Abstract
Trematodes of the genus Metagonimus Katsurada, 1912 (Digenea: Heterophyidae) are zoonotic parasites that cause infections in humans, with most cases reported in Southeast Asia. Larvae from the second intermediate host, called metacercariae, of one of human-infecting species, M. yokogawai (Katsurada, 1912), have been reported from cyprinoid fish in Europe. In the present study, we provided DNA-based evidence that metacercariae of Metagonimus, which are commonly found in the scales of various cyprinoids in Central Europe (Danube River in Hungary) do not belong to M. yokogawai. Sequence analysis of the ITS region, 28S rDNA, and cox1 genes showed that this species is clearly distinct from all Asian species, including M. yokogawai, which probably does not occur in Europe. Metacercariae from cyprinoids might belong to Metagonimus romanicus (Ciurea, 1915), an insufficiently known species described from Romania.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trematodes of the family Heterophyidae are common parasites of birds and mammals (Pearson, 2008). Some species of the so-called small intestinal flukes are causative agents of fish-borne diseases in humans, with human cases reported from Southeast Asia (Chai et al. 2005; Chai and Jung 2017). Five species of the heterophyid genus Metagonimus Katsurada, 1912 have also been detected in humans (Chai 2015), with most cases caused by M. yokogawai (Katsurada, 1912), the type species of the genus. This species was described by Katsurada (1912) in Japan as Heterophyes yokogawai and then reported from other Asian countries (South Korea, Taiwan, and India), as well as Russia, Israel and several European countries (Bulgaria, Czech Republic, Hungary, Serbia, and Spain) (Yu and Mott 1994; Rácz and Zemankovics 2002; Chai et al. 2009; Pornruseetairatn et al. 2016). However, the global distribution of M. yokogawai has yet to be confirmed, as other species of Metagonimus have a limited geographical distribution, with most species described from East Asia (Shimazu 1999, 2002; Chai and Lee 2002; Shimazu and Urabe 2002; Kino et al. 2006; Shumenko et al. 2017; Tatonova et al. 2018; Nakao et al. 2022).
Metagonimus metacercariae have been reported from freshwater fish in eastern and central Europe, especially from the Danube River basin (Vojtek 1959; Žitňan 1969; Kulišić and Lepojev 1994). In Hungary, Prettenhoffer (1930) reported metacercariae identified as M. romanicus (Ciurea 1915), a species described insufficiently by Ciurea (1915) from Romania. In contrast, Molnár (1969), Rácz and Zemankovics (2002), and Molnár and Baska (2017) identified metacercariae found in cyprinoids as M. yokogawai.
In the present work, we provide molecular evidence that Metagonimus metacercariae commonly found in cyprinoids in Hungary and adults from experimental infections do not belong to M. yokogawai, suggesting that this trematode infecting humans is not present in Europe as previously thought.
Material and methods
Examination and collection of metacercariae of Metagonimus sp.
In the present study, 131 cyprinoid fish (Cyprinoidei: Leuciscidae) were collected from the Danube River at Zebegény (47°48'N, 18°55'E) and Szentendre (47°40' N, 19°4'E). Between 2015 and 2017, 9 white bream (Blicca bjoerkna), 26 freshwater bream (Abramis brama), 42 bleak (Alburnus alburnus), 4 ide (Leuciscus idus), 31 common nase (Chondrostoma nasus) and 5 chub (Squalius cephalus) were collected. Between 2020 and 2021, only 4 common nase and 6 chub were taken from the Danube, while two chub and two vimba bream (Vimba vimba) were caught in 2022. In addition, 19 European perch (Perca fluviatilis) were taken from Lake Balaton between 2020 and 2021, one of which was also infected with Metagonimus metacercariae (Table 1). The fish were caught with a 15-m seine net and brought to the laboratory alive in oxygenated plastic bags.
