Abstract
Phlebotomine sand flies are dipterans of relevance due to their role as vectors of several pathogens worldwide. Bacteria in the gut of sand flies possibly affect their vectorial capacity and competence to transmit parasites. A retrospective study was performed in sand fly specimens that had previously been collected in four localities of the state of Chiapas during the period 2009–2011 to detect Wolbachia and Bartonella and their possible coinfection with Leishmania. For the molecular detection of bacteria, we used primers and conditions that had previously been reported. A total of 531 sand fly specimens of 10 species were analyzed. Four Wolbachia strains were detected in five sand fly species, showing a prevalence of 8.6%. All the Wolbachia strains had previously been reported in other taxa. In one sand fly species, we also detected a new lineage of Bartonella evidenced by a phylogenetic analysis. No sand fly specimens showed coinfections of these bacteria and Leishmania. The bacteria found in the phlebotomine sand flies are possibly transmitted by plant-mediated horizontal transmission and during blood meal feeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phlebotomine sand flies (Diptera: Psychodidae, Phlebotominae) are vectors of several pathogens such as Leishmania, Bartonella, and some arboviruses that affect human health worldwide (Akhoundi et al. 2016). In recent years, comprehensive studies have revealed basic aspects of the life cycle of this vector, helping to establish efficient prevention and control strategies to avoid the transmission of these pathogens.
The analysis of the diversity of bacteria in the gut of sand fly species provides insights into their vectorial capacity and competence to transmit parasites (Sallum et al. 2019; Vivero et al. 2019). For instance, the detection of the α-proteobacteria Wolbachia is relevant, since this bacterium spreads quickly and manipulates the reproductive success of its insect host, in order to guarantee its own propagation, causing reproductive alterations such as cytoplasmic incompatibility (CI), induction of parthenogenesis, and feminization or death of males (Werren 1997; Pimentel et al. 2020). These effects represent a possible strategy for the biological control of sand flies. The presence of Wolbachia in some mosquitoes of the genus Culex and Aedes has been reported to confer protection against nematodes and virus transmission, such as in dengue, preventing its replication (Karimian et al. 2018; Pimentel et al. 2020).
For that reason, it has been proposed that the association between Wolbachia and sand flies probably interferes with the establishment of Leishmania species. In the American continent, only 11 sand fly species have been associated with Wolbachia strains in Brazil, Colombia, Mexico, and Panama, although the role of these associations remains unclear (Ono et al. 2001; Azpurua et al. 2010; Mikery-Pacheco et al. 2012; Monteiro et al. 2016; Kelly et al. 2017; Vivero et al. 2017; Lozano-Sardaneta et al. 2021a, b; Lozano-Sardaneta et al. 2022). Recent studies propose that Wolbachia in sand flies might induce cytoplasmic incompatibility that reduces the genetic variability, causing speciation (Kassem et al. 2003; Azpurua et al. 2010). In Mexico, only Lutzomyia cruciata and Psathyromyia shannoni have been reported to be infected with the strain Wolbachia wWhi in the states of Chiapas, Veracruz, and Tabasco (Mikery-Pacheco et al. 2012; Lozano-Sardaneta et al. 2021a, b; Lozano-Sardaneta et al. 2022). This is relevant, since both sand flies are considered to be vectors of Leishmania mexicana and Leishmania infantum in Mexico (Pech-May et al. 2010; Lozano-Sardaneta et al. 2020).
Another bacterium transmitted by a sand fly is Bartonella, a hemotropic bacterium that causes chronic intraerythrocytic infections in their hosts (Chomel et al. 2009). The only confirmed species causing bartonellosis transmitted by sand flies are Bartonella bacilliformis and Bartonella grahamii, which are endemic species in the Andean valleys (Lozano-Sardaneta et al. 2019). Uncultured Bartonella spp. have recently been recorded in association with Lu. cruciata and Pa. shannoni in the states of Veracruz and Tamaulipas, Mexico, although it remains to be established whether the sand flies are vectors of these bacteria and if these Bartonella species cause emerging diseases (Lozano-Sardaneta et al. 2019; Lozano-Sardaneta et al. 2021a).
