Abstract
In the family of fruit bats, Pteropodidae Gray, 1821, as in the third most diverse group of bats (Chiroptera), the bacterium of the genus Bartonella was detected in several species as well as in a few species of their insect ectoparasites in some tropical and sub-tropical regions of the Old World. The Egyptian fruit bat, Rousettus aegyptiacus (Geoffroy, 1810), is one of the most widespread fruit bats, occurring between South Africa, Senegal, and Pakistan. In this bat species, Candidatus Bartonella rousetti has been detected in three African populations in Nigeria, Kenya, and Zambia. This fruit bat, however, also occurs in the Palaearctic, an area isolating the species geographically and phylogenetically from the Afrotropical part of its distribution range. We screened the blood-sucking bat flies (family Nycteribiidae) from R. aegyptiacus for the presence of the Bartonella bacteria. A rich material of bat fly Eucampsipoda aegyptia (Macquart, 1850), a monoxenous ectoparasite of the Egyptian fruit bats, was collected at 26 localities in seven countries (Egypt, Iran, Jordan, Lebanon, Oman, United Arab Emirates, and Yemen) of the Middle East in 2007–2013. The DNA isolates from the bat flies were subjected to a three-marker (gltA, ssrA, and intergenic spacer region, ITS) multilocus sequence analysis. Based on the amplification of the fragment of ssrA gene by a real-time PCR, 65 E. aegyptia samples from 19 localities in all seven countries were positive for the bacteria. One to five Bartonella-positive individuals of E. aegyptia were collected per one individual of R. aegyptiacus. An analysis of the ITS and gltA genes indicated the presence of an uncultured Bartonella sp., belonging to the Cand. B. rousetti genogroup, identified from populations of the Egyptian fruit bat in Africa. These results support the hypothesis that Bartonella’s diversity corresponds to its host’s diversity (and phylogenetic structure). Specific lineages of pathogens are present in specific phylogenetic groups of bats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Bartonella Strong, Tyzzer, Brues et Sellards, 1915 (Hyphomicrobiales: Bartonellaceae) comprises phylogenetically diversified facultative intracellular Gram-negative α-proteobacteria that infect mainly the erythrocytes and endothelial cells of mammals (Eicher and Dehio 2012). These bacteria are distributed worldwide and transmitted predominantly by blood-feeding arthropods (Chomel et al. 2009).
McKee et al. (2021) suggested bats (Chiroptera) are a group of mammals that have a crucial role in the origin and spread of the Bartonella bacteria among geographical regions and other mammal groups. In bats, several taxa of blood-feeding arthropods can be found, which can help in the dispersal of this bacterium (Marshall 1982; Szubert-Kruszyńska and Postawa 2008). The available results demonstrated that the genus Bartonella used to be found most frequently in one group of these arthropods, in the family of bat flies, Nycteribiidae (Szentiványi et al. 2019). Identical genotypes of Bartonella are often reported from bats and their bat flies, which suggest that bat flies act as vectors for the spreading of this bacterium among bats and perhaps also other mammals (cf. Kamani et al. 2014; Brook et al. 2015; Moskaluk et al. 2018; Judson et al. 2015; Dietrich et al. 2016; Qiu et al. 2020).
In the last decade, several new strains/genotypes of Bartonella were detected in bats and their flies throughout the world (see Han et al. 2022: 2, Fig. 1). However, the taxonomic diversity of Bartonella is only poorly manifested among bat taxa and/or populations. Due to this, our understanding of whether and how particular species of Bartonella are shared among related bat species is somewhat limited.
A map of the bat fly Eucampsipoda aegyptia collection sites used in this study (circles); a circle with an asterisk marks the pathogen presence, and the numbers correspond with locality numbers in Material and methods. The dark grey area shows the distribution range of Rousettus aegyptiacus in the Middle East reconstructed after Benda et al. (2011, 2023)
The family of fruit bats, Pteropodidae Gray, 1821, is one of the richest bat groups concerning species diversity, broadly distributed in the tropics and subtropics of the Old World. Despite this, reports on the prevalence of Bartonella in these bats and their bat flies are available only from a few localities (Bai et al. 2018; Kamani et al. 2014; Dietrich et al. 2016; Brook et al. 2015; Wilkinson et al. 2016; Qiu et al. 2020; Fagre et al. 2023). The Egyptian fruit bat, Rousettus aegyptiacus (Geoffroy, 1810), ranks among fruit bats with extensive geographical distribution—it is the only fruit bat species occurring in two continents; it lives in most of Africa and southwestern Asia. It occupies areas around the Gulf of Guinea from Senegal to Angola, in southern and eastern Africa from the Cape to Eritrea, and the southwest part of the Palaearctic, from Egypt, Yemen, and Turkey to Pakistan (Kwiecinski and Griffiths 1999; Benda et al. 2011). The distribution range in the Middle East (comprising wholes or parts of Egypt, Sudan, Turkey, Cyprus, Levant, Arabia, Iran, and Pakistan, see Benda et al. 2011 for a review, with additions by Benda et al. 2012, 2023, Judas et al. 2018, Benda and Ševčík 2020) is an occurrence spot isolated both geographically and phylogenetically from the Afro-tropical populations (Střibná et al. 2019).