Fish were anaesthetized by clove oil and then decapitated. The body surface and scales were examined under a dissecting microscope. Metacercariae were isolated with a fine needle or by tissue digesting in 0.5% pepsin solution (2 l tap water, 10 g pepsin based on 1: 10,000 NF powder (Molar Chemicals, Halásztelek, Hungary) and 16 ml 25% hydrochloric acid (HCl) at a temperature of 40 °C with stirring. Encysted metacercariae were fixed in 70% ethanol. They were excysted from their capsules using a solution of 50 ml distilled water, 2.5 g pancreatin, and 0.25 g NaHCO3 (Fried, 1994). Excystation performed at 27 °C for 5–10 min, and then the metacercariae were placed in 0.9% physiological saline to avoid overdigestion. After treatment, the released metacercariae were observed under the microscope and kept alive for further use, including experimental infections.
Experimental infection for recovery of adult trematodes
To obtain adults, chicks, ducks, and Syrian hamsters were experimentally infected (permit number PEI/001/1004–4/2015, PEI/001/1792–4/2014) with metacercariae of Metagonimus sp. (50 cysts per animal) from cyprinoid fish in 4 experiments (Table 2). Experimental animals were necropsied in accordance with European animal welfare regulations. Adult Metagonimus specimens were collected from both the duodenum and faeces of the animals after decantation. The isolated trematodes were considered as adults when they were oviparous. Adult trematodes found were measured and fixed in molecular-grade ethanol for DNA sequencing.
Molecular methods
For DNA extraction, samples preserved in 80% ethanol were centrifuged at 8,000 g for 5 min, then the ethanol was removed with a pipette and/or by evaporation with a vacuum centrifuge. DNA was extracted using a Geneaid DNA Mini Kit (Geneaid, Taipei City, Taiwan) and eluted into 100 μl of AE buffer according to the manufacturer’s recommendations. The ITS region (part of 18S rDNA, ITS1, 5.8S rDNA, ITS2, and part of 28S rDNA) was amplified by nested PCR. Primers S18 (5′-TAACAGGTCTGTGATGCC-3′) and L3T (5′-CAACTTTCCCTCACGGTACTTG-3′) (Jousson et al. 1999) were used in the first run. The reaction mixture consisted of 14.4 μl nuclease-free water, 2.5 μl of 10 × DreamTaq buffer (Thermo Scientific, Vilnius, Lithuania), 0.1 μl of DreamTaq Polymerase (1 U; Thermo Scientific), 0.2 mM dNTPs (Thermo Scientific), 0.5 μM of each primer and 2 μl of the extracted DNA in a final volume of 25 μl. The PCR profile consisted of an initial denaturation step at 95 °C for 3 min, followed by 40 cycles at 95 °C for 30 s, 50 °C for 30 s and 72 °C for 2 min, and was terminated with a terminal extension at 72 °C for 5 min and then stored at 4 °C. Primers D1 (5′-AGGAA-TTCCTGGTAAGTGCAA-3′) and D2 (5′-CGTTACTGAGGGAATCCTGGT-3′) (Galazzo et al. 2002) were used in the second run, in 50 μl reaction mixture with the same concentrations as in the first round. 1 μl template form the first PCR round was added to each sample. The second round of PCR consisted of an initial denaturation step at 95 °C for 3 min, followed by 30 cycles at 95 °C for 30 s, 56 °C for 30 s, 72 °C for 2 min, and a final extension step at 72 °C for 5 min, followed by storage at 4 °C. The 28S rRNA gene was amplified using the following primers: DIG12 (5′-AAGCATATCACTAAGCGG-3′), 1500R (5′-GCT ATC CTGAGGGAAACTTCG-3′) according to the protocol of Tkach et al. 2003. The cox1 gene was amplified and sequenced with primers JB3 (5′-TTTTTTGGGCATCCTGAGGTTTAT-3′) and JB4.5 (5′-TAAAGAAAGAACATAATGAAAATG-3′) (Bowles and McManus 1994). The conditions used were identical to those for ribosomal markers, except for the annealing temperature (52 °C).
