Abstract
Avian haemosporidians are vector-borne parasites, infecting a great variety of birds. The order Passeriformes has the highest average infection probability; nevertheless, some common species of Passeriformes have been rather poorly studied. We investigated haemosporidians in one such species, the Eurasian jay Garrulus glandarius (Corvidae), from a forest population in Hesse, Central Germany. All individuals were infected with at least one haemosporidian genus (overall prevalence: 100%). The most common infection pattern was a mixed Haemoproteus and Leucocytozoon infection, whereas no Plasmodium infection was detected. Results on lineage diversity indicate a rather pronounced host-specificity of Haemoproteus and Leucocytozoon lineages infecting birds of the family Corvidae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vector-borne haemosporidian blood parasites (phylum Apicomplexa, order Haemosporidia), including avian malaria (genus Plasmodium) and malaria-like pathogens (genera Leucocytozoon and Haemoproteus), are widespread and infect a great variety of avian host species (Valkiūnas 2005). The application of molecular techniques for detecting and characterising haemosporidians is providing an increasingly growing and accurate picture of their host range, (host-specific) prevalence, distribution, and diversity (Rivero and Gandon 2018). The intensified research has revealed an extensive genetic diversity of haemosporidian lineages (> 4800 lineages, MalAvi 2023), which seems to be matched by an equally rich phenotypic diversity, such as lineages differing in their host range or within-host effects or virulence (Rivero and Gandon 2018; Ágh et al. 2022). At the avian host-species level, differences in host life history and behavioural traits, e.g. preferred foraging habitat or nest type, can influence rates of dipteran vector exposure, which can lead to heterogeneous infection probabilities across hosts (Fecchio et al. 2021). In a global study, including 141 avian families, those with the highest average Leucocytozoon infection probabilities belonged to the order Passeriformes, e.g. Paridae, Corvidae (Fecchio et al. 2021). Also, the highest Plasmodium lineage diversity across all continents was found in Passeriformes (Rivero and Gandon 2018). Nevertheless, in species of the order Passeriformes, many lineages and lineage-host relations likely were not found yet, particularly in hosts that have not been intensively sampled. For instance, there are 23 entries, covering around 60 individuals sampled in Africa, Asia and Europe, for Eurasian jays Garrulus glandarius, (Linnaeus, 1758) in the MalAvi database (274 entries in total for Corvidae), comprising seven Haemoproteus, two Plasmodium and five Leucocytozoon lineages (MalAvi 2023). The Palearctic-oriental distributed Eurasian jay is in Central Europe a resident, mainly forest-dwelling, open-nesting species. In some years, the species, predominantly individuals from Scandinavian breeding grounds, shows eruptive movements in autumn, which seem to be related to population density and the variation in acorn availability (Selås 2017). In this study, we aimed to (a) assess the Haemosporidia infection status in Eurasian jays from one forest population in Central Germany, (b) investigate the lineage diversity, and (c) compare our results with known lineages in Eurasian jays from other breeding sites.
Material and methods
Blood sample collection
We sampled 16 Eurasian jays from May to June 2020 and from May to July 2021. By limiting the sampling time to the spring–summer period, it was ensured that Eurasian jays from the local population and no migratory individuals were sampled. We captured the birds using walk-in traps placed in the Marburg Open Forest, a 250-ha managed forest, consisting of mixed stands dominated by common beech Fagus sylvatica and common oak Quercus robur, in Hesse, Central Germany (50°50′ N, 8°39′ E). In and around the forest site are smaller streams and temporary standing water bodies, which, amongst others, represent an ecological determinant of haemosporidian infections, as water bodies are necessary for the development of dipteran vectors (Ferraguti et al. 2018). We obtained the blood samples by puncture of the brachial vein (Table 1) and stored them on Whatman FTA cards (Whatman®, UK). Additionally, we prepared two blood smears per individual, which we fixed with methanol (100%) for 30 s and stained with Giemsa in a work solution prepared with buffer pH 7.0 (ratio 1:5) for 30 min.
For DNA isolation, we cut a 3 × 3-mm piece of the samples in the FTA card to extract DNA (ammonium-acetate protocol; Martínez et al. 2009). We determined DNA concentration and quality using a NanoDrop2000c UV–Vis spectrophotometer (NanoDrop Technologies, USA).
