Abstract
Canine vector-borne diseases (CVBDs) are highly prevalent in tropical and subtropical countries, mainly due to favorable climate conditions and reduced adoption of preventive measures. This study aimed to provide a comprehensive overview on the prevalence of CVBDs in Iran and Pakistan where limited data are available. Blood samples were collected from 403 dogs from six provinces in Iran and Pakistan to assess the presence of pathogen DNA (i.e., Anaplasma spp., Coxiella burnetii, Ehrlichia spp., Rickettsia spp., Babesia spp., Hepatozoon spp., filarioids, and Leishmania spp.). Sera were also screened by an immunofluorescence antibody test for the detection of antibodies against Leishmania infantum. In total, 46.9% of dogs scored positive to Hepatozoon canis being the most frequently detected (41.4%), followed by Anaplasma platys (6.4%), Ehrlichia canis (3.4%), Rickettsia spp. (2.2%), Babesia vogeli (1.0%), and L. infantum (0.3%). A seroprevalence of 9.6% to anti-L. infantum IgG was also recorded. Data reported herein demonstrate that dogs from Iran and Pakistan are at a high risk of CVBDs, particularly of canine hepatozoonosis. Effective control strategies are advocated for minimizing the risk of infection in animals and humans, also in consideration of the zoonotic potential of some pathogens detected.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vector-borne diseases (VBDs) are of growing concern to people across the world and their increasing incidence has been attributed to several factors, such as climate change and animal movements (Ogden and Lindsay 2016). Their distribution depends on a complex combination of biotic and abiotic factors, making their control extremely difficult (Dantas-Torres et al. 2012; Otranto et al. 2017). The impact of VBDs is heavier on tropical and subtropical countries, where the climate is more suitable for various arthropod vectors and where people and animals have limited access to healthcare services, including diagnosis and treatment of these diseases (Otranto et al. 2017; Colella et al. 2019).
Dogs serve as blood source to arthropod vectors and are suitable reservoirs of many vector-borne pathogens (VBPs; Otranto 2018). For example, dogs are well recognized as the primary reservoirs of Leishmania infantum, the causative agent of zoonotic visceral leishmaniasis and the target of integrated control strategies (Travi et al. 2018; Dantas-Torres et al. 2019). Also, dogs are primary reservoirs of some mosquitoes-transmitted filarial helminths (e.g., Dirofilaria immitis and Dirofilaria repens), which may pose a risk to humans in areas where dogs are highly infected, including municipal shelters (Panarese et al. 2020). In the latter case, dog relocation projects may also contribute to increase pathogen circulation in previously non-endemic regions, and eventually their transmission, when a proper vector is present (Otranto et al. 2017; Mendoza-Roldan et al. 2020).
Dogs also contribute to the circulation of certain tick species, such as Rhipicephalus sanguineus sensu lato (s.l.), as well as other more generalist tick species (e.g., Ixodes ricinus) which are well recognized as vectors of pathogens to animals and humans (Otranto et al. 2009a).
Therefore, canine vector-borne diseases (CVBDs) caused by a wide range of pathogens, including viruses, bacteria, protozoa, and helminths, are of veterinary importance and may represent a public health issue (Otranto et al. 2009a, 2017). Data on the occurrence of CVBDs are reported in a few countries of the Middle East such as Iraq (Otranto et al. 2019), Turkey (Kirkan et al. 2013), and Qatar (Alho et al. 2017), making their impact on animal and human populations difficult to estimate, which ultimately impairs the implementation of preventive strategies for minimizing the risk of infections. For example, the occurrence of VBPs has been documented only in a few regions and on limited number of dogs in Iran and Pakistan (Khazeni et al. 2013; Khedri et al. 2014; Motaghipisheh et al. 2016; Ahmad et al. 2018a; Mohebali et al. 2018; Barati and Razmi 2018; Azari-Hamidian et al. 2019). In this perspective, the aim of this study was to investigate the prevalence of VBDs in sheltered and owned dogs from five Iranian provinces and from Punjab in Pakistan in order to fill gaps in the knowledge of VBPs in these areas.