PCR products were electrophoresed in 1.0% agarose gels in Tris–acetate-EDTA (TAE) buffer gel, stained with 1% ethidium bromide, and then purified using an EZ-10 Spin Column PCR Purification Kit (Bio Basic Inc., Markham, Canada). The purified PCR products from the ITS region, 28S rDNA, and coxI were sequenced with the PCR primers and two additional internal primers 5.8Sr (5′-TGTCGATGAAGAGCGCAGC-3′) and 5.8S2 (5′-TAAGCCGACCCTCGGACAGG-3′) (Tkach et al. 2003) for the ITS region and with 1200R (5′-GCATAGTTCACCATCTTTCGG-3′) (Shumenko et al. 2017) for 28S rDNA. ABI BigDye Terminator v3.1 Cycle Sequencing Kit was used for sequencing, and sequences were read at the MTA SZBK Sequencing Platform in Szeged, Hungary, using an ABI Prism 3100 Genetic Analyser (Thermo Fisher Scientific, Waltham, USA).
Phylogenetic analysis
Sequence fragments were assembled using MEGA X (Kumar et al. 2018), and ambiguous bases were manually corrected based on the ABI chromatogram. Reference sequences were downloaded from GenBank (see below) with the program BLAST and then aligned with the program MEGA X (Kumar et al. 2018) using the algorithm CLUSTAL W (Thompson et al. 1994). Pairwise distance estimates representing genetic distances were determined using the p-distance model. Phylogenetic analysis was performed using the Maximum Likelihood (ML) method. The best-fitting nucleotide substitution model defined by the Akaike Information Criteria (AIC) was used to analyse the data set. The robustness of the ML phylogenetic tree was determined using bootstrap values based on 1000 resampled datasets. Bayesian Inference (BI) analysis was performed using Geneious Prime® 2019.2.3 software with the MrBayes (Huelsenbeck and Ronquist 2001) plug-in. Posterior probabilities (PP) were estimated over 1,000,000 generations by two independent runs of four simultaneous MCMCMC chains, with every 100th tree saved. The first 25% of the sampled trees were discarded as ‘burn-in’. Phylogenetic trees were visualised using the Tree Explorer of MEGA X.
Sequences of the following species were used for comparison: M. ciureanus (AY245702), M. hakubaensis (LC576458, LC576462, KM061388–90), M. katsuradai (KM061391–93), M. kinoi (LC5999533-34, LC666755–56, LC666627–29), M. kogai (LC666749–50), M. miyatai (LC375946, HQ832633, KM061409–11), M. otsurui (KM061394–96, KM061421–23), M. pusillus (MF406209–10, MF407172–73), M. saitoi (LC666745–46), M. shimazui (LC666753-54), M. suifunensis (KX387456, KX387520–24, KX387459–60, MK736844, MK736869, MN116490, MN116492), M. takahashii (HQ832636), M. yokogawai (AB470519, HQ832639, KC330755, KJ631740, KM061412, KX832895, KX857497, OK166789) and Metagonimus sp. (LC422948, LC422951).
Sequences of Metagonimus sp. were deposited in the GenBank under the accession numbers OQ281688-OQ281703, OQ286093-OQ286097, OQ286071-OQ286088 and OQ308609 (Table 3).
