipulations in fungi. Academic, San Diego, pp. 441–447
Berges T, Barreau C, Peberdy JF, Boddy LM (1993) Cloning of an Aspergillus niger invertase gene by expression in Trichoderma reesei. Curr Genet 24:53–59
Boddy LM, Berges T, Barreau C, Vainstein C, Dobson MJ, Balance DJ, Peberdy JF (1993) Purification and characterization of an Aspergillus niger invertase and its DNA sequence. Curr Genet 24:60–66
Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ, Zelter A, Kalkman ER, O’Rourke R, Bowring F, Yeadon J, Ishii C, Suzuki K, Sakai W, Pratt R (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68:1–108
Bos CJ, Debets AJ, Swart K, Huybers A, Kobus G, Slakhorst SM (1988) Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet 14:437–443
Burger G, Strauss J, Scazzocchio C, Lang BF (1991) nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. Mol Cell Biol 11:5746–5755
Cazelle B, Pokorska A, Hull E, Green PM, Stanway G, Scazzocchio C (1998) Sequence, exon–intron organization, transcription and mutational analysis of prnA, the gene encoding the transcriptional activator of the prn gene cluster in Aspergillus nidulans. Mol Microbiol 28:355–370
Crabeel M, de Rijcke M, Seneca S, Heimberg H, Pfeiffer I, Matisova A (1995) Further definition of the sequence and position requirements of the arginine control element that mediates repression and induction by arginine in Saccharomyces cerevisiae. Yeast 11:1367–1380
de Ruiter-Jacobs YM, Broekhuijsen M, Unkles SE, Campbell EI, Kinghorn JR, Contreras R, Pouwels PH, van den Hondel CA (1989) A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae. Curr Genet 16:159–163
El Alami M, Feller A, Pierard A, Dubois E (2000) Characterization of a tripartite nuclear localization sequence in the regulatory protein Lys14 of Saccharomyces cerevisiae. Curr Genet 38:78–86
Flynn PJ, Reece RJ (1999) Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol Cell Biol 19:882–888
Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ (1998) Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol 143:1471–1484
Geber A, Williamson PR, Rex JH, Sweeney EC, Bennett JE (1992) Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J Bacteriol 174:6992–6996
Gielkens MM, Dekkers E, Visser J, de Graaff LH (1999) Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol 65:4340–4345
Gomez D, Cubero B, Cecchetto G, Scazzocchio C (2002) PrnA, a Zn2Cys6 activator with a unique DNA recognition mode, requires inducer for in vivo binding. Mol Microbiol 44:585–597
Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, Iimura Y (2000) Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci Biotechnol Biochem 64:816–827
Goosen C, Yuan XL, van Munster JM, Ram AF, van der Maarel MJ, Dijkhuizen L (2007) Molecular and Biochemical characterization of a novel intracellular invertase from Aspergillus niger with transfructosylating activity. Eukaryot Cell 6:674–681
Gupta A K, Kaur N (1997) Fructan storing plants—a potential source of high fructose syrups. J Sci Ind Res 56:447–452
Hasper AA, Trindade LM, van der Veen D, van Ooyen AJ, de Graaff LH (2004) Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiology 150:1367–1375
Inoue JC, Nojima H, Okayama H (1990) High efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28
Ito T, Tani S, Itoh T, Tsukagoshi N, Kato M, Kobayashi T (2004) Mode of AmyR binding to the CGGN8AGG sequence in the Aspergillus oryzae taaG2 promoter. Biosci Biotechnol Biochem 68:1906–1911
Kaur N, Gupta AK (2002) Applications of inulin and oligofructose in health and nutrition. J Biosci 27:703–714
Kelly R, Kwon-Chung KJ (1992) A zinc finger protein from Candida albicans is involved in sucrose utilization. J Bacteriol 174:222–232
Kirkpatrick CR, Schimmel P (1995) Detection of leucine-independent DNA site occupancy of the yeast Leu3p transcriptional activator in vivo. Mol Cell Biol 15:4021–4030
Kohlhaw GB (2003) Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev 67:1–15
Kolar M, Punt PJ, van den Hondel CA, Schwab H (1988) Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62:127–134
Kulmburg P, Sequeval D, Lenouvel F, Mathieu M, Felenbok B (1992) Identification of the promoter region involved in the autoregulation of the transcriptional activator ALCR in Aspergillus nidulans. Mol Cell Biol 12:1932–1939
Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science 262:208–214
L’Hocine L, Wang W, Jiang B, Xu S (2000) Purification and partial characterization of fructosyltransferase and invertase from Aspergillus niger AS0023. J Biotechnol 81:73–84
Mewes HW, Albermann K, Heumann K, Liebl S, Pfeiffer F (1997) MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res 25:28–30