Materials and methods
Sample collection
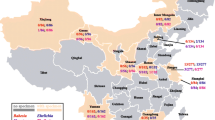
From October 2018 to November 2019, blood samples (1.5–5 ml) were collected from cephalic or saphenous vein of 403 dogs (i.e., 357 sheltered and 46 privately owned dogs) from 5 cities in 5 provinces of Iran including Amol in the North (n = 75), Hamedan (n = 81) and Kermanshah (n = 51) in the west, Yazd in the center (n = 78), Ahvaz in the south-west (n = 69), and from Bahawalpur in the eastern province of Punjab, Pakistan (n = 49) (Fig. 1). Animal data (i.e., age, sex, breed, neutering status, and province) were recorded. All dogs stayed outside overnight. Dogs were grouped according to age in ≤1 year (G1), >1 to <5 years (G2), and ≥ 5 years (G3). The blood of dogs was collected with permission of the Ethical Committee of Hamedan University of Medical Sciences, Iran (code: IR.UMSHA.REC.1398.124).
Serological testing
Serum samples from 354 out of 403 dogs were tested for anti-L. infantum antibodies by using an immunofluorescence antibody test (IFAT), as described elsewhere (Otranto et al. 2009c). Promastigotes of L. infantum zymodeme MON-1 were used as antigen. Samples were scored as positive when they produced a clear cytoplasmic and membrane fluorescence of promastigotes at a cut-off dilution of 1:80. Positive sera were titrated by serial dilutions until negative results were obtained.
DNA extraction, PCR protocols, and sequencing
Genomic DNA was extracted from 200 μl aliquots of EDTA-treated blood samples using GenUP DNA Kit (Biotechrabbit, Berlin, Germany) following the manufacturer’s instructions. All samples were tested for the presence of Anaplasma spp., Ehrlichia spp., Rickettsia spp., Babesia spp., Hepatozoon spp., and filarioid DNA by using conventional PCR (cPCR). In particular, for the detection of Rickettsia spp., DNA samples were firstly tested by PCR targeting a partial sequence of the gene citrate synthase (gltA) and positive samples were further tested by a second PCR targeting a fragment of the outer membrane protein A (ompA) gene. All primers and cPCR protocols used for the detection of VBPs are summarized in Table 1. Leishmania spp. and Coxiella burnetii were detected by using real-time PCR (qPCR) and positive samples were further tested by cPCR. For all reactions, DNA of pathogen-positive blood samples served as a positive control. Amplified PCR products were visualized in 2% agarose gel stained with GelRed (VWR International PBI, Milan, Italy) by GelLogic 100 gel documentation system (Kodak, NY, USA). The cPCR amplicons were sequenced using the Big Dye Terminator v.3.1 chemistry in a 3130 Genetic Analyzer (Applied Biosystems, CA, USA). Nucleotide sequences were edited, aligned, and analyzed using the Geneious platform version 7.0 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. 2012) and compared with those available in the GenBank® database using the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic analysis
For phylogenetic analyses, the 18S rRNA and gltA representative sequence types (STs) of Hepatozoon spp. and Rickettsia spp., respectively, and the corresponding sequences available from the GenBank® database were included. Phylogenetic relationships were inferred using the maximum likelihood (ML) method based on Hasegawa-Kishino-Yano model (Hasegawa et al. 1985) for Hepatozoon canis analysis and Tamura 3-parameter model (Tamura 1992) for Rickettsia spp. Gamma distribution (+G) was used to model evolutionary rate differences among sites selected by best-fit model (Nei and Kumar 2000). Evolutionary analyses were conducted on 8000 bootstrap replications using the MEGA X software (Kumar et al. 2018). Homologous sequences from Adelina bambarooniae (accession nos. AF494058) as well as Rickettsia typhi and Rickettsia prowazekii (accession nos. U59714 and U59715) were used as outgroups for Hepatozoon spp. and Rickettsia spp. phylogenetic trees, respectively.
Statistical analysis
Exact binomial 95% confidence intervals (CIs) were established for proportions. The chi-square and Fisher tests were used to compare proportions with a probability p value < 0.05 regarded as statistically significant. Analyses were done using the GraphPad Prism version 8.0.0 (GraphPad Software, San Diego, CA, USA).