Results
Occurrence of metacercariae of Metagonimus sp. in fish
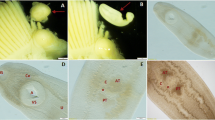
Metacercariae of Metagonimus sp. were found only in the scales of 121 cyprinoid fish of the following species: Abramis brama, Alburnus alburnus, Blicca bjoerkna, Chondrostoma nasus, Leuciscus idus, Squalius cephalus, and Vimba vimba from the Danube (see Table 1). In addition, a European perch (Perca fluviatilis) from Lake Balaton was also infected with metacercariae. The metacercariae were encysted, located on the inner side of the scales, and the cysts appeared as small, pearly, roundish structures 180–240 µm long and 140–190 µm wide (Fig. 1a-d).
a: Metagonimus sp. metacercaria embedded in the scales of chub (Squalius cephalus) b: released after artificial digestion c, Metagonimus sp. metacercaria in the scales of perch (Perca fluviatilis) d: and close up e: Metagonimus sp. adult recovered from a Syrian hamster experimentally infected with metacercariae in this study in ventral view f: dorsal view
Morphology of metacercariae of Metagonimus sp.
Cysts (n = 30) round, 215 ± 14 (205–225), or oval 207 ± 23 (180–240) long by 160 ± 21 (214–190) wide. Encysted metacercariae with clearly visible oral sucker, pharynx, and excretory vesicle. Excysted metacercariae (n = 15) 471 ± 34 (427–507) long, 240 ± 32 (196–280) wide. Body covered with numerous spines. Oral sucker spherical, 67 ± 3.1 (63–70) in diameter. Prepharynx short; pharynx somewhat wider than longer, 33 ± 2 (31–35) long and 37 ± 2 (35–39) wide; oesophagus short, 72 ± 2 (70–74) long. Intestinal caeca narrow, long, ending at level of testes. Ventral sucker elliptical, 30 ± 5 (26–32) long and 28 ± 5 (24–30) wide. Excretory bladder Y-shaped, filled with fine granules, lateral branches 110 ± 5 (105–115) µm long.
Adult trematodes (Fig. 1 e–f) were recovered from experimentally infected chicks, ducks, and Syrian hamsters fed with metacercariae of Metagonimus sp. from 4 species of fish (Table 2.). Selected biometrical characteristics of experimentally obtained adults are in Table 4.
Molecular characterisation of metacercariae and adults of Metagonimus sp.
For the 28S rRNA gene, the final alignment consisted of 782 bp with 657 conserved and 125 variable positions. Amplification of the ITS region yielded products of approximately 1,400 bp long. The alignment of ITS was 1,132 bp long and contained 801 conservative and 328 variable positions. The final alignment of cox1 sequences was 347 bp long and consisted of 186 conservative and 157 variable positions.
The ML and BI phylogenetic analysis of the ITS region, 28S rDNA and cox1 genes revealed that the samples from Hungary formed a single clade distinct from other species of Metagonimus, all from East Asia and Far East (Fig. 2).
Maximum likelihood tree of the samples of Metagonimus spp. from the present study (a: 28S rDNA, b: ITS region, c: coxI) in relation to other heterophyid and opisthorchiid sequences deposited in GenBank. Bootstrap values are indicated at the nodes; posterior probabilities for Bayesian inference are indicated after the bootstrap values. Samples from the present study are in bold. The scale bar indicates the expected number of substitutions per site
Results from the 28S rDNA gene also showed that the sequences from metacercariae and the sequences of the adult specimens of Metagonimus sp. formed a clade distinct from other species of Metagonimus. The clade was located in a basal position in relation to the Asian species. It should be noted that a single metacercaria from European perch was also placed into the same clade as other isolates from the Danube. Within-group patterns of Metagonimus sp. were almost identical (mean distance within groups = 0.001), which was also true for the reference sequences of Metagonimus species from Asia. The genetic distance (the mean distance between groups) between our samples and Asian Metagonimus samples varied between 1.9 and 3.5%, including M. yokogawai (2.6%); the difference between individual East Asian species was smaller, ranging from 0.2% to 2.7%.