The search for Bartonella in the populations of Rousettus aegyptiacus brought detection of the genotype Candidatus Bartonella rousetti from the bat fly Eucampsipoda africana Theodor, 1955, a frequent ectoparasite of this fruit bat. This strain was found in this bat fly collected in Nigeria (Bai et al. 2018) and Zambia (Szentiványi et al. 2022) and was also confirmed in the blood of R. aegyptiacus from Kenya (Kosoy et al. 2010). The knowledge of Bartonella in the Egyptian fruit bat is thus limited to a few sites in tropical Africa. At the same time, this pathogen has not been screened in the Palaearctic populations.
The absence of any evidence of Bartonella in the Palaearctic populations of the Egyptian fruit bat and its monoxenous arthropod parasite was an impulse for a more detailed study of the bat fly Eucampsipoda aegyptia (Macquart, 1850), an obligatory ectoparasite of Rousettus aegyptiacus. The screening for the presence of Bartonella was done in almost the entire known distribution range of this bat fly in the Middle East, from Egypt to Iran. Our study finally brought new evidence of the Bartonella presence from a large part of the Palaerctic range of the Egyptian fruit bat and, thus, of its wide distribution and relatively common occurrence.
Material and methods
The material examined
Our study comprises the populations of the Egyptian fruit bat, Rousettus aegyptiacus, from the Middle East, including its NE African part. The bat flies Eucampsipoda aegyptia were collected from the fruit bats at the following 26 sites situated in seven countries (Fig. 1): Egypt: (1) Aswan, 24°07′N, 32°54′E, 92 m a. s. l. (24 January 2010, 10 January 2011); (2) Bahariya Oasis, Bawiti, 28°21′N, 28°52′E, 98 m a. s. l. (18 January 2010, 30 December 2010, 2 January 2011); (3) El A’aqab, 24°16′N, 32°54′E, 96 m a.s.l. (25 January 2010); (4) El Qahirah, Gezira Island, 30°03′N, 31°13′E, 20 m a. s. l. (29 January 2010); Iran: (5) Bishapur, Sasan Cave, 29°47′N, 51°35′E, 860 m a. s. l. (6 October 2011); (6) Jahrom, Sang Eshkan, 28°29′N, 53°35′E, 1102 m a. s. l. (8 October 2011); (7) Zangard, 27°13′N, 54°38′E, 493 m a. s. l. (9 October 2011); Jordan: (8) Iraq Al Amir, Wadi As Sir, 31°55′N, 35°45′E, 515 m a. s. l. (2 July 2010); (9) Jufat Al Qafrayn, 31°53′N, 35°37′E, 235 m a. s. l. (15 July 2010); (10) Nahla, 32°17′N, 35°51′E, 728 m a. s. l. (13 July 2010); (11) An Nuzha, Wadi Al Wala, 31°33′N, 35°44′E, 335 m a. s. l. (11 July 2010); Lebanon: (12) Dahr El Mghara, Aaonamie Cave, 33°40′N, 35°27′E, 255 m a. s. l. (19 January 2008); (13) Trablous, Matal El Azraq Cave, 34°25′N, 35°50′E, 15 m a. s. l. (21 January 2007); Oman: (14) Ain Sahalnawt, 17°09′N, 54°11′E, 123 m a. s. l. (27 March 2012); (15) Al War, Wadi Khabbah, 22°56′N, 58°51′E, 406 m a. s. l. (5 April 2011); (16) Bidbid, Wadi Dabaum, 23°25′N, 58°08′E, 205 m a. s. l. (26 March 2011); (17) Misfah, 23°14′N, 57°08′E, 1196 m a. s. l. (9 April 2011); (18) Mithqub, Wadi Harabein, 23°04′N, 58°59′E, 52 m a. s. l. (2 November 2009); (19) Mudhai, 17°29′N, 53°21′E, 542 m a. s. l. (25 March 2012); (20) Shihayt, Wadi Darbat, 17°09′N, 54°28′E, 326 m a. s. l. (28 March 2012); (21) Tayma, 22°31′N, 59°20′E, 196 m a. s. l. (3 April 2011); (22) Wadi Hannah, 17°03′N, 54°37′E, 310 m a. s. l. (30 March 2012); United Arab Emirates: (23) Al Khari, Shawka Dam, 25°06′N, 56°03′E, 295 m a. s. l. (29 October 2013); (24) Shis, 25°17′N, 56°15′E, 363 m a. s. l. (30 October 2013); Yemen: (25) Mashgab, Ash Shamshara, 13°21′N, 43°57′E, 1170 m a. s. l. (26 October 2007); (26) Wadi Zabid, Al Mawkir, 14°10′N, 43°30′E, 270 m a. s. l. (30 October 2007).