Results
Out of 403 dogs tested, 189 (46.9%; 95% CI 42.1–51.9) scored positive to at least one pathogen. Data on sex, age, and location of animals are reported in Table 2 along with number and percentage of dogs positive to H. canis, the most frequently detected pathogen species (41.4%; 95% CI 36.7–46.3) and seropositive to antibodies against L. infantum (9.6%; 95% CI 6.9–13.1). The prevalence of infection by other pathogens including Anaplasma platys, Ehrlichia canis, spotted-fever group (SFG) Rickettsia spp., Babesia spp., and L. infantum is reported in Table 3. No DNA of filarioids or C. burnetii was amplified. Co-infections with H. canis and A. platys were the most frequent (4.9%, n = 20), followed by H. canis and E. canis (1.4%, n = 3); H. canis and Rickettsia spp. (1.4%, n = 3); H. canis and L. infantum (0.3%, n = 1); and H. canis, E. canis, and Rickettsia spp. (0.3%, n = 1). The ompA gene was not amplified in any of the samples positive to Rickettsia gltA gene. Out of 354 serum samples, 34 (9.6%) were positive to anti-L. infantum antibodies with titers of 1:80 (n = 14), 1:160 (n = 15), 1:320 (n = 3), 1:640 (n = 1), and 1:1280 (n = 1).
The risk of H. canis infection in dogs was significantly associated with their keeping conditions and the geographical areas, whereas the risk of L. infantum exposure only with the geographical areas. In particular, sheltered dogs were related to higher risk of H. canis infection than owned dogs (χ2 = 29.4, df = 1, p < 0.00001). Based on location, the highest prevalence of H. canis infection (i.e., 78.4%) was recorded in Kermanshah in Iran followed by Bahawalpur in Pakistan (63.3%) and both percentages were significantly higher than those recorded in other cities (p < 0.05). Dogs from Amol in Iran were more exposed to L. infantum than those in the other Iranian provinces (p < 0.05) (Table 2). No statistical association was found between H. canis infection or L. infantum exposure in dogs and their sex and age. The region with the highest pathogen diversity was Ahvaz in Iran where a high prevalence of Rickettsia spp. infection was recorded (13%) (Table 3).
Consensus sequences for each detected pathogen displayed 99–100% nucleotide identity with those available in the GenBank® database. In particular, Babesia vogeli (n = 4) nucleotide similarity was 99.3–99.6% with MT499357. In addition, five STs were identified for H. canis (ST1, n = 6, 100% identity with MN393911; ST2, n = 10, 99.7% with MK673827; ST3, n = 24, 100% with KX880505; ST4, n = 39, 100% with MT499354; ST5, n = 88, 100% with MK673850) and L. infantum 100% identity was recorded with MK496879. Anaplasma platys sequences (n = 26) and E. canis (n = 14) had 100% nucleotide identity with MN922611 and MN922610, respectively. As far as Rickettsia spp., four samples had 100% nucleotide identity with Rickettsia monacensis (KC993860) and two had 99.7% identity with Rickettsia helvetica (U59723). One sample from a dog in Pakistan was 100% identical to Rickettsia conorii (U59730)/Rickettsia honei (U59726)/Rickettsia raoultii (CP019435), and two other samples from dogs in Ahvaz (Iran) were 100% identical to Rickettsia heilongjiangensis (CP002912)/Rickettsia raoultii (DQ365803)/Rickettsia slovaca (U59725).
Molecular identification of nucleotide sequences for Rickettsia spp. and H. canis was supported by the distinct separation of species-specific clades inferred from the phylogenetic analyses. The ML tree of H. canis grouped all representative STs in a large clade supported by high bootstrap value (i.e., 98%), to the exclusion of other species of Hepatozoon (Fig. 2). The ML tree based on the partial gltA gene sequences of Rickettsia spp. showed that all sequences, herein detected, clustered in well-supported clades with other SFG rickettsiae (Fig. 3). Representative sequences of pathogens detected in this study were deposited in the GenBank® database under the accession numbers MW019628 and MW019629 for B. vogeli, MW019630-MW019643 for H. canis, MW019670 and MW019671 for A. platys, MW019672 and MW019673 for E. canis, MW039480 for R. helvetica, MW039481 for R. monacensis, MW039482 and MW039483 for Rickettsia spp., and MW074300 for L. infantum.