Chiapas is the Mexican state that harbors the highest number of sand fly species (36 of 52) and is also considered an endemic area of leishmaniasis (Ibáñez-Bernal et al. 2015). Therefore, the aim of this study was to conduct a retrospective study focused on the molecular detection of the bacteria of the genus Wolbachia and Bartonella in sand flies and their possible coinfection with Leishmania and determine their prevalence in specimens collected near to the Chiapas-Guatemala border. Obtaining information on the microbiome of this phlebotomine fauna possibly helps to evaluate its possible use as a biological control method for sand flies, helping to prevent the transmission of leishmaniasis.
Material and method
Study area and specimens analyzed
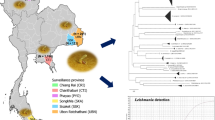
We analyzed sand fly specimens that had previously been collected in areas with an elevated risk of transmission of leishmaniasis in the state of Chiapas, near the border between Mexico and Guatemala, during the period 2009–2011. The localities that were analyzed included (1) San Antonio Buenavista (16.1523 N; −91.6497 W), (2) Tziscao (16. 0812 N; −91. 6670 W), (3) Guadalupe Miramar (16. 1562 N; −91.2792 W), and (4) Loma Bonita (16. 1980 N; -91.2078 W) (Fig. 1).
All analyzed specimens were collected using CDC light traps (Mod. 512) and modified Magoon traps baited with bite-protected humans. The traps were located in five houses (indoors and peridomicile) in four transects ranging 500 m from the edge of the houses into the surrounding vegetation. During the winter months (October–March) traps were used between 18:00 and 06:00 h and in the summer months (April–September), the traps were active between 19:00 and 07:00 h (Ibáñez-Bernal et al. 2015). The collected sand flies were stored in 70% ethanol, the head was dissected for morphological identification, and the remaining parts of the body were used for molecular analysis. Permanent mounting was done following published protocols (Ibáñez-Bernal 2005), and the identification and classification of specimens were based on Galati (2019) proposal. We use the abbreviation system proposed by Marcondes (2007).
DNA extraction and polymerase chain reaction (PCR) conditions
DNA was only extracted from the females, using a plasmid extraction protocol modified by Pech-May et al. (2013). Since only small amounts of DNA were available, we first analyzed sand fly DNA pools of 10 specimens of the same species and the same localities. If these tested positive, the specimens of the pools were individually analyzed. For molecular detection of Wolbachia strains, we amplified a fragment of the surface protein (wsp) gene of ~ 600 bp using the primers wsp 81F (5′ TGG TCC AAT AAG TGA TGA AGA AAC 3′) and wsp 691R (5′ AAA AAT TAA ACG CTA CTC CA 3′) (Braig et al. 1998). The PCR was performed with an initial denaturation at 95 °C for 5 min, followed by 35 cycles at 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min; with a final extension at 72 °C for 5 min. For the detection of Bartonella species, we amplified a segment of ~ 378 bp of the citrate synthase (gltA) gene, using the primers BhCS871.p (5′-GGG GAC CAG CTC ATG GTG G -3′) and BhCS1137.n (5′-AAT GCA AAA AGA ACA GTA AAC A-3′) (Norman et al. 1995). The PCR was performed under the following conditions: initial denaturation at 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, 51 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 7 min (Rubio et al. 2014). Additionally, for the analysis of a possible coinfection of Leishmania with the bacteria, we amplified a ~ 350 bp fragment of the gene ITS1 for the detection of Leishmania, using the primers LITSR (5′-CTG GAT CAT TTT CCG ATG—3′) and L5.8S (5′-TGA TAC CAC TTA TCG CAC TT—3′), using previously reported PCR conditions (El Tai et al. 2001; Lozano-Sardaneta et al. 2020).
The reaction mixture was prepared in a final volume of 25 μl containing 12.5 μl GoTaq® Green Master Mix 2X Promega Corporation (Madison, WI, USA), 1 μl of each primer (100 ng each), 5 μl DNA template (~50 ng/μl), and 5.5 μl nuclease-free water. The negative control consisted of ultrapure water instead of DNA. The PCR reactions were performed in a Veriti 96 Well Thermal Cycler (Applied BiosystemsTM, Thermo Fisher Scientific, USA). The amplified products were analyzed by electrophoresis in 2% agarose gels stained with 0.4µL of Midori Green Advance (Nippon genetics). The positive PCR products were purified and sequenced at Laboratorio de Secuenciación Genómica de la Biodiversidad y de la Salud, Instituto de Biología, UNAM.