Parasite detection
We determined the presence or absence of avian haemosporidians through a nested PCR protocol targeting a 479-base pair (bp) region of the cytochrome b gene (cyt b; for detailed PCR protocol see Hellgren et al. 2004). In each PCR run, we included DNA from birds with known haemosporidian infection and deionised water as positive and negative controls, respectively. We rerun each sample resulting in a negative PCR reaction to confirm the parasite absence. We visualised the PCR amplicons using QIAxcel Advanced (QIAGEN) high-resolution capillary gel electrophoresis. Subsequently, we sent all PCR products of samples rendering a clear band during gel electrophoresis for Sanger bidirectional sequencing at Microsynth-Seqlab (Sequence Laboratories Goettingen GmbH, Germany). Using CLC Main Workbench 7.6.4 (CLC Bio, Qiagen, Denmark), we assembled and trimmed forward and reverse sequences. To identify lineages, we aligned the sequences with sequences deposited in the MalAvi database using BLASTN 2.3.0 + (Bensch et al. 2009). If the sequencing quality did not allow a clear determination, we repeated the PCR and sequencing. However, regardless of this, we could not clearly determine the lineage of one Haemoproteus-positive sample (Table 1).
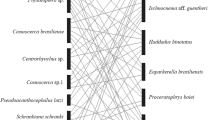
We constructed a haplotype network of haemosporidian lineages (Fig. 1), using the medium joining network method implemented in PopART 1.7 (Leigh and Bryant 2015). The network covers all sequences clearly assigned to a lineage from this study (n = 29) and sequences found in Eurasian jays deposited in MalAvi (n = 31). However, we had to exclude two sequences from MalAvi, corresponding to the lineages GAGLA01 and GAGLA04, due to being short (< 476 bp) and containing undetermined nucleotides.
Median-joining network of mitochondrial cytochrome b lineages (476 bp, n = 60 sequences) of haemosporidian parasites found in Eurasian jays Garrulus glandarius. Circle size is proportional to the lineage frequency. Lineage names are noted at the associated circles together with one exemplary GenBank association number in parentheses. One hatch mark represents one mutation. Sample origins are represented by different colours, ‘MalAvi’ referring to sequences from other studies deposited in the MalAvi database (MalAvi 2023). Morphospecies names for GAGLA07 (H. homopicae) and TURDUS2 (H. minutus) are not provided within the figure
We examined the stained blood smears at × 1000 magnification (light microscope PrimoStar Zeiss, Germany) for at least 10,000 monolayered erythrocytes to calculate the infection intensity based on the number of intraerythrocytic gametocytes of Haemoproteus and Leucocytozoon. No gametocytes of Plasmodium were detected during blood smear examination.
Results and discussion
The overall infection rate in the Eurasian jay population was 100%, i.e. each individual showed a positive PCR result for at least one haemosporidian genus. Interestingly, we could detect no Plasmodium infections, while Haemoproteus and Leucocytozoon infections reached a high prevalence (94% and 88%, respectively), and often occurred as mixed infections (Table 1). Mixed infections, of either different species, genetic lineages or both in the same host, of haemosporidian parasites are common (Bernotienė et al. 2016). A recent study on Eurasian sparrowhawks Accipiter nisus showed that the prevalence of Haemoproteus was higher for hosts infected with Leucocytozoon and vice versa, i.e. a positive Haemoproteus × Leucocytozoon association (Svobodová et al. 2023). However, further investigations would be needed to elucidate if this represents a general pattern and an explanation of the high prevalence of mixed Haemoproteus-Leucocytozoon infections in our study.
Plasmodium infections (lineages GRW11 and SGS1, both P. relictum) have been found in a few Eurasian jays from Asia and Europe (Beadell et al. 2006; Dimitrov et al. 2010; Drovetski et al. 2014; Fig. 1), however other studies, including this one, did not find Plasmodium infections in this host species (Stanković et al. 2019; Šujanová et al. 2021). These differences might partly be due to a seasonal variation in the Plasmodium prevalence in temperate regions. For instance, Šujanová et al. (2021) report a higher-than-expected prevalence of Plasmodium-positive samples in autumn and Cosgrove et al. (2008) could show a clear pattern of seasonal variation of P. circumflexum, including an autumn peak. However, P. relictum showed a relatively stable seasonal pattern of prevalence (Cosgrove et al. 2008) and Neto et al. (2020) demonstrated the existence of a different seasonality of the same Plasmodium lineage between European countries (Bulgaria, Poland, Spain and Sweden). Generally, the entries on Eurasian jays in the MalAvi database included less than 10 jay individuals, except from the studies of Šujanová et al. (2021, n = 10, Slovakia) and Valkiūnas et al. (2019, n = 21, Lithuania), thus a general estimation of the variation in prevalence in this host is difficult with the available data. Both studies compared to our results observed a lower overall prevalence (30% and 5%, respectively).
Matching our result, a community analysis, including 29 avian species, from the same study site in Hesse found low Plasmodium infection prevalence (8%), compared to a higher Haemoproteus (68%) and Leucocytozoon (60%) prevalence (Strehmann et al. 2023).