Phylogenetic relationship of Hepatozoon spp. sequences isolated in this study (in bold) to other Hepatozoon spp. based on a partial sequence (327 bp) of the 18S rRNA gene. The analyses were performed using a maximum likelihood with Hasegawa-Kishino-Yano model. A gamma distribution was used to model evolutionary rate differences among sites. Homologous sequence from Adelina bambarooniae (accession nos. AF494058) was used as the outgroup
Phylogenetic relationship of Rickettsia spp. sequences isolated in this study (in bold) to other Rickettsia strains based on a partial sequence (345 bp) of the gltA gene. The analyses were performed using a maximum likelihood method with Tamura 3-parameter model. A gamma distribution was used to model evolutionary rate differences among sites. Homologous sequences from Rickettsia typhi (accession nos. U59714) and Rickettsia prowazekii (accession nos. U59715) were used as the outgroups
Discussion
The high frequency of CVBD-causing pathogens reported herein indicates that dog populations from Iran and Pakistan are exposed to multiple pathogens, including some of zoonotic importance, thus posing a risk not only to dogs, but also to public health. Many of the herein detected pathogens (i.e., A. platys, E. canis, B. vogeli, and H. canis) are transmitted by Rh. sanguineus s.l. (Latrofa et al. 2014), the most common tick species found on dogs in Iran and Pakistan (Mosallanejad et al. 2012; Cabezas-Cruz et al. 2019). The high prevalence of H. canis in all the provinces of Iran as well as from Bahawalpur in Pakistan suggests that Rh. sanguineus s.l. is highly prevalent in these areas. After the first detection of H. canis gametocytes in a blood smear of an Iranian dog (Khoshnegah et al. 2009), the prevalence of this infection was reported only in dogs from northern half of Iran ranging from 1.6 to 23% (Amoli et al. 2012; Dalimi et al. 2017; Soltani and Dalimi 2018; Barati and Razmi 2018). Conversely, the prevalence herein recorded for H. canis in Pakistan (i.e., 41.4%) was similar to that of farm dogs recorded in Punjab districts (45.5%, Ahmad et al. 2018a). Scientific data indicates that under certain circumstances, such as in poor socioeconomic settings, the high environmental infestation of arthropod vectors and the absence of preventive strategies in dogs and in the environment concur to increase the incidence of CVBD (Otranto et al. 2017). Indeed, similar high prevalence of H. canis infections has been reported in dogs (37.8%) and in Rh. sanguineus s.l. ticks (42.5%) from India (Manoj et al. 2020) as well as in dogs from Iraq (33%) (Otranto et al. 2019). Although Rh. sanguineus s.l., Rhipicephalus bursa and Rhipicephalus annulatus are the common species infesting dogs in the studied area (Jamshidi et al. 2012; Akhtardanesh et al. 2016; Ali et al. 2019; Cabezas-Cruz et al. 2019), the circulation of other competent tick vectors cannot be ruled out (Murata et al. 1995; Rubini et al. 2009; Najm et al. 2014; Giannelli et al. 2017). In addition, the significantly higher occurrence of dogs infected by H. canis in Kermanshah in Iran (78.4%) and Bahawalpur in Pakistan (63.3%) could be related to the high population density of tick vectors in these shelters observed during the dog sampling. The risk for sheltered dogs to be more infected by H. canis than owned dogs suggests that in this environment, the animals are more exposed to the vectors. Indeed, preventive measures such as ectoparasite repellents are not commonly employed in sheltered dogs in Iran and Pakistan because of the limitation in budget of non-governmental organization which mainly aim to feed and protect dogs from culling program (personal observations of AS and MMA). Conversely, B. vogeli was found in only few dog samples (i.e., 1%) as recorded in other studies from Iran (Alborzi et al. 2013; Akhtardanesh et al. 2016; Bigdeli and Namavari 2017) and Pakistan (Ahmad et al. 2018b). Also, Babesia gibsoni was reported causing canine babesiosis in these two countries (Akhtardanesh et al. 2016; Ahmad et al. 2018b).