The Egyptian fruit bats were caught using standard methods like mist or hand nets. The bats’ whole bodies were checked for the presence of ectoparasites. The bat flies’ maximum individuals were removed, collected using tweezers, and preserved in 96% ethanol.
The fixed bat fly specimens were studied with a microscope without additional interventions. The species and sex determinations of the Eucampsipoda aegyptia specimens were carried out using the key by Theodor (1967: 413–416). The taxonomy and nomenclature follow Maa (1965).
Only a part (176/196) of the collected bat fly material was used to detect pathogens. The examined samples of extracted DNA are stored at the Institute of Virology, Biomedical Research Center, Slovakian Academy of Sciences, Bratislava, Slovakia. The other bat fly E. aegyptia material not screened for a pathogen presence remains in a private collection of Martin Ševčík, Nitra, Slovakia.
DNA extraction, molecular genetic, and statistical analyses
The ethanol-fixed bat fly specimens were washed with sterile water, dried, and crushed with a sterile scalpel. Following the manufacturer’s protocol, their DNA was extracted using the QIAamp DNA Mini kit (Qiagen). Thirty additional samples from Egypt were stored as dry specimens, and their DNA was extracted by chelex (Walsh et al. 1991). The quantity and quality of the DNA were assessed by Nano Photometer Pearl (Implen, Germany), and the extracted DNA was used as a template for the PCR amplification to determine the presence of Bartonella, with the following species identification (Table 1). The Bartonella positive amplicons were purified and then analyzed by sequencing in both directions with the same primers as for the PCR amplification by Macrogen Inc. (Amsterdam, The Netherlands). The obtained partial sequences of ITS and gltA genes were compared with those available in the GenBank using the Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov).
Phylogenetic analyses were conducted in MEGA11 (Tamura et al. 2021) and the phylogenetic tree based on ITS region was constructed using the neighbor-joining method (Saitou and Nei 1987) with the Kimura 2-parameter method (Kimura 1980). Partial gltA genes and ITS region sequences for representative samples were submitted to the GenBank under the accession numbers OR553951–OR553952 for the gltA gene, and OQ058984–OQ058989 and OR523867–OR523871 for the ITS region, respectively.
Statistical analyses testing the geographical and sexual differences in the presence of Bartonella species in Eucampsipoda aegyptia specimens have been done using Fisher’s exact test with an online calculator (http://www.socscistatistics.com). The p value < 0.05 was considered as proof of significant difference, and 95% confidence intervals (CI) were calculated using an online calculator (http://epitools.ausvet.com.au).
Results
Bat flies, localities, and Bartonella presence
Altogether, 176 individual bat flies Eucampsipoda aegyptia were analyzed for the presence of Bartonella; these flies were collected from 68 individuals of Rousettus aegyptiacus (Egypt: 4 sites, 44 flies; Iran: 3 sites, 37 flies; Jordan: 4 localities, 13 flies; Lebanon: 2 sites, 5 flies; Oman: 9 sites, 68 flies; UAE: 2 sites, 5 flies; Yemen: 2 sites, 4 flies). Based on the real-time PCR analysis, a total of 65 of the bat fly DNA samples (36.9% of the 176 samples analyzed; 95% confidence interval (next CI) 29.80–44.06) were found positive for Bartonella (Table 2), and further characterized. The Bartonella DNA was found both in females (40.91%; 95% CI 29.05–52.77) and males (34.55%; 95% CI 25.66–43.43). The difference between the presence depending on sex was not significant (p = 0.423). The Bartonella-positive bat flies were collected from 37 individuals of R. aegyptiacus, with a frequency between one and five positive bat flies per fruit bat individual. The DNA of Bartonella spp. was detected in the samples in all countries from where the samples were examined (Table 2).