In the phylogenetic analysis of the ITS region, metacercariae isolated from the Danube (MM2, DPM6) and three adult specimens isolated from experiments (ME1, ME2, ME3) formed a distinct clade. Similarly to the 28S rDNA analysis, Metagonimus sp. took a basal position within a monophyletic clade containing all analyses species of Metagonimus. Sequences of all isolates of Metagonimus sp. from the Danube were identical. Metagonimus sp. was most closely related in its 28S rDNA sequences with Metagonimus ciureanus (Witenberg 1929) (KX387520), a species of uncertain taxonomic status, which was originally described as Dexiogonimus ciureanus in Israel.
The analysis of mitochondrial cox1 gene corresponded to the results of analyses of sequences of nuclear markers. Metagonimus sp. samples formed a most basal clade of the monophyletic Metagonimus, distinct from other Metagonimus species. Mean distance of isolates of Metagonimus sp. was only 0.001. Genetic distances between these samples and other Metagonimus species ranged from 20.5% to 27.3%. The present specimens were most closely related to Metagonimus kinoi (LC666627–29), but similarity was only 20.5% (similarity with M. yokogawai was 23.5%). The previously described Asian Metagonimus species showed 10.0–25.5% interspecific distances to each other.
Discussion
In the present study, as many as seven species of cyprinoid fish from the Danube (Blicca bjoerkna, Abramis brama, Alburnus alburnus, Chondrostoma nasus, Leuciscus idus, Squalius cephalus and Vimba vimba) were heavily infected with Metagonimus metacercariae. Earlier, metacercariae most likely conspecific with those found recently in the Danube fish were identified as M. romanicus by Prettenhoffer (1930) and M. yokogawai by the other authors, such as Žitňan (1969) from Slovakia, Francová et al. (2011) from the Czech Republic, Cakić et al. (2007) and Djikanovic et al. (2011) from Serbia, Nachev and Sures (2009) and Ondračková et al. (2012) as M. yokogawai from Bulgaria.
Genotyping of these metacercariae and adults from experimentally infected definitive hosts showed that they are conspicuously distinct all East Asian species analysed, including M. yokogawai. However, the identification of trematodes from Hungary to species level was not possible due to poor quality of adults obtained (specimens were not properly fixed and thus are not suitable for a reliable morphological and biometrical comparison). In addition, comparison of selected metrical data of Metagonimus sp. from Hungary and M. romanicus has revealed some differences, especially in the size of the oral sucker and pharynx (Table 4). Therefore, specimens from Hungary are tentatively identified as Metagonimus sp.
The occurrence of Metagonimus sp. in European perch from Lake Balaton is noteworthy because there were only two previous reports of metacercariae of Metagonimus in percids in Europe: Cojocaru (2006) found two infected perch in Romania and Bykhovskaya-Pavlovskaya (1962) reported one infected perch in the U.S.S.R.. However, perch is probably only incidental host because prevalence of its infection in Hungary was low and only a single perch was infected with a single metacercaria.
It is possible that Metagonimus sp. from the Danube in Hungary is conspecific with M. romanicus, which was found in fish from the same river basin in Romania (Ciurea 1915, 1933). However, there are no molecular data on M. romanicus, and the available material of Metagonimus sp., including adults from experimentally infected hamsters and birds, does not allow us reliable comparison of the two species. In fact, the present study was focused on genotyping metacercariae and adults and insufficient attention was paid to proper processing of trematodes from experimentally infected hosts. In addition, there are some metrical differences between M. romanicus and Metagonimus sp., especially the size of the oral sucker and pharynx (Table 4). Therefore, it is necessary to obtain properly fixed material of Metagonimus sp. and compare it with specimens from Romania, provided they are available. However, there is no doubt that Metagonimus sp. from Hungary and M. yokogawai are different species because of nucleotide difference between our samples and M. yokogawai (ITS: 6.4–6.5%; 28S rRNA: 2.0–3.4%, cox1: 21–22%).