The intensity of infection (parasitaemia) varied between 0 and 6.8%, however, most individuals (81%) had an intensity < 1.0% (Table 1). Generally, low-intensity infections (connected with mainly chronic infections) can persist in the hosts without direct visible, overt symptoms (Schoenle et al. 2017; Krams et al. 2022). This fits the observation that none of the sampled Eurasian jays showed any clinical signs of disease. However, low-intensity infections may have long-term detrimental effects on hosts, such as accelerated senescence or reduced reproductive success (Asghar et al. 2015; Schoenle et al. 2017).
We were able to assign the detected Haemoproteus infections to two lineages: GAGLA02 (prevalence: 56%, GenBank accession number: OR069477) and CIRCUM05 (38%, OR069478), whereby one Eurasian jay individual was infected with both lineages (Table 1). Both lineages already have been proved in Eurasian jays from other sampling locations in Europe (Fig. 1). Considering the lineage network, the GAGLA-Haemoproteus lineages cluster together (Fig. 1). The clustering lineages GAGLA02, GAGLA03 and GAGLA07 (Haemoproteus (Parahaemoproteus) homopicae) have so far only been detected in Eurasian jays, the lineage GAGLA05 also in another member of the Corvidae family, the common raven Corvus corax (Linnaeus, 1758; MalAvi 2023). This suggests a certain host-specificity of the found Haemoproteus lineages, which is in line with other findings proposing Haemoproteus to be rather host-specific (e.g. Ellis et al. 2020; Strehmann et al. 2023). The Leucocytozoon lineages GAGLA06 (OR069481) and COCOR02 (OR069479) were so far found in Eurasian jays and common ravens only, whereas the Leucocytozoon lineage EUSE1 (OR069480) up to now was detected in common ravens only (MalAvi 2023). We found four Eurasian jay individuals to be infected with EUSE1 (Table 1), constituting a new host-lineage interaction. The results show that similar to the Haemoproteus lineages, some Leucocytozoon lineages may be quite host-specific to Corvidae species. Therefore, this avian family, which has been rather poorly studied with respect to haemosporidian infections, might be a good model group for further research on haemosporidian host specificity.
Data availability
Sequences are deposited in GenBank (accession numbers OR069477/81) and will be submitted to the MalAvi database. The (raw) data of this article is freely available for download from PANGAEA at https://doi.org/10.1594/PANGAEA.961154, https://doi.org/10.1594/PANGAEA.961255, and https://doi.org/10.1594/PANGAEA.961493. A PrePrint can be found at bioRxiv: https://doi.org/10.1101/2023.06.20.545710.
References
Ágh N, Csörgő T, Szöllősi E (2022) Delay in arrival: lineage-specific influence of haemosporidians on autumn migration of European robins. Parasitol Res 121:2831–2840. https://doi.org/10.1007/s00436-022-07621-5
Asghar M, Hasselquist D, Hansson B, Zehtindjiev P, Westerdahl H, Bensch S (2015) Hidden costs of infection: chronic malaria accelerates telomere degradation and senescence in wild birds. Science 347:436–438. https://doi.org/10.1126/science.126112
Beadell JS, Ishtiaq F, Covas R et al (2006) Global phylogeographic limits of Hawaii’s avian malaria. Proc R Soc B 273:2935–2944. https://doi.org/10.1098/rspb.2006.3671
Bensch S, Hellgren O, Peréz-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x
Bernotienė R, Palinauskas V, Iezhova T, Murauskaitė D, Valkiūnas G (2016) Avian haemosporidian parasites (Haemosporida): a comparative analysis of different polymerase chain reaction assays in detection of mixed infections. Exp Parasitol 163:31–37. https://doi.org/10.1016/j.exppara.2016.01.009
Cosgrove CL, Wood MJ, Day KP, Sheldon BC (2008) Seasonal variation in Plasmodium prevalence in a population of blue tits Cyanistes caeruleus. J Anim Ecol 77:540–548. https://doi.org/10.1111/j.1365-2656.2008.01370.x
Dimitrov D, Zehtindjiev P, Bensch S (2010) Genetic diversity of avian blood parasites in SE Europe: Cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Bulgaria. Acta Parasitol 55:201–209. https://doi.org/10.2478/s11686-010-0029-z
Drovetski SV, Aghayan SA, Mata VA et al (2014) Does the niche breadth or trade-off hypothesis explain the abundance–occupancy relationship in avian Haemosporidia? Mol Ecol 23:3322–3329. https://doi.org/10.1111/mec.12744
Ellis VA, Huang X, Westerdahl H et al (2020) Explaining prevalence, diversity and host specificity in a community of avian haemosporidian parasites. Oikos 129:1314–1329. https://doi.org/10.1111/oik.07280
Fecchio A, Clark NJ, Bell JA et al (2021) Global drivers of avian haemosporidian infections vary across zoogeographical regions. Glob Ecol Biogeogr 30:2393–2406. https://doi.org/10.1111/geb.13390