Data analysis
The gltA and wsp electropherograms were visualized and edited in the software Chromas. Each sequence was compared with all the sequences available at the NCBI database, using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) as a preliminary confirmation.
The retrieved sequences were aligned with other reference sequences deposited on GenBank using MEGA version X (Kumar et al. 2018). For the phylogenetic analysis of the sequences of both genes, we used a maximum likelihood (ML) reconstruction performed in MEGA X, with 10,000 bootstraps, using the Tamura 3 parameters (T92)+ Gamma distribution substitution model, showing a BIC score of 5117.294 (for gltA in Bartonella) and BIC 8424.828 (for wsp of Wolbachia). The genetic distances were calculated in MEGA X. For the wsp gene, we translated the sequences into amino acids to facilitate the alignment (Zhou et al. 1998). The obtained sequences were deposited in GenBank under the following accession numbers Wolbachia wsp OP618079-OP618084 and Bartonella gltA OP618073-OP618079.
Results
Sand fly specimens analyzed
A total of 531 sand fly specimens were recovered (Table 1), belonging to six genera and 10 species. The most abundant species were Psychodopygus panamensis, Pintomyia ovallesi, and Dampfomyia deleoni. Guadalupe Miramar was the locality with the highest numbers of specimens available for molecular analysis.
Wolbachia detection
Wolbachia strains that were detected in the sand flies showed a prevalence of 8.6% (46/531). Wolbachia was detected in Ps. corossoniensis, Lu. cruciata, Pi. ovallesi, Ps. panamensis, and Ny. ylephiletor from Guadalupe Miramar and Loma Bonita. The prevalences were Ps. corossoniensis (3/7; 42.8%), Lu. cruciata (4/25; 16%), Pi. ovallesi (2/108; 1.85%), Ps. panamensis (24/286; 8.4%), and Ny. ylephiletor (13/39; 33.33%). The highest number of infected specimens was found in Guadalupe Miramar (32/46).
The sequences showed similarities with other strains previously reported in sand flies. Thus, Wolbachia detected in Lu. cruciata was 99.81% similar to Wolbachia endosymbiont of Pa. shannoni (MT533592.1), and the Wolbachia detected in Ps. panamensis was 100% identical to Wolbachia endosymbiont of Micropygomyia stewarti (KJ174699.1).
Additionally, we observed similarities with other strains detected in mosquitoes of the genus Culex sp. and the parasitoid wasp. Thus, the Wolbachia detected in Ny. ylephiletor was 99% similar to Wolbachia of Belonocnema treatea (wasp) (MG252474.1), and the Wolbachia strain detected in Ps. corosoniensis and Ps. panamensis was 100% similar to Wolbachia endosymbiont of Culex quinquefasciatus (LC276757.1) and Culex pipiens (KT964225.1), respectively.
According to the ML analysis (Fig. 2), we detected one Wolbachia strain belonging to the supergroup A. This Wolbachia strain wWhi was detected in Lu. cruciata species (collected in Loma Bonita). It had previously been recorded in Ny. intermedia, Pa. shannoni, and Ny. whitmani from Brazil, Mexico, and Colombia, showing a bootstrap value of 98%, and a genetic distance ranging from 0 to 2.5%. Although an earlier study had reported Wolbachia in Lu. cruciata from Chiapas (Mikery-Pacheco et al. 2012), the molecular identity of this strain had not been shown.
We also detected three strains belonging to the supergroup B: (1) the wTre4 strain detected in Ny. ylephiletor (collected in Guadalupe Miramar). It had previously been reported in the parasitoid wasp of oak trees Belonocnema treatea, showing a bootstrap value of 100%; (2) the wSte strain detected in Ps. panamensis (collected in Guadalupe Miramar). It had previously been reported in Micropygomyia stewarti, showing a bootstrap value of 97%; and (3) the strain wPip detected in Ps. panamensis (collected in Loma Bonita and Guadalupe Miramar) and in Ps. corossoniensis (collected in Guadalupe Miramar) showing a bootstrap value of 99%. This species had also been reported in mosquitoes of the genus Culex from India (LC276757.1) and Tunisia (AF020061.1) (Fig. 2).