In addition to the genetic differences between Metagonimus sp. from Hungary and the Asian species of the genus, they also differ significantly in the site of infection of the metacercariae. Those of Metagonimus sp. are localised exclusively in the scales, whereas the metacercariae of all Asian species can be found in the muscles, intestines, gills and scales as well (Kino et al. 2006; Nakao et al. 2022). This site of infection of European specimens makes them less important from an epidemiological point of view, as whole fish with scales are rarely consumed in Europe. In contrast, successful experimental infections of hamsters with Metagonimus sp. provide evidence that this species can mature in mammals. Therefore, the potential risk of zoonotic infection with Metagonimus sp. from consumption of raw and undercooked fish from the Danube River cannot be ruled out entirely.
Conclusions
Metacarcariae of the genus Metagonimus were found in large numbers in the scales of cyprinoid fishes from the Hungarian Danube. Sequence data from three different loci (28S rDNA, ITS region and cox1) show that they do not belong to the East Asian M. yokagawai, which is very unlikely to occur in Europe despite previous records in the literature. The species found in Hungary may be conspecific with insufficiently known M. romanicus described by Ciurea (1915) in Romania. Successful infections of hamsters with metacercariae from fish demonstrate the zoonotic potential of this species.
Data availability
The sequence data generated during the current study are available in the GenBank repository under the accession numbers OQ281688-OQ281703, OQ286093-OQ286097, OQ286071-OQ286088 and OQ308609.
References
Braun M (1925) Die Tierischen Parasiten des Menschen. VI. Auflage, Leipzig, p 608
Bykhovskaya-Pavlovskaya IE, Kulakovskaya AP (1987) Class Trematoda Rudolphi, 1808. In: Bauer ON (ed) Key to Determination of the Parasites of Freshwater Fishes of the USSR, vol 3. Nauka, Leningrad, pp 77–198 (in Russian)
Bykhovskaya-Pavlovskaya IE (1962) Class Trematoda Rudolphi, 1808. In: Bykhowski BE (ed) [keys to the parasites of freshwater fishes of the USSR.] Akademii Nauk SSSR, Moscow–Leningrad, Russia. 776 pp (in Russian)
Cakić P, Paunovic M, Stojanovic B, Djikanovic V, Kulisic Z (2007) Metagonimus yokogawai (Katsurada, 1912), a new parasitic Trematoda species in ichtyoparasitofauna of the Serbia. Acta Vet 57(5–6):537–543. https://doi.org/10.2298/AVB0706537C
Chai JY, Jung BK (2017) Fishborne zoonotic heterophyid infections: An update. Food Waterborne Parasitol 8–9:33–63. https://doi.org/10.1016/j.fawpar.2017.09.001
Chai JY, Lee SH (2002) Food-borne intestinal trematode infections in the Republic of Korea. Parasitol Int 51:129–154. https://doi.org/10.1016/S1383-5769(02)00008-9
Chai JY, Murrell KD, Lymbery AJ (2005) Fish-borne parasitic zoonoses: status and issues. Int J Parasitol 35:1233–1254. https://doi.org/10.1016/j.ijpara.2005.07.013
Chai JY, Shin EH, Lee SH, Rim HJ (2009) Foodborne intestinal flukes in Southeast Asia. Korean J Parasitol 47:69–102. https://doi.org/10.3347/kjp.2009.47.S.S69
Chai JY (2015) Metagonimus. (Xiao L, Ryan U, Feng Y (eds), Biology of Foodborne Parasites. CRC Press, Taylor & Francis Group, Boca Raton, Florida, pp. 427-443. https://doi.org/10.1201/b18317
Ciurea J (1915) Über einige neue Distomen aus dem Darm unserer Haustiere und des Pelikans, für welche die Fische als Infektionsquelle zu betrachten sind. Z Infektionskr Parasitäre Krankh Hyg Haustiere 16:445–458 (in German)