In Iran, the first study on leishmaniosis in carnivores was conducted in 1982 with a seroprevalence of 2.4% in jackals and 3% in dogs from the northern part (Hamidi et al. 1982). Afterwards, L. infantum infection was reported in domestic and wild animals in most of the regions of Iran with differences in frequency probably related to geographic regions, environmental conditions, and dog population (Shokri et al. 2017; Mohebali et al. 2018). The seropositivity to L. infantum in dogs from Iran herein detected (9.6%) confirms the circulation of infected sand fly vector (Yaghoobi-Ershadi 2016), thus the risk of dogs as well as humans to be exposed to their bites and to be infected. Differences in climatic factors may affect the sand fly population (Cheghabalaki et al. 2019) in terms of species composition and abundance, thus leading to a higher risk of infection in dogs living in some provinces. Indeed, the highest seroprevalence (25.3%) was recorded in dogs from Amol, located in the north of Iran with a Mediterranean climate vs no infected dog in Yazd and Kermanshah which are regions characterized by hot and dry climate. The detection of L. infantum DNA only in one dog may also be related to the use of the blood, which is a convenient type of sample but poor as a parasite source (Maia et al. 2009; Otranto et al. 2009c).
Canine monocytic ehrlichiosis by E. canis and canine cyclic thrombocytopenia by A. platys affect dogs worldwide, varying from asymptomatic infections or with mild symptoms to a severe illness (Otranto et al. 2009b). Although the prevalence of these infections is scantly reported in the examined areas, the finding of E. canis in dogs from Iran and Pakistan provinces is not surprising since this bacterium is transmitted by Rh. sanguineus s.l. A few seroprevalence studies (Akhtardanesh et al. 2010) as well as molecular detection of E. canis (Motaghipisheh et al. 2016; Malik et al. 2018) in blood samples confirm that dogs from the examined areas are exposed to the pathogen. Conversely, the first report of A. platys in both countries with higher prevalence than E. canis (6.4% vs 3.4%) indicates that this pathogen may affect dogs living mainly in rural area and in shelters. While the unsuccessful amplification of the ompA gene may represent a limitation of the study, some sequences obtained from dogs from Iran had high (> 99%) sequence identity with R. helvetica and R. monacensis. Although the pathogenicity of R. helvetica and R. monacensis in dogs is unknown, both species may cause severe diseases in humans as reported in Europe and Southeast Asia for R. helvetica (Nilsson et al. 1999; Fournier et al. 2004; Nilsson et al. 2010) and in Spain and Italy for R. monacensis (Jado et al. 2007; Madeddu et al. 2012). Data on SFG rickettsiae in dogs and their ectoparasites in the Middle East are scant (Baneth et al. 1998; Harrus et al. 2011; Levin et al. 2011; Kirkan et al. 2013; Parola et al. 2013; Orkun et al. 2014). Our findings suggest the occurrence of SFG rickettsiae in dogs from Iran and Pakistan, but further studies are needed to confirm the identity of these microorganisms and also to understand the potential risks for public health.
In this study, none of the tested dogs scored positive to C. burnetii although this bacteria was serologically diagnosed in a domestic dog in Ahvaz, Iran (Rezaei et al. 2016). Indeed, Q fever is endemic in Iran and Pakistan with high prevalence among human patients and domestic ruminants (Ullah et al. 2019; Esmaeili et al. 2019; Mohabbati Mobarez et al. 2017). In fact, it is acknowledged that ruminants (cattle, sheep, and goats) are the most important reservoirs of C. burnetii for human infection (Angelakis and Raoult 2010).
In conclusion, the herein reported data provide more knowledge of CVBDs in these countries suggesting that different pathogens as well as arthropod vectors circulate among animal populations. Of the detected pathogens, SFG rickettsiae are of great relevance for their pathogenicity to humans. Considering the occurrence of infection by zoonotic pathogens in dogs and their close relationship with humans, effective control strategies are advocated for minimizing the risk of infection in animals as well as in humans.