Bartonella detection
The bacterium Bartonella sp. was detected in 27/531 specimens, showing a prevalence of 5.08%. The only positive species was the sand fly Pi. ovallesi from Loma Bonita, Chiapas. The sequences were 98% similar to each other and showed 83% similarity with a Bartonella sp. of Lutzomyia sp. from Mexico (MN325839.1). The ML analysis (Fig. 3) showed that the sequences correspond to a new lineage of Bartonella sp. associated with Pi. ovallesi, showing a bootstrap support value of 99% and genetic distances ranging 0 to 3.4%. It clustered in a clade with another lineage that had previously been recorded in sand flies of Mexico, showing a bootstrap value of 60% and genetic distances ranging from 25 to 29% with regard to other Uncultured Bartonella sp. from the states of Veracruz and Tamaulipas. This clade seems to be separated from the other pathogenic Bartonella species (Fig. 3). Since there are only few records on Bartonella species associated with sand flies outside of endemic areas, more studies are necessary to delimit and characterize these bacterial species associated with phlebotomine sand flies.
Although we analyzed the sand flies for infections with Leishmania spp., none of the specimens tested positive. Thus, our study ruled out the coinfection of these bacteria and Leishmania in sand flies of Chiapas.
Discussion
Phlebotomine sand flies have a relevant role in vector-borne diseases worldwide. Gaining insights into their microbiome helps reveal biological aspects of their cycle including reproduction, immune system, vectorial capacity, fitness, survival, and competence (Vivero et al. 2019). The gut microbiota of sand flies is involved in a wide range of biological and physiological processes, which could hamper or facilitate pathogen transmission, since some bacteria can negatively affect pathogen colonization (Louradour et al. 2017). The microbiome is closely related to the nutrition of sand flies, which can acquire the microorganisms from the soil, plants, and blood during their development and feeding. Variations of the intestinal microbiota can play a role in the survival and colonization of some parasites in the gut of sand flies (Louradour et al. 2017; Vivero et al. 2019).
In Mexico, studies on microorganisms associated with sand flies are scarce. Yet, Wolbachia and Bartonella had previously been reported in sand flies (Mikery-Pacheco et al. 2012; Lozano-Sardaneta et al. 2019, 2021a, b; Lozano-Sardaneta et al. 2021a, 2022). This led us to retrospectively analyze the possible presence of bacteria in sand flies species that had previously been collected in Chiapas, Mexico, a state with a high prevalence of leishmaniasis.
We now detected four Wolbachia strains (wWhi, wTre4, wPip, and wSte) in five sand fly species from Chiapas. These Wolbachia strains had previously been reported in other sand fly species and other insects, showing a prevalence of 8.6%. This is higher compared to other studies carried out in Mexico, where a prevalence of 0.98% has been reported (Mikery-Pacheco et al. 2012; Lozano-Sardaneta et al. 2021a, b; Lozano-Sardaneta et al. 2022). Generally, Wolbachia strains are widely distributed in wild populations of arthropods, causing reproductive alterations to ensure their own propagation by vertical transmission, thereby infecting a high number of specimens (Ono et al. 2001).
The ML analysis showed that the Wolbachia strain wPip was detected in Ps. panamensis in two distant localities (Guadalupe Miramar and Loma Bonita) and in Ps. corossoniensis. This strain has also been reported in species of mosquitoes of the genus Culex, where it provides protection against viral infections (Fraser et al. 2020). Additionally, we detected the Wolbachia wSte strain in Ps. panamensis. This Wolbachia strain had previously been reported in the sand fly Mi. stewarti in the USA and shown to confer protection to the sand fly against Plasmodium (Hughes et al. 2014). The role of these strains in species of the genus Psychodopygus is unknown, yet is likely that they could prevent the establishment of some viruses. Although the Wolbachia wWhi strain has previously been reported in Nyssomyia intermedia, Nyssomyia whitmani, and Pa. shannoni and from Brazil, Colombia, and Mexico (Ono et al. 2001; Lozano-Sardaneta et al. 2022), this is the first time that this strain is recorded in Lu. cruciata. Interestingly, in this study, this strain was not found in Pa. shannoni, a sand fly where it has previously been reported in Mexico (Lozano-Sardaneta et al. 2021a, b; Lozano-Sardaneta et al. 2022). The transmission of the Wolbachia wWhi strain is generally occurs by horizontal transmission between closely related species, which excludes the sand fly Lu. cruciata, since it belongs to another genus (Ono et al. 2001; Karimian et al. 2018).