Ciurea J (1933) Les vers parasites de l’homme, des mammifères, et des oiseaux provenant des poissons du Danube et de la Mer Noire. Premier mémoire. Trématodes, famille Heterophyidae Odhner, avec un essai de classification des trématodes de la superfamille Heterophyoidea Faust. Arch Roum Pathol Exp Microbiol 6(1–2):5–134 (in French)
Cojocaru C (2006) Infection with Metagonimus yokogawai in fish from Romania. Author content. https://www.researchgate.net/publication/289524801_Infection_with_Metagonimus_yokogawai_in_fish_from_Romania Accessed 1 March 2023
Djikanovic V, Paunovic M, Nikolic V, Simonovic P, Cakic P (2011) Parasitofauna of freshwater fishes in the Serbian open waters: a checklist of parasites of freshwater fishes in Serbian open waters. Rev Fish Biol Fish 22:297–324. https://doi.org/10.1007/s11160-011-9226-6
Francová K, Ondračková M, Polačik M, Jurajda P (2011) Parasite fauna of native and non-native populations of Neogobius melanostomus (Pallas, 1814) (Gobiidae) in the longitudinal profile of the Danube River. J Appl Ichthyol 27(3):879–886. https://doi.org/10.1111/j.1439-0426.2010.01582.x
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–7555. https://doi.org/10.1093/bioinformatics/17.8.754
Isaitshikov IM (1925) To the fauna of parasitic worms of domestic carnivorous animals of Crimea. I. Parasitic worms of the dog. Uchebn Trudi Sibirsk Vet Inst 6:132–170 (in Russian)
Kino H, Suzuki T, Oishi H, Suzuki S, Yamagiwa S, Ishiguro M (2006) Geographical distribution of Metagonimus yokogawai and M. miyatai in Shizuoka Prefecture, Japan, and their site preferences in the sweetfish, Plecoglossus altivelis, and hamsters. Parasitol Int 55:201–206. https://doi.org/10.1016/j.parint.2006.05.001
Kulišić Z, Lepojev O (1994) Trematodes of wild duck (Anas platyrhynchos L.) in the Belgrade area. Acta Vet 44:323–328
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Molnár K (1969) Beiträge zur Kenntnis der Fischparasitenfauna Ungarns IV. Trematoden Parasitol Hung 2:119–136
Molnár K, Baska F (2017) Fish Diseases (Halbetegségek). MÁOK Kft, Budapest, p 167 (in Hungarian)
Morozov FN (1952) Superfamily Heterophyoidea Faust, 1929. In: Skrjabin KI (ed) Trematodes of Animals and Men. Osnovy Trematologii. AN SSSR, Moskva, 6:153–601. (in Russian)
Nachev N, Sures B (2009) The endohelminth fauna of barbel (Barbus barbus) correlates with water quality of the Danube River in Bulgaria. Parasitology 136:545–552. https://doi.org/10.1017/S003118200900571X
Nakao M, Ishikawa T, Hibino Y, Ohari Y, Taniguchi R, Takeyama T, Nakamura S, Kakino W, Ikadai H, Sasaki M (2022) Resolution of cryptic species complexes within the genus Metagonimus (Trematoda: Heterophyidae) in Japan, with descriptions of new species. Parasitol Int 90:102605. https://doi.org/10.1016/j.parint.2022.102605
Ondračková M, Slovačkova I, Trichkova T, Polačik M, Jurajda P (2012) Shoreline distribution and parasitic infection of black-striped pipefish Syngnathus abaster Risso, 1827 in the low River Danube. J Appl Ichthyol 28(4):590–596. https://doi.org/10.1111/j.1439-0426.2012.01967.x
Pornruseetairatn S, Kino H, Shimazu T, Nawa Y, Scholz T, Ruangsittichai J, Saralamba NT, Thaenkham U (2016) A molecular phylogeny of Asian species of the genus Metagonimus (Digenea) – small intestinal flukes – based on representative Japanese populations. Parasitol Res 115:1123–1130. https://doi.org/10.1007/s00436-015-4843-y