References
Ahmad AS, Saeed MA, Rashid I et al (2018a) Molecular characterization of Hepatozoon canis from farm dogs in Pakistan. Parasitol Res 117:1131–1138. https://doi.org/10.1007/s00436-018-5790-1
Ahmad AS, Rashid I, Ashraf K et al (2018b) Molecular occurrence of canine babesiosis in rural dog population in Pakistan. Trop Biomed 35:593–603
Akhtardanesh B, Ghanbarpour R, Blourizadeh H (2010) Serological evidence of canine monocytic ehrlichiosis in Iran. Comp Clin Pathol 19:469–474. https://doi.org/10.1007/s00580-009-0889-5
Akhtardanesh B, Saberi M, Nurollahifard SR, Aghazamani M (2016) Molecular detection of Babesia spp. in tick-infested dogs in southeastern Iran. J Dis Glob Heal 8:72–77
Alborzi AR, Avizeh R, Mosallanejad B et al (2013) Babesia infection in urban and rural dogs in Ahvaz district, Southwest of Iran. Arch Razi Inst 68:37–42. https://doi.org/10.7508/ari.2013.01.006
Alho AM, Lima C, Latrofa MS et al (2017) Molecular detection of vector-borne pathogens in dogs and cats from Qatar. Parasit Vectors 10:1–5. https://doi.org/10.1186/s13071-017-2237-y
Ali A, Khan MA, Zahid H et al (2019) Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front Physiol 10:1–15. https://doi.org/10.3389/fphys.2019.00793
Amoli AR, Khoshnegah J, Razmi G (2012) A preliminary parasitological survey of Hepatozoon spp. infection in dogs in Mashhad, Iran. Iran J Parasitol 7:99–103
Angelakis E, Raoult D (2010) Q fever. Vet Microbiol 140:297–309. https://doi.org/10.1016/j.vetmic.2009.07.016
Azari-Hamidian S, Norouzi B, Harbach RE (2019) A detailed review of the mosquitoes (Diptera: Culicidae) of Iran and their medical and veterinary importance. Acta Trop 194:106–122. https://doi.org/10.1016/j.actatropica.2019.03.019
Baneth G, Breitschwerdt EB, Hegarty BC et al (1998) A survey of tick-borne bacteria and protozoa in naturally exposed dogs from Israel. Vet Parasitol 74:133–142. https://doi.org/10.1016/S0304-4017(97)00149-0
Barati A, Razmi GR (2018) A parasitologic and molecular survey of Hepatozoon canis infection in stray dogs in Northeastern Iran. J Parasitol 104:413–417. https://doi.org/10.1645/17-105
Bigdeli M, Namavari M (2017) Evaluation of PCR assay using specific primers in diagnosis of canine ehrlichiosis and babesiosis: a study on herd and stray dogs in Shiraz. J Altern Vet Med 1:1–15
Cabezas-Cruz A, Allain E, Ahmad AS et al (2019) Low genetic diversity of Ehrlichia canis associated with high co-infection rates in Rhipicephalus sanguineus (s.l.). Parasit Vectors 12:1–13. https://doi.org/10.1186/s13071-018-3194-9
Casiraghi M, Anderson TJC, Bandi C et al (2001) A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology 122:93–103. https://doi.org/10.1017/S0031182000007149
Cheghabalaki ZZ, Yarahmadi D, Karampour M, Shamsipour A (2019) Spatial dynamics of a phlebotomine sand flies population in response to climatic conditions in Bushehr Province of Iran. Ann Glob Heal 85:1–11. https://doi.org/10.5334/aogh.30
Colella V, Hodžić A, Iatta R et al (2019) Zoonotic Leishmaniasis, Bosnia and Herzegovina. Emerg Infect Dis 25:385–386. https://doi.org/10.3201/eid2502.181481
Dalimi A, Jameie F, Mohammadiha A et al (2017) Molecular detection of Hepatozoon canis in dogs of Ardabil Province, Northwest of Iran. Arch Razi Inst 72:197–201. https://doi.org/10.22034/ari.2017.108389
Dantas-Torres F, Chomel BB, Otranto D (2012) Ticks and tick-borne diseases: a one health perspective. Trends Parasitol 28:437–446. https://doi.org/10.1016/j.pt.2012.07.003