The Wolbachia strain wTre was detected in the sand fly Ny. ylephyletor. This Wolbachia strain has also been associated with the parasitoid wasp B. treatea in the USA and could have been acquired by horizontal transmissions from the oak tree (Fagaceae: Quercus) or other parasitoids, causing reproductive isolation in their host (Schuler et al. 2018).
Although the vertical transmission of Wolbachia strains is common, in some cases a horizontal transmission also occurs, such as across parasitoids, invertebrate predators, ectoparasitic mites, and host plants or food sources (Vavre et al. 1999; Li et al. 2017). In the case of sand flies, the plant-mediated horizontal transmission in sand fly species seems a plausible potential transmission pathway. It has been confirmed that when different insect taxa feed on the same plant and share resting sites, they can acquire the same endosymbionts (Sintupachee et al. 2006; Li et al. 2017). Since phlebotomine sand fly males and females feed on sugar from plants (nectar and/or phloem sap), as well as on sugars excreted by “honeydew” aphids (Lima et al. 2016; Abbasi et al. 2018), this could explain why the same Wolbachia strains show widespread distributed in other taxa, and in phlebotomine sand flies from different localities from Chiapas, Mexico. It is therefore important to take the horizontal transmission route into account, since it could introduce new phenotypes in the different hosts. This could enable different fitness benefits, such as increasing the resistance against particular pathogens or inducing speciation, depending on the sand fly species (Monteiro et al. 2016; Li et al. 2017; Abbasi et al. 2018; Schuler et al. 2018). Therefore, complementary studies are necessary to test the relevance of horizontal transmission of Wolbachia strains in sand flies.
We now also detected, for the first time, a new lineage of Bartonella sp. associated to Pi. ovallesi in Loma Bonita, Chiapas. The sand fly Pi. ovallesi is anthropophilic and considered to be a vector for Leishmania spp. in Belize, Colombia, Panama, and Venezuela. In Mexico, this sand fly was recently found to be infected with Leishmania and is now regarded as a potential vector for the parasite (Lozano-Sardaneta et al. 2022). According to the ML analysis, the sequences obtained for the Bartonella sp. correspond to a new lineage that grouped into a clade with other Uncultured Bartonella spp. associated with sand flies in Mexico (Lozano-Sardaneta et al. 2019; Lozano-Sardaneta et al. 2021b). Even though the ML analysis showed a genetic difference of 29%, this lineage is included in the genus Bartonella. A Bartonella species can be classified as new, if it exhibits less than ≥ 96.0% nucleotide similarity using the gltA gene with other validated species (La Scola et al. 2003). In general, Bartonella species have host specificity, which suggests that specific adaptations are involved in the successful establishment and survival in a new arthropod vector or mammal host (Chomel et al. 2009). It is probable that the new Bartonella lineage detected in our study is specific for Pi. ovallesi and could have been acquired during the blood feeding.
The fact that we did not observe the presence of Leishmania in any of the analyzed sand fly species is consistent with other records of Mexico. None of the studies has shown the coinfection of Bartonella with Leishmania in sand flies (Lozano-Sardaneta et al. 2021b). Undoubtedly, more studies are needed to determine whether infection of sand flies with this bacterium possibly avoids the establishment of Leishmania in the same sand fly.
Although the detection of bacteria is relevant for the study of phlebotomine sand flies due to their potential use as biological controls to prevent the transmission of Leishmania, it is difficult to characterize all the species. We now report for the first time the presence of four Wolbachia strains associated to sand flies from Chiapas, Mexico, showing a high prevalence. It is probable that Wolbachia may confer some benefits when it is transmitted horizontally, possibly by decreasing the transmission of parasites and viruses. Furthermore, we now report a new lineage of Bartonella in sand flies, although we cannot confirm if the sand flies are vectors of this bacterium, since to date all infected sand fly species have shown to harbor different Bartonella lineage. Clearly, further studies are required to elucidate the possible role of sand fly species as potential vectors of other Bartonella species, to help understand their transmission pathway, and to improve their molecular identification.