Ferraguti M, Martínez-de la Puente J, Bensch S et al (2018) Ecological determinants of avian malaria infections: An integrative analysis at landscape, mosquito and vertebrate community levels. J Anim Ecol 87:727–740. https://doi.org/10.1111/1365-2656.12805
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90:797–802. https://doi.org/10.1645/GE-184R1
Krams R, Krama T, Elferts D et al (2022) High blood parasite infection rate and low fitness suggest that forest water bodies comprise ecological traps for pied flycatchers. Birds 3(2):221–233. https://doi.org/10.3390/birds3020014
Leigh JW, Bryant D (2015) PopART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210X.12410
MalAvi (2023) Tables ‘Database Summary Report’ and ‘Hosts And Sites Table’ downloaded on 16.05.2023. https://130.235.244.92/Malavi/
Martínez J, Martínez-de La Puente J, Herrero J et al (2009) A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: on the inefficiency of general primers for detection of mixed infections. Parasitol 136:713722. https://doi.org/10.1017/S0031182009006118
Neto MJ, Mellinger S, Halupka L, Marzal A, Zehtindjiev P, Westerdahl H (2020) Seasonal dynamics of haemosporidian (Apicomplexa, Haemosporida) parasites in house sparrows Passer domesticus at four European sites: comparison between lineages and the importance of screening methods. Int J Parasitol 50:523–532. https://doi.org/10.1016/j.ijpara.2020.03.008
Rivero A, Gandon S (2018) Evolutionary ecology of avian malaria: past to present. Trends Parasitol 34:712–726. https://doi.org/10.1016/j.pt.2018.06.002
Schoenle LA, Kernbach M, Haussmann MF, Bonier F, Moore T (2017) An experimental test of the physiological consequences of avian malaria infection. J Anim Ecol 86:1483–1496. https://doi.org/10.1111/1365-2656.12753
Selås V (2017) Autumn irruptions of Eurasian Jay (Garrulus glandarius) in Norway in relation to acorn production and weather. Ornis Fenn 94:92–100
Stanković D, Jönsson J, Raković M (2019) Diversity of avian blood parasites in wild passerines in Serbia with special reference to two new lineages. J Ornithol 160:545–555. https://doi.org/10.1007/s10336-019-01628-z
Strehmann F, Becker M, Lindner K et al (2023) Half of a forest bird community infected with haemosporidian parasites. Front Ecol Evol 11:1107736. https://doi.org/10.3389/fevo.2023.1107736
Šujanová A, Špitalská E, Václav R (2021) Seasonal dynamics and diversity of Haemosporidians in a natural woodland bird community in Slovakia. Diversity 13:439. https://doi.org/10.3390/d13090439
Svobodová M, Čepička I, Zídková L et al (2023) Blood parasites (Trypanosoma, Leucocytozoon, Haemoproteus) in the Eurasian sparrowhawk (Accipiter nisus): diversity, incidence and persistence of infection at the individual level. Parasit Vectors 16:15. https://doi.org/10.1186/s13071-022-05623-x
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia, 2nd edn. CRC Press, Boca Raton
Valkiūnas G, Ilgūnas M, Bukauskaitė D et al (2019) Molecular characterization of six widespread avian haemoproteids, with description of three new Haemoproteus species. Acta Trop 197:105051. https://doi.org/10.1016/j.actatropica.2019.105051
Acknowledgements
We thank Sabine Wagner, Anna Wahle and Wiebke Schäfer for supporting the laboratory work, and Kim Lindner and Sascha Rösner for assistance with the fieldwork.
Funding
Open Access funding enabled and organized by Projekt DEAL. This project was part of the LOEWE priority project Nature 4.0—Sensing Biodiversity funded by the Hessen State Ministry for Higher Education, Research and the Arts.
Author information
Authors and Affiliations
Contributions
YRS, PQ and JFM conceptualized the study and carried out fieldwork. PQ acquired the funding. YRS and NLP performed laboratory work. YRS analysed the data and wrote the first draft of the manuscript. All authors contributed to manuscript revision.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Animal handling, including ethical approval according to the national animal protection laws, was carried out under the permit of the Regierungspräsidium Gießen, Hesse, Germany (permit number G10/2019).
Consent for publication
All the authors have read the manuscript and approved its publication.
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Leonhard Schnittger
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schumm, Y.R., Lederer-Ponzer, N., Masello, J.F. et al. High prevalence of haemosporidian parasites in Eurasian jays. Parasitol Res 123, 182 (2024). https://doi.org/10.1007/s00436-024-08170-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00436-024-08170-9