Genetic diversity
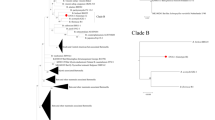
Fifty-four good-quality sequences of fragments of the ITS region (n = 39) and gltA (n = 15) gene were obtained and analyzed. The sequence analysis of the partial ITS region of all 39 samples revealed the presence of Bartonella strains of the Ca. B. rousetti genogroup (Fig. 2). Twenty samples (4 females from 4 bats from Lebanon; 1 female and 2 males from 1 bat from Yemen; 2 males from 2 bats from Jordan; 3 males from 3 bats from Egypt; 4 females and 4 males from 5 bats from Iran) were identical with the uncultured Bartonella clone 84 deposited in the GenBank (OR523867)— > 99% sequence identity to Bartonella sp. R-191 (KM382255) was detected in the blood of Rousettus aegyptiacus from Kenya (Kosoy et al. 2010). Bartonella rousetii is a name for Bartonella sp. R-191 proposed by Bai et al. (2018). Another four male samples of bat flies collected from R. aegyptiacus in Jordan, UAE, Egypt, and Iran, were identical with the uncultured Bartonella clone 136 deposited in the GenBank (OR523868)— > 96% sequence identity to Ca. B. rousetti (KM382255). Other five samples of Eucampsipoda aegyptia, 1 male and 2 females from 1 bat from Egypt, and 1 male and 1 female from 1 bat from Iran were identical to the uncultured Bartonella clone 175 deposited in the GenBank (OR523869)— > 97% sequence identity to Ca. B. rousetti (KM382255). Sequences from 1 male and 1 female from 2 bats from Iran were identical to the uncultured Bartonella clone 91 deposited in the GenBank (OR523870)— > 98% sequence identity to Ca. B. rousetti (KM382255). The Bartonella ITS region sequences derived from 7 individuals of E. aegyptia, 3 males (OQ058984–OQ058986) and 3 females (OQ058987–OQ058989) from Jordan, and 1 female from Iran (OR523871) shared > 94% similarity with Ca. B. rousetti (KM382255).
Phylogenetic relationship of Bartonella strains based on the internal transcribed spacer sequences (ITS). The neighbour-joining method by the Kimura 2-parameter distance and bootstrap calculation was conducted with 500 replicates for phylogenetic analysis. GenBank Accession Numbers are provided for all sequences
In total, 15 bat flies positive for the gltA gene were further analyzed, and the analysis revealed two types of the gltA sequences; sequence type 1 (OR553951) from a male of Eucampsipoda aegyptia collected from the fruit bat from Egypt showed 99% identity to two clones, the uncultured Bartonella clone YNBS/BF03 (OP433671) and clone YNBS/BF06 (OP433673) identified previously from E. africana from China (Kuang et al. 2022), and 85% identity to Ca. B. rousetti (HM363764). The maximum likelihood tree of the genus Bartonella based on the gltA, ftsZ, and rpoB genes using the MLSA approach showed Bartonella strain R-191 closely clustered with the fly-associated strain YNBS/BF03 (Kuang et al. 2022).
In total, 14 DNA samples of Bartonella of the gltA genotype 1 were identical, and their ITS regions represented three sequences (OQ058984, OR523870, OR523871). They originated from 3 females of E. aegyptia collected from 2 fruit bats from Oman, four samples of E. aegyptia from Iran, three from Lebanon, and one from Yemen. The nucleotide sequences of the gltA genotype 2 originated from a female of E. aegyptia collected from a fruit bat from Jordan (OR553952) showed 96% identity to Bartonella clone Batfly-3 (LC461051) previously found in E. africana from Zambia (Qiu et al. 2020). The sequence of the ITS region from this bat fly was unusable. For the comparison of Bartonella sequences amplified from E. aegyptia with selected sequences obtained from the GenBank via the BLAST query, see Table S1.
Discussion
Our study presents screening results for Bartonella presence in one of the obligatory parasitic species of Egyptian fruit bat colonies in the Middle East. We detected the presence of Bartonella sp. in the bat fly Eucampsipoda aegyptia collected from Rousettus aegyptiacus in this region for the first time. Moreover, we found this bacterium to be distributed over almost the whole Palaearctic range of the Egyptian fruit bat, except for the northernmost countries of the range, like Turkey, Cyprus, and Syria.
The analysis of the ITS region revealed the presence of eleven strains of Bartonela belonging to the genogroup Ca. B. rousetti. Twenty positive samples coming from a large area comprising Egypt, Lebanon, Jordan, Yemen, and Iran showed almost hundred percent identity to Bartonella R-191 that was identified in the Egyptian fruit bat population in Africa, i.e., in the blood samples of R. aegyptiacus from Kenya and the bat fly Eucampsipoda africana collected from the same species in Nigeria (Kosoy et al. 2010; Bai et al. 2018). The remaining detected sequences of the ITS region were also similar to this region of Ca. B. rousetti, and our strains of uncultured Bartonella were similar to the uncultured Bartonella clones also identified in the African populations of the Egyptian fruit bat, from the samples of Eucampsipoda africana collected from these bats in Nigeria (Bai et al. 2018). On the other hand, the analysis of the partial gltA gene sequences of uncultured Bartonella sp. from our study showed that they are identical to the uncultured Bartonella clones identified in Eucampsipoda africana from Yunnan, China (Kuang et al. 2022).