Data availability
The sequences generated and analyzed in this study have been deposited in the GenBank database under the accession numbers: OP618073-OP618084.
References
Abbasi I, Lopo T, de Queiroz A, Kirstein OD et al (2018) Plant-feeding phlebotomine sand flies, vectors of leishmaniasis, prefer Cannabis sativa. Proc Natl Acad Sci U S A 115:11790–11795. https://doi.org/10.1073/pnas.1810435115
Akhoundi M, Kuhls K, Cannet A et al (2016) Historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis 10:e0004349. https://doi.org/10.1371/journal.pntd.0004349
Azpurua J, de la Cruz D, Valderama A, Windsor D (2010) Lutzomyia sand fly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis 4:e627. https://doi.org/10.1371/journal.pntd.0000627
Braig HR, Zhou W, Dobson SL, O’ Neill SL (1998) Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol 180:2373–2378. https://doi.org/10.1128/JB.180.9.2373-2378.1998
Chomel BB, Boulouis HJ, Breitschwerdt EB et al (2009) Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res 40:1–22. https://doi.org/10.1051/vetres/2009011
El Tai NO, EL Fari M, Mauricio I et al (2001) Leishmania donovani: Intraspecific polymorphisms of Sudanese isolates revealed by PCR-based analyses and DNA sequencing. Exp Parasitol 97:35–44. https://doi.org/10.1006/expr.2001.4592
Fraser JE, O’Donnell TB, Duyvestyn JM et al (2020) Novel phenotype of Wolbachia strain wPip in Aedes aegypti challenges assumptions on mechanisms of Wolbachia-mediated dengue virus inhibition. PLoS Pathog 16:e1008410. https://doi.org/10.1371/journal.ppat.1008410
Galati E (2019) Morfologia e terminologia de Phlebotominae (Diptera: Psychodidae). Classificação e identificação de táxons das Américas. In: Vol I. Apostila da Disciplina Bioecologia e Identificação de Phlebotominae do Programa de Pós-Graduação em Saúde Pública. Faculdade de Saúde Pública da Universidade de São Paulo, São Paulo, 1–133
Hughes GL, Samuels SK, Shaikh K et al (2014) Discrimination of the Plasmodium mexicanum vectors Lutzomyia stewarti and Lutzomyia vexator by a PCR-RFLP assay and Wolbachia infection. J Vector Ecol 39:224–227. https://doi.org/10.1111/j.1948-7134.2014.12092.x
Ibáñez-Bernal S (2005) Phlebotominae (Diptera: Psychodidae) de México. V. Clave ilustrada para la identificación de los machos de Lutzomyia França. Fol Entomol Mex 44:49–66
Ibáñez-Bernal S, Muñoz J, Rebollar-Téllez E et al (2015) Phlebotomine sand flies (Diptera: Psychodidae) of Chiapas collected near the Guatemala border, with additions to the fauna of Mexico and a new subgenus name. Zootaxa 3994:151–186. https://doi.org/10.11646/zootaxa.3994.2.1
Karimian F, Vatandoost H, Rassi Y et al (2018) wsp-based analysis of Wolbachia strains associated with Phlebotomus papatasi and P. sergenti (Diptera: Psychodidae) main cutaneous leishmaniasis vectors, introduction of a new subgroup wSerg. Pathog Glob Health 112:152–160. https://doi.org/10.1080/20477724.2018.1471438
Kassem HA, Hassan AN, Abdel-Hamid I et al (2003) Wolbachia infection and the expression of cytoplasmic incompatibility in sandflies (Diptera: Psychodidae) from Egypt. Ann Trop Parasitol 97:639–644. https://doi.org/10.1179/000349803225001391
Kelly P, Bahr S, Serafim T et al (2017) The gut microbiome of the vector Lutzomyia longipalpis is essential for survival of Leishmania infantum. mBio 18:e01121-16. https://doi.org/10.1128/mBio.01121-16
Kumar S, Stecher G, Li M et al (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
La Scola B, Zeaiter Z, Khamis A, Raoult D (2003) Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol 11:318–321. https://doi.org/10.1016/S0966-842X(03)00143-4