Dantas-Torres F, Miró G, Baneth G et al (2019) Canine leishmaniasis control in the context of one health. Emerg Infect Dis 25:E1–E4. https://doi.org/10.3201/eid2512.190164
de Almeida M, Steurer F, Koru O et al (2011) Identification of Leishmania spp. by molecular amplification and DNA sequencing analysis of a fragment of rRNA internal transcribed spacer 2. J Clin Microbiol 49:3143–3149. https://doi.org/10.1128/JCM.01177-11
Esmaeili S, Bagheri Amiri F, Mokhayeri H et al (2019) Seroepidemiological study of Q fever, brucellosis and tularemia in butchers and slaughterhouses workers in Lorestan, western of Iran. Comp Immunol Microbiol Infect Dis 66:101322. https://doi.org/10.1016/j.cimid.2019.06.003
Fournier P-E, Allombert C, Supputamongkol Y et al (2004) Aneruptive fever associated with antibodies to Rickettsia helvetica in Europe and Thailand. J Clin Microbiol 42:816–818. https://doi.org/10.1128/jcm.42.2.816-818.2004
Francino O, Altet L, Sánchez-Robert E et al (2006) Advantages of real-time PCR assay for diagnosis and monitoring of canine leishmaniosis. Vet Parasitol 137:214–221. https://doi.org/10.1016/j.vetpar.2006.01.011
Giannelli A, Lia RP, Annoscia G et al (2017) Rhipicephalus turanicus, a new vector of Hepatozoon canis. Parasitology 144:730–737. https://doi.org/10.1017/S003118201600250X
Hamidi AN, Nadim A, Edrissian GH et al (1982) Visceral leishmaniasis of jackals and dogs in northern Iran. Trans R Soc Trop Med Hyg 76:756–757. https://doi.org/10.1016/0035-9203(82)90100-6
Harrus S, Perlman-Avrahami A, Mumcuoglu KY et al (2011) Molecular detection of Rickettsia massiliae, Rickettsia sibirica mongolitimonae and Rickettsia conorii israelensis in ticks from Israel. Clin Microbiol Infect 17:176–180. https://doi.org/10.1111/j.1469-0691.2010.03224.x
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. https://doi.org/10.1007/BF02101694
Jado I, Oteo JA, Aldámiz M et al (2007) Rickettsia monacensis and human disease, Spain. Emerg Infect Dis 13:1405–1407. https://doi.org/10.3201/eid1309.060186
Jamshidi S, Maazi N, Ranjbar-Bahadori S et al (2012) A survey of ectoparasite infestation in dogs in Tehran, Iran. Rev Bras Parasitol Vet 21:326–329. https://doi.org/10.1590/s1984-29612012000300030
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Khazeni A, Telmadarraiy Z, Oshaghi MA et al (2013) Molecular detection of Ehrlichia canis in ticks population collected on dogs in Meshkin-Shahr, Ardebil Province, Iran. J Biomed Sci Eng 6:1–5
Khedri J, Radfar MH, Borji H et al (2014) Canine heartworm in Southeastern of Iran with review of disease distribution. Iran J Parasitol 9:560–567
Khoshnegah J, Mohri M, Movassaghi AR, Mehrjerdi HK (2009) The first report of Hepatozoon canis infection of a dog in Iran. Comp Clin Pathol 18:455–458. https://doi.org/10.1007/s00580-008-0794-3
Kirkan Ş, Savaşan S, Erbaş G, Parin U (2013) Prevalence of Rickettsia rickettsii infection in dogs from the urban and rural areas of western Turkey. Ankara Üniv Vet Fak Derg 60:165–169
Klee SR, Tyczka J, Ellerbrok H et al (2006) Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol 6:2. https://doi.org/10.1186/1471-2180-6-2
Kumar S, Stecher G, Li M et al (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Labruna MB, Whitworth T, Horta MC et al (2004) Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of São Paulo, Brazil, where Brazilian spotted fever is endemic. J Clin Microbiol 42:90–98. https://doi.org/10.1128/jcm.42.1.90-98.2004