Prettenhoffer Z (1930) Experimental investigations on the Hungarian occurrence of larval trematodes infecting Danube fishes (Kísérleti vizsgálatok dunai halakban élősködő trematoda-lárvák hazai előfordulásáról). Közlemények Az Összehasonlító Élet-És Kórtan Köréből 24:71–83 (in Hungarian)
Rácz OZ, Zemankovics E (2002) Survival of metacercariae of Metagonimus yokogawai (Digenea: Heterophyidae) on fish from River Danube. Hung Vet J 124(7):437–444 (in Hungarian)
Shimazu T (1999) Metagonimus hakubaensis sp. n. Digenea, Heterophyidae from Nagano, Japan: morphology and life cycle. Bull Nat Sci Museum 25:87–99
Shimazu T (2002) Life cycle and morphology of Metagonimus miyatai (Digenea: Heterophyidae) from Nagano, Japan. Parasitol Int 51:271–280. https://doi.org/10.1016/S1383-5769(02)00038-7
Shimazu T, Kino H (2015) Metagonimus yokogawai (Trematoda: Heterophyidae): from discovery to designation of a neotype. Korean J Parasitol 53:627–639. https://doi.org/10.3347/kjp.2015.53.5.627
Shimazu T, Urabe M (2002) Morphology and life cycle of Metagonimus otsurui (Digenea, Heterophyidae) from Nara, Japan. Bull Nat Sci Museum 28:21–28
Shumenko PG, Tatonova YV, Besprozvannykh VV (2017) Metagonimus suifunensis sp. n. (Trematoda: Heterophyidae) from the Russian Southern Far East: Morphology, life cycle, and molecular data. Parasitol Int 66(1):982–991. https://doi.org/10.1016/j.parint.2016.11.002
Tatonova Y, Shumenko P, Besprozvannykh V (2018) Description of Metagonimus pusillus sp. nov. (Trematoda: Heterophyidae): Phylogenetic relationships within the genus. J Helminthol 92(6):703–712. https://doi.org/10.1017/S0022149X17001146
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Vojtek J (1959) Data to the knowledge of the helminthofauna of fish around Komarno. Publ Fac Sci Univ Brno Czechoslovakia 207:437–465 (in Czech)
Witenberg G (1929) Studies on the trematode-family Heterophyidae. Ann Trop Med Parasitol 23:131–239. https://doi.org/10.1080/00034983.1929.11684600
Yu SH, Mott KE (1994) Epidemiology and morbidity of food-borne intestinal trematode infections. Trop Dis Bull 91:125–152
Žitňan R (1969) Zur Helminthenfauna der Fische in der Kleinen Donau. Helminthologia 10:313–320 (in German)
Acknowledgements
The authors thank two anonymous reviewers for their helpful suggestions. We are also thankful for Mr. Gergely Lajos Zöldi for his contribution in fish sampling and Dr. László Egyed for his help in the experimental infections. We would like to express our special thanks to Dr. Tomáš Scholz, who significantly contributed to the success of the publication with his professional advices during the review process.
Funding
Open access funding provided by Veterinary Medical Research Institute. This project has received funding from the Hungarian Scientific Research Fund (NKFI OTKA FK 140350).
Author information
Authors and Affiliations
Contributions
All authors have contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Gábor Cech, Martina Gyöngy, Diána Sándor, Kálmán Molnár, Boglárka Sellyei, Ádám Varga and Csaba Székely. The first draft of the manuscript was written by Gábor Cech and Martina Gyöngy; all authors commented on previous versions of it. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Guillermo Salgado-Maldonado
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cech, G., Gyöngy, M., Sándor, D. et al. Molecular evidence of the absence of Metagonimus yokogawai (Katsurada, 1912) in Europe: report of Metagonimus sp. in cyprinoid fish from the River Danube in Hungary. Parasitol Res 122, 2325–2334 (2023). https://doi.org/10.1007/s00436-023-07932-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-023-07932-1