Li SJ, Ahmed MZ, Lv N et al (2017) Plant-mediated horizontal transmission of Wolbachia between whiteflies. ISME J 11:1019–1028. https://doi.org/10.1038/ismej.2016.164
Lima LHGM, Mesquita MR, Skrip L et al (2016) DNA barcode for the identification of the sand fly Lutzomyia longipalpis plant feeding preferences in a tropical urban environment. Sci Rep 6:1–6. https://doi.org/10.1038/srep29742
Louradour I, Monteiro CC, Inbar E et al (2017) The midgut microbiota plays an essential role in sand fly vector competence for Leishmania major. Cell Microbiol 19:1–13. https://doi.org/10.1111/cmi.12755
Lozano-Sardaneta YN, Colunga-Salas P, Sánchez-Montes S et al (2019) First report of Bartonella sp. in sand flies (Diptera: Psychodidae: Phlebotominae) from southern Mexico. J Am Mosq Control Assoc 35:224–227
Lozano-Sardaneta YN, Sánchez-Montes S, Sánchez-Cordero V et al (2020) Molecular detection of Leishmania infantum in sand flies (Diptera: Psychodidae: Phlebotominae) from Veracruz, Mexico. Acta Trop 207:1–7. https://doi.org/10.1016/j.actatropica.2020.105492
Lozano-Sardaneta YN, Valderrama A, Sánchez-Montes S et al (2021a) Rickettsial agents detected in the genus Psathyromyia (Diptera:Phlebotominae) from a Biosphere Reserve of Veracruz, Mexico. Parasitol Int 82:1–7. https://doi.org/10.1016/j.parint.2021.102286
Lozano-Sardaneta YN, Soto-Olguín NJ, Rodríguez-Rojas JJ et al (2021b) Molecular detection of Bartonella sp. in Psathyromyia shannoni and Lutzomyia cruciata from Northeastern Mexico. Front Trop Dis 2:1–6. https://doi.org/10.3389/fitd.2021.780808
Lozano-Sardaneta YN, Jacobo-Olvera E, Ruiz-Tovar K et al (2022) Detection of Wolbachia and Leishmania DNA in sand flies (Diptera: Psychodidae, Phlebotominae) from a focus of cutaneous leishmaniasis in Tabasco, Mexico. Parasitol Res 121:513–520. https://doi.org/10.1007/s00436-021-07412-4
Marcondes B (2007) A proposal of generic and subgeneric abbreviations for phlebotomine sandflies (Diptera: Psychodidae: Phlebotominae) of the world. Entomol News 118:351–356. https://doi.org/10.3157/0013-872X(2007)118[351:APOGAS]2.0.CO;2
Mikery-Pacheco O, Marina-Fernández C, Ibáñez-Bernal S et al (2012) Infección natural de Lutzomyia cruciata (Diptera: Psychodidae, Phlebotominae) con Wolbachia en cafetales de Chiapas, México. Acta Zool Mex 28:401–413
Monteiro C, Villegas L, Campolina T et al (2016) Bacterial diversity of the American sand fly Lutzomyia intermedia using high-throughput metagenomic sequencing. Parasit Vectors 31:480. https://doi.org/10.1186/s13071-016-1767-z
Norman F, Regnery R, Jameson P et al (1995) Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol 33:1797–1803
Ono M, Braig HR, Munstermann LE et al (2001) Wolbachia infections of phlebotomine sand flies (Diptera: Psychodidae). J Med Entomol 38:237–241. https://doi.org/10.1603/0022-2585-38.2.237
Pech-May A, Escobedo-Ortegón F, Berzunza-Cruz M, Rebollar-Téllez E (2010) Incrimination of four sandfly species previously unrecognized as vectors of Leishmania parasites in Mexico. Med Vet Entomol 24:150–161. https://doi.org/10.1111/j.1365-2915.2010.00870.x
Pech-May A, Marina C, Vázquez-Domínguez E et al (2013) Genetic structure and divergence in populations of Lutzomyia cruciata, a phlebotomine sand fly (Diptera: Psychodidae) vector of Leishmania mexicana in southeastern Mexico. Infect Genet Evol 16:254–262. https://doi.org/10.1016/j.meegid.2013.02.004
Pimentel AC, Cesar CS, Martins M, Cogni R (2020) The antiviral effects of the symbiont bacteria Wolbachia in insects. Front Immunol 11:626329. https://doi.org/10.3389/fimmu.2020.626329