These results correspond with the results of previous studies, which demonstrated that monoxenous parasites, like Eucampsipoda aegyptia, express a lower diversity of infection by Bartonella over polyxenous species but a higher prevalence of these bacteria (Sándor et al. 2018). Simultaneously, they support the hypothesis that several very similar lineages of Bartonella occur in different geographical regions. Thus, the range size of the host distribution cannot be the main drive of Bartonella diversification. Hence, Bartonella’s diversity corresponds to its host’s diversity (and phylogenetic structure). Specific lineages of pathogens are present in specific phylogenetic groups of bats (species groups, families, etc.; cf. McKee et al. 2016). Besides this, the increasing phylogenetic distances among hosts decrease the probability of the pathogen transfers between them (McKee et al. 2016). This suggests that the determinant of the Bartonella distribution is its hosts’ diversity rather than its geographical distribution. On the other hand, evidence of Bartonella transfer between phylogenetically distant species increases (including that between wild and domestic animals; Frank et al. 2018).
The presence of Bartonella was surveyed in the bat flies Eucampsipoda aegyptia, whose distribution range corresponds closely with that of its primary host, Rousettus aegyptiacus. This bat fly is a monoxenous parasite (see above) whose life cycle occurs on the host’s body or in its proximity. Therefore, the parasitation of any other host species is exceptional or rather excluded (see Kock and Nader 1979). This close relation explains the presence of Bartonella in the colonies of R. aegyptiacus, but the role of the insect parasite in the spread of Bartonella remains to be elucidated. Numerous authors (Morse et al. 2012a, b; Dick and Dittmar 2014; Olival et al. 2015; Wilkinson et al. 2016; Han et al. 2017) suggested that bat flies of the family Nycteribiidae act as vectors of the Bartonella bacteria. Some of them (Olival et al. 2015; Wilkinson et al. 2016) suggested that bat flies act as reservoirs of these bacteria. McKee et al. (2021) hypothesized that Bartonella species evolved from symbionts found in blood-feeding ectoparasites because these arthropods depended on symbionts for additional nutrients (Husnik 2018). Bat flies are obligate ectoparasites of bats and contain endosymbiotic prokaryotes whose role is poorly understood. However, they are assumed to establish a symbiotic relationship with mutualistic bacteria (Morse et al. 2013). The blood-sucking dipterans of the superfamily Hippoboscoidea require milk secretion for larval development, and certain bacteria, such as Bartonella and Wolbachia, can be vertically transmitted during this process. These bacteria can also be transmitted horizontally through parasitoids or contact with contaminated saliva (de Bruin et al. 2015; Heath et al. 1999). However, horizontal transmission has not been recorded in the nycteribiid bat flies or other hippoboscoids.
Our study unveils Bartonella’s geographical distribution and genetic diversity within the Palaearctic population of Rousettus aegyptiacus in its almost complete range. However, the calculation of prevalency that could indicate some aspects of Bartonella biology remains omitted. The reason is that it requires a different way of data and material collection than was used in our study, i.e., to be taken from several colonies during and throughout the year. Although the biology of Eucampsipoda aegyptia is partly known from Egypt (Hafez et al. 1978), these data are insufficient regarding the range size and habitat diversity of the Middle East. Besides the detailed description of the Eucampsipoda life cycle and biology in various segments of its distribution range, for the complex picture of Bartonella biology, it is also necessary to monitor the bat host, consider the pathogen outbreak and other details like the changes in colony behavior or roost switches. So, extensive additional research is still necessary to describe the complete role of bat flies in the Bartonella transfer and its biology in bats.
In a recent study, antibodies against bat-associated Ca. B. rousetti were detected in humans (Bai et al. 2018). It indicates that bat-associated bacteria can potentially infect humans. However, antibodies against Bartonella tend to be highly cross-reactive within the genus and with other non-Bartonella agents. The DNA of Bartonella sp. was detected in the bat saliva, urine, and guano (Dietrich et al. 2017; Veikkolainen et al. 2014). Thus, the possibility of transmission of Bartonella to humans does not represent a direct and natural way.
On the other hand, even an accidental visit to a fruit bat roost could be potentially dangerous. Since the Egyptian fruit bat frequently uses anthropogenous roosts, it thus remains in close contact with humans. Eventually, humans could represent one of the connecting links of the bacterium transmission and spread.
Data availability
No datasets were generated or analysed during the current study.