Moriyama S, Tanaka H, Uwataki M, Muguruma M, Ohta K (2003) Molecular cloning and characterization of an exoinulinase gene from Aspergillus niger strain 12 and its expression in Pichia pastoris. J Biosci Bioeng 96:324–331
Moriyama S, Muguruma M, Ohta K (2006) Quantitative expression analysis of inulinase gene cluster of Penicillium sp. strain TN-88. J Biosci Bioeng 101:277–279
Ohta K, Akimoto H, Matsuda S, Toshimitsu D, Nakamura T (1998) Molecular cloning and sequence analysis of two endoinulinase genes from Aspergillus niger. Biosci Biotechnol Biochem 62:1731–1738
Ni BF, Needleman RB (1990) Identification of the upstream activating sequence of MAL and the binding sites for the MAL63 activator of Saccharomyces cerevisiae. Mol Cell Biol 10:3797–3800
Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d’Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wösten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25:221–231
Petersen KL, Lehmbeck J, Christensen T (1999) A new transcriptional activator for amylase genes in Aspergillus. Mol Gen Genet 262:668–676
Pina C, Goncalves P, Prista C, Loureiro-Dias MC (2004) Ffz1, a new transporter specific for fructose from Zygosaccharomyces bailii. Microbiology 150:2429–2433
Punt PJ, van den Hondel CA (1992) Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol 216:447–457
Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA (1987) Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117–124
Punt PJ, Strauss J, Smit R, Kinghorn JR, van den Hondel CA, Scazzocchio C (1995) The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol Cell Biol 15:5688–5699
Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, Mewes HW (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32:5539–5545
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schjerling P, Holmberg S (1996) Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res 24:4599–4607
Tani S, Katsuyama Y, Hayashi T, Suzuki H, Kato M, Gomi K, Kobayashi T, Tsukagoshi N (2001a) Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr Genet 39:10–15
Tani S, Itoh T, Kato M, Kobayashi T, Tsukagoshi N (2001b) In vivo and in vitro analyses of the AmyR binding site of the Aspergillus nidulans agdA promoter; requirement of the CGG direct repeat for induction and high affinity binding of AmyR. Biosci Biotechnol Biochem 65:1568–1574
Todd RB, Andrianopoulos A (1997) Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol 21:388–405
Tsuji G, Kenmochi Y, Takano Y, Sweigard J, Farrall L, Furusawa I, Horino O, Kubo Y (2000) Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol Microbiol 38:940–954
van Hartingsveldt W, Mattern I E, van Zeijl CM, Pouwels PH, van den Hondel CA (1987) Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet 206:71–75
van Peij NN, Visser J, de Graaff LH (1998a) Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol 27:131–142
van Peij NN, Gielkens MM, de Vries RP, Visser J, de Graaff LH (1998b) The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol 64:3615–3619
Vankuyk PA, Diderich JA, MacCabe AP, Hererro O, Ruijter GJ, Visser J (2004) Aspergillus niger mstA encodes a high-affinity sugar/H+ symporter which is regulated in response to extracellular pH. Biochem J 379:375–383
Vienken K, Scherer M, Fischer R (2005) The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits sexual development under low-carbon conditions and in submersed culture. Genetics 169:619–630
Walton JD (2000) Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: a hypothesis. Fungal Genet Biol 30:167–171
Woloshuk CP, Foutz KR, Brewer JF, Bhatnagar D, Cleveland TE, Payne GA (1994) Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol 60:2408–2414
Yu J, Chang P, Bhatnagar D, Cleveland TE (2000) Cloning of a sugar utilization gene cluster in Aspergillus parasiticus. Biochim Biophys Acta 1493:211–214
Yuan XL, Goosen C, Kools H, van der Maarel MJ, van den Hondel CA, Dijkhuizen L, Ram AF (2006) Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology 152:3061–3073
Acknowledgments
We thank Dr. J. Visser and Dr. P. vanKuyk for helpful discussions and comments on the manuscript. This work was financially supported by SenterNovem in the framework of the Innovative Onderzoeks Programma (IOP) Genomics program (project IGE1021).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Perez-Martin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Yuan, XL., Roubos, J.A., van den Hondel, C.A.M.J.J. et al. Identification of InuR, a new Zn(II)2Cys6 transcriptional activator involved in the regulation of inulinolytic genes in Aspergillus niger . Mol Genet Genomics 279, 11–26 (2008). https://doi.org/10.1007/s00438-007-0290-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-007-0290-5