Rubio AV, Ávila-Flores R, Osikowicz LM et al (2014) Prevalence and genetic diversity of Bartonella strains in rodents from Northwestern Mexico. Vector-Borne Zoonotic Dis 14:838–845. https://doi.org/10.1089/vbz.2014.1673
Sallum MAM, Conn JE, Bergo ES et al (2019) Vector competence, vectorial capacity of Nyssorhynchus darlingi and the basic reproduction number of Plasmodium vivax in agricultural settlements in the Amazonian Region of Brazil. Malar J 18:117. https://doi.org/10.1186/s12936-019-2753-7
Schuler H, Egan SP, Hood GR et al (2018) Diversity and distribution of Wolbachia in relation to geography, host plant affiliation and life cycle of a heterogonic gall wasp. BMC Evol Biol 18:37. https://doi.org/10.1186/s12862-018-1151-z
Sintupachee S, Milne JR, Poonchaisri S et al (2006) Closely related Wolbachia strains within the pumpkin arthropod community and the potential for horizontal transmission via the plant. Microb Ecol 51:294–301. https://doi.org/10.1007/s00248-006-9036-x
Vavre F, Fleury F, Lepetit D et al (1999) Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol Biol Evol 16:1711–1723. https://doi.org/10.1093/oxfordjournals.molbev.a026084
Vivero R, Cadavid-Restrepo G, Moreno-Herrera CX, Uribe-Soto SI (2017) Molecular detection and identification of Wolbachia in three species of the genus Lutzomyia on the Colombian Caribbean coast. Parasit Vectors 10:1–9. https://doi.org/10.1186/s13071-017-2031-x
Vivero RJ, Villegas-Plazas M, Cadavid-Restrepo GE et al (2019) Wild specimens of sand fly phlebotomine Lutzomyia evansi, vector of leishmaniasis, show high abundance of Methylobacterium and natural carriage of Wolbachia and Cardinium types in the midgut microbiome. Sci Rep 9:17746. https://doi.org/10.1038/s41598-019-53769-z
Werren J (1997) Biology of Wolbachia. Annu Rev Entomol 42:587–609. https://doi.org/10.1146/annurev.ento.42.1.587
Zhou W, Rousset F, O’Neil S (1998) Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci 265:509–515. https://doi.org/10.1098/rspb.1998.0324
Acknowledgements
We thank M. Sc. Vicente Viveros-Santos for his support in designing the map, and Ph.D. student Uriel Garduño Montes de Oca for his advice in registering the sequences in GenBank. We are grateful to M. Sc. Laura Marquez Valderrama and Nelly López Ortiz for their support in sequencing PCR products in the Instituto de Biologia, UNAM. We are indebted to José Muñoz-Reyez (deceased), Magne Rúbito Roblero-Díaz, Jesús Aníbal Velazco-Durán, Nohemí Cigarroa-Toledo, and Angélica Pech-May for their technical assistance in sampling and species identification (CRISP-INSP).
Funding
Yokomi N. Lozano Sardaneta is currently a postdoctoral researcher funded by the project CONACyT: 6682. This work was supported by CONACyT: 6682, PAPIIT: IG201221, and CRISP-CONACYT-FOSSIS-69530.
Author information
Authors and Affiliations
Contributions
Yokomi N. Lozano-Sardaneta: conceptualization, methodology, formal analyses, visualization, writing (reviewing and editing), and project administration. Carlos F. Marina: methodology, review, and editing. Jorge A. Torres-Monzón: methodology, review, and editing. Víctor Sánchez-Cordero: methodology, review, and editing. Ingeborg Becker: visualization, resources, review and editing, and project administration. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Van Lun Low
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lozano-Sardaneta, Y.N., Marina, C.F., Torres-Monzón, J.A. et al. Molecular detection of Wolbachia and Bartonella as part of the microbiome of phlebotomine sand flies from Chiapas, Mexico. Parasitol Res 122, 1293–1301 (2023). https://doi.org/10.1007/s00436-023-07829-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-023-07829-z