References
Bai Y, Osinubi MOV, Osikowicz L, McKee C, Vora NM, Rizzo MR, Recuenco S, Davis L, Niezgoda M, Ehimiyein AM, Kia GSN, Oyemakinde A, Adeniyi OS, Gbadegesin YH, Saliman OA, Ogunniyi A, Ogunkoya AB, Kosoy MY, IBFIT (2018) Human exposure to novel Bartonella species from contact with fruit bats, Nigeria. Emerg Infect Dis 24:2317–2323. https://doi.org/10.3201/eid2412.181204
Benda P, Ševčík M (2020) Bats (Mammalia: Chiroptera) of the Eastern Mediterranean and Middle East. Part 16. Review of the distribution and taxonomy of bats in Egypt. Acta Soc Zool Bohem 84:115–279
Benda P, Abi-Said M, Bartonička T, Bilgin R, Faizolahi K, Lučan RK, Nicolaou H, Reiter A, Shohdi W, Uhrin M, Horáček I (2011) Rousettus aegyptiacus (Pteropodidae) in the Palaearctic: list of records and revision of the distribution range. Vespertilio 15:3–36
Benda P, Faizolâhi K, Andreas M, Obuch J, Reiter A, Ševčík M, Uhrin M, Vallo P, Ashrafi S (2012) Bats (Mammalia: Chiroptera) of the Eastern Mediterranean and Middle East. Part 10. Bat fauna of Iran. Acta Soc Zool Bohem 76:163–582
Benda P, Reiter A, Uhrin M (2023) On the distribution of the Egyptian Fruit Bat Rousettus aegyptiacus in Saudi Arabia (Mammalia: Chiroptera). Zool Middle East 63:299–308. https://doi.org/10.1080/09397140.2023.2266911
Brook CE, Bai Y, Dobson AP, Osikowicz LM, Ranaivoson HC, Zhu Q, Kosoy YM, Dittmar K (2015) Bartonella spp. in fruit bats and blood-feeding ectoparasites in Madagascar. PLoS Negl Trop Dis 9(2):e0003532. https://doi.org/10.1371/journal.pntd.0003532
Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, Koehler JE, Dehio C (2009) Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res 40(2):29. https://doi.org/10.1051/vetres/2009011
de Bruin A, van Leeuwen AD, Jahfari S, Takken W, Foldvari M, Dremmel L, Sprong H, Földvári G (2015) Vertical transmission of Bartonella schoenbuchensis in Lipoptena cervi. Parasit Vectors 8:176. https://doi.org/10.1186/s13071-015-0764-y
Dick CW, Dittmar K (2014) Parasitic bat flies (Diptera: Streblidae and Nycteribiidae): host specificity and potential as vectors. In: Klimpel S, Mehlhorn H (eds) Bats (Chiroptera) as vectors of diseases and parasites. Parasitology Research Monographs, vol 5. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-39333-4_6
Dietrich M, Tjale MA, Weyer J, Kearney T, Seamark ECJ, Nel LH, Monadjem A, Markotter W (2016) Diversity of Bartonella and Rickettsia spp. in bats and their blood-feeding ectoparasites from South Africa and Swaziland. PLoS ONE 11:e0152077. https://doi.org/10.1371/journal.pone.0152077
Dietrich M, Kearney T, Seamark ECJ, Markotter W (2017) The excreted microbiota of bats: evidence of niche specialisation based on multiple body habitats. FEMS Micr Lett 364:1–7. https://doi.org/10.1093/femsle/fnw284
Eicher SC, Dehio C (2012) Bartonella entry mechanisms into mammalian host cells. Cell Microb 14(8):1166–1173. https://doi.org/10.1111/j.1462-5822.2012.01806.x
Fagre AC, Islam A, Reeves WK, Kading RC, Plowright RK, Gurley ES, McKee C (2023) Bartonella infection in fruit bats and bat flies, Bangladesh. Microb Ecol 86:2910–2922. https://doi.org/10.1007/s00248-023-02293-9
Frank HK, Boyd SD, Hadly EA (2018) Global fingerprint of humans on the distribution of Bartonella bacteria in mammals. PLoS Negl Trop Dis 12(11):e0006865. https://doi.org/10.1371/journal.pntd.0006865
Hafez M, Hilali M, Faouad M (1978) Ecological studies on Eucampsipoda hyrtlii (Kolenati, 1856) (Diptera: Nycteribiidae) infesting fruit-eating bats in Egypt. J Egypt Soc Parasitol 7:205–218
Han HJ, Wen HL, Zhao L, Liu JW, Luo LM, Zhou CM, Qin XR, Zhu YL, Zheng XX, Yu XJ (2017) Novel Bartonella species in insectivorous bats, Northern China. Plos One 12:e0167915. https://doi.org/10.1371/journal.pone.0167915
Han H-J, Li Z-M, Li X, Liu J-X, Peng Q-M, Wang R, Gu X-L, Jiang Y, Zhou C-M, Li D, Xiao X, Yu X-J (2022) Bats and their ectoparasites (Nycteribiidae and Spinturnicidae) carry diverse novel Bartonella genotypes, China. Trans Emerg Dis 69:e845–e858. https://doi.org/10.1111/tbed.14357
Heath BD, Butcher RDJ, Whitfield WGF, Hubbard SF (1999) Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol 9:313–316. https://doi.org/10.1016/S0960-9822(99)80139-0
Husnik F (2018) Host–symbiont–pathogen interactions in blood-feeding parasites: nutrition, immune cross-talk and gene exchange. Parasitology 145:1294–1303. https://doi.org/10.1017/S0031182018000574
Judas J, Csorba G, Benda P (2018) The bat fauna (Mammalia: Chiroptera) of the United Arab Emirates: a review of published records and museum specimens with conservation notes. J Threat Taxa 10:11379–11390. https://doi.org/10.11609/jott.3096.10.3.11379-11390
Judson SD, Frank HK, Hadly EA (2015) Bartonellae are prevalent and diverse in Costa Rican bats and bat flies. Zoonoses Public Health 62:609–617. https://doi.org/10.1111/zph.12188
Kamani J, Baneth G, Mitchell M, Mumcuoglu KY, Gutiérrez R, Harrus S (2014) Bartonella species in bats (Chiroptera) and bat flies (Nycteribiidae) from Nigeria, West Africa. Vector Borne Zoonotic Dis 14:625–632. https://doi.org/10.1089/vbz.2013.1541
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kock D, Nader IA (1979) Two bat flies from the Kingdom of Saudi Arabia, their nomenclature, host specifity and zoogeography (Insecta: Diptera: Nycteribiidae). Senck Zool 60:65–73
Kosoy M, Bai Y, Lynch T, Kuzmin IV, Niezgoda M, Franka R, Agwanda B, Breiman RF, Rupprecht CE (2010) Bartonella spp. in bats. Kenya Emerg Infect Dis 16:1875–1881. https://doi.org/10.3201/eid1612.100601
Kuang G, Zhang J, Yang W, Pan H, Han X, Yang L, Wang J, Yang T, Song Z, Feng Y, Liang G (2022) Molecular detection and phylogenetic analyses of diverse Bartonella species in bat ectoparasites collected from Yunnan Province China. Pathogens 11:1283. https://doi.org/10.3390/pathogens11111283
Kwiecinski GG, Griffiths TA (1999) Rousettus Egyptiacus. Mammalian Species 611:1–9
Maa TC (1965) An interim world list of batflies: (Diptera: Nycteribiidae and Streblidae). J Med Ent 1(4):377–386. https://doi.org/10.1093/jmedent/1.4.377
Maggi RG, Kosoy M, Mintzer M, Breitschwerdt EB (2009) Isolation of Candidatus Bartonella melophagi from human blood. Emerg Infect Dis 15:66–68. https://doi.org/10.3201/eid1501.081080
Mardosaitė-Busaitienė D, Radzijevskaja J, Balčiauskas L, Bratchikov M, Jurgelevičius V, Paulauskas A (2019) Prevalence and diversity of Bartonella species in small rodents from coastal and continental areas. Sci Rep 9:12349. https://doi.org/10.1038/s41598-019-48715-y
Marshall AG (1982) Ecology of insects ectoparasitic on bats. In: Kunz, T.H. (eds) Ecology of Bats. Springer, Boston, MA. https://doi.org/10.1007/978-1-4613-3421-7_10
McKee CD, Hayman DTS, Kosoy MY, Webb CT (2016) Phylogenetic and geographic patterns of bartonella host shifts among bat species. Inf Gen Evol 44:382–394. https://doi.org/10.1016/j.meegid.2016.07.033
McKee CD, Bai Y, Webb CT, Kosoy MY (2021) Bats are key hosts in the radiation of mammal-associated Bartonella bacteria. Inf Gen Evol 89:04719. https://doi.org/10.1016/j.meegid.2021.104719
Morse SF, Dick CW, Patterson BD, Dittmar K (2012a) Some like it hot: evolution and ecology of novel endosymbionts in bat flies of cave-roosting bats (Hippoboscoidea, Nycterophiliinae). Appl Environ Microbiol 78:8639–8649. https://doi.org/10.1128/AEM.02455-12
Morse SF, Olival KJ, Kosoy M, Billeter S, Patterson BD, Dick CW, Dittmar K (2012b) Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae). Infect Genet Evol 12:1717–1723. https://doi.org/10.1016/j.meegid.2012.06.009
Morse SF, Bush SE, Patterson BD, Dick CW, Gruwell ME, Dittmar K (2013) Evolution, multiple acquisition, and localization of endosymbionts in bat flies (Hippoboscoidea, Streblidae and Nycteribiidae). App Envir Micr 79:2952–2961. https://doi.org/10.1128/AEM.03814-12
Moskaluk AE, Stuckey MJ, Jaffe DA, Kasten RW, Aguilar-Setién A, Olave-Leyva JI, Galvez-Romero G, Obregón-Morales C, Salas-Rojas M, García-Flores MM, Aréchiga-Ceballos N, García-Baltazar A, Chomel BB (2018) Molecular detection of Bartonella species in blood-feeding bat flies from Mexico. Vector Borne Zoonotic Dis 18:258–265. https://doi.org/10.1089/vbz.2017.2213
Norman AF, Regnery R, Jameson P, Greene C, Krause DC (1995) Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol 33(7):1797–1803. https://doi.org/10.1128/jcm.33.7.1797-1803.1995
Olival KJ, Dittmar K, Bai Y, Rostal MK, Lei BR, Daszak P, Kosoy M (2015) Bartonella spp. in a Puerto Rican bat community. J Wildl Dis 51:274–278. https://doi.org/10.7589/2014-04-113
Qiu Y, Kajihara M, Nakao R, Mulenga E, Harima H, Hangombe BM, Eto Y, Changula K, Mwizabi D, Sawa H, Higashi H, Mweene A, Takada A, Simuunza M, Sugimoto C (2020) Isolation of Candidatus Bartonella rousetti and other bat-associated Bartonellae from bats and their flies in Zambia. Pathogens 9(6):E469. https://doi.org/10.3390/pathogens9060469
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sándor AD, Földvári M, Krawczyk AI, Sprong H, Corduneanu A, Barti L, Görföl T, Estók P, Kováts D, Szekeres S, Laszló Z, Hornok S, Földvári G (2018) Eco-epidemiology of novel Bartonella genotypes from parasitic flies of insectivorous bats. Micr Ecol 76:1076–1088. https://doi.org/10.1007/s00248-018-1195-z
Střibná T, Romportl D, Demjanovič J, Vogeler A, Tschapka M, Benda P, Horáček I, Juste J, Goodman SM, Hulva P (2019) Pan African phylogeography and palaeodistribution of rousettine fruit bats: ecogeographic correlation with Pleistocene climate vegetation cycles. J Biogeogr 46(10):2336–2349. https://doi.org/10.1111/jbi.13651
Szentiványi T, Christe P, Glaizot O (2019) Bat flies and their microparasites: current knowledge and distribution. Front Vet Sci 6:115. https://doi.org/10.3389/fvets.2019.00115
Szentiványi T, Heintz A-C, Markotter W, Wassef J, Christe P, Glaizot O (2022) Vector-borne protozoan and bacterial pathogen occurrence and diversity in ectoparasites of the Egyptian Rousette bat. Med Vet Ent 37:189–194. https://doi.org/10.1111/mve.12639
Szubert-Kruszyńska A, Postawa T (2008) Parasitic mites (Acari) of bats (Chiroptera) – a review of trophical and topical relations to the host. In: Hutson AM, Lina PHC (eds) XIth European bat research symposium, Babeş-Bolyai University, Romanian Bat Protection Association & Emil Racoviţă Speleological Institute, Cluj-Napoca, pp 151
Tamura K, Stecher G, Kumar S (2021) MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Theodor O (1967) An illustrated catalogue of the Rothschild collection of Nycteribiidae (Diptera) in the British Museum (Natural History). Publication No. 655. British Museum (Natural History), London
Veikkolainen V, Vesterinen EJ, Lilley TM, Pulliainen AT (2014) Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emer Inf Dis 20:960–967. https://doi.org/10.3201/eid2006.130956
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Wilkinson DA, Duron O, Cordonin C, Gomard Y, Ramasindrazana B, Mavingui P, Goodman SM, Tortosa P (2016) The bacteriome of bat flies (Nycteribiidae) from the Malagasy region: a community shaped by host ecology, bacterial transmission mode, and hostvector specificity. Appl Environ Microbiol 82:1778–1788. https://doi.org/10.1128/AEM.03505-15
Acknowledgements
We thank Michal Andreas, Robert Černý, Ivan Horáček, Radek Lučan, Antonín Reiter (Czech Republic), Ján Obuch, and Marcel Uhrin (Slovakia) for their help in the fieldwork, and Peter Kabát for help with extraction by chelex.
Funding
Open access funding provided by The Ministry of Education, Science, Research and Sport of the Slovak Republic in cooperation with Centre for Scientific and Technical Information of the Slovak Republic This work was financially supported by the Scientific Grant Agency of the Ministry of Education and Slovakian Academy of Sciences (# VEGA1/0298/19, 2/0021/21), the Slovakian Research and Development Agency (# APVV-19–0066), and the Ministry of Culture of the Czech Republic (# DKRVO 2024–2028/6.I.a,00023252).
Author information
Authors and Affiliations
Contributions
EŠ, methodology, PCR test and sequencing, phylogenetic analyses, Genbank processing, supervision and contribution to molecular analyses, manuscript writing, funding acquisition. MŠ, conceptualization, study design, resources, determination of samples, formal analysis, (draft) manuscript writing. YYP, PCR test and sequencing. PB, sample collection, study design, review and editing manuscript, funding acquisition.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Boris Krasnov
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Eva Špitalská and Martin Ševčík contributed equally as first authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Špitalská, E., Ševčík, M., Peresh, YY. et al. Bartonella in bat flies from the Egyptian fruit bat in the Middle East. Parasitol Res 123, 144 (2024). https://doi.org/10.1007/s00436-024-08165-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00436-024-08165-6