Latrofa MS, Dantas-Torres F, Giannelli A, Otranto D (2014) Molecular detection of tick-borne pathogens in Rhipicephalus sanguineus group ticks. Ticks Tick Borne Dis 5:943–946. https://doi.org/10.1016/j.ttbdis.2014.07.014
Levin ML, Killmaster LF, Zemtsova GE (2011) Domestic dogs (Canis familiaris) as reservoir hosts for Rickettsia conorii. Vector-Borne Zoonotic Dis 12:28–33. https://doi.org/10.1089/vbz.2011.0684
Madeddu G, Mancini F, Caddeo A et al (2012) Rickettsia monacensis as cause of Mediterranean spotted fever-like illness, Italy. Emerg Infect Dis 18:702–704. https://doi.org/10.3201/eid1804.111583
Maia C, Ramada J, Cristóvão JM et al (2009) Diagnosis of canine leishmaniasis: conventional and molecular techniques using different tissues. Vet J 179:142–144. https://doi.org/10.1016/j.tvjl.2007.08.009
Malik MI, Qamar M, Ain Q et al (2018) Molecular detection of Ehrlichia canis in dogs from three districts in Punjab (Pakistan). Vet Med Sci 4:126–132. https://doi.org/10.1002/vms3.94
Manoj RRS, Iatta R, Latrofa MS et al (2020) Canine vector-borne pathogens from dogs and ticks from Tamil Nadu, India. Acta Trop 203:105308. https://doi.org/10.1016/j.actatropica.2019.105308
Martin AR, Brown GK, Dunstan RH, Roberts TK (2005) Anaplasma platys: an improved PCR for its detection in dogs. Exp Parasitol 109:176–180. https://doi.org/10.1016/j.exppara.2004.11.007
Mendoza-Roldan J, Benelli G, Panarese R et al (2020) Leishmania infantum and Dirofilaria immitis infections in Italy, 2009 – 2019: changing distribution patterns. Parasit Vectors 13:193. https://doi.org/10.1186/s13071-020-04063-9
Mohabbati Mobarez A, Bagheri Amiri F, Esmaeili S (2017) Seroprevalence of Q fever among human and animal in Iran: a systematic review and meta-analysis. PLoS Negl Trop Dis 11:e0005521. https://doi.org/10.1371/journal.pntd.0005521
Mohebali M, Moradi-Asl E, Rassi Y (2018) Geographic distribution and spatial analysis of Leishmania infantum infection in domestic and wild animal reservoir hosts of zoonotic visceral leishmaniasis in Iran: a systematic review. J Vector Borne Dis 55:184–188. https://doi.org/10.4103/0972-9062.249125
Mosallanejad B, Alborzi A, Katvandi N (2012) A survey on ectoparasite infestations in companion dogs of Ahvaz district, south-west of Iran. J Arthropod Borne Dis 6:70–78
Motaghipisheh S, Akhtardanesh B, Ghanbarpour R et al (2016) Ehrlichiosis in household dogs and parasitized ticks in Kerman-Iran: preliminary zoonotic risk assessment. J Arthropod Borne Dis 10:246–252
Murata T, Inoue M, Taura Y et al (1995) Detection of Hepatozoon canis oocyst from ticks collected from the infected dogs. J Vet Med Sci 57:111–112
Najm N-A, Meyer-Kayser E, Hoffmann L et al (2014) Hepatozoon canis in German red foxes (Vulpes vulpes) and their ticks: molecular characterization and the phylogenetic relationship to other Hepatozoon spp. Parasitol Res 113:2679–2685. https://doi.org/10.1007/s00436-014-3923-8
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press
Nilsson K, Lindquist O, Påhlson C (1999) Association of Rickettsia helvetica with chronic perimyocarditis in sudden cardiac death. Lancet 354:1169–1173. https://doi.org/10.1016/S0140-6736(99)04093-3
Nilsson K, Elfving K, Pahlson C (2010) Rickettsia helvetica in patient with meningitis, Sweden, 2006. Emerg Infect Dis 16:490–492. https://doi.org/10.3201/eid1603.090184
Ogden NH, Lindsay LR (2016) Effects of climate and climate change on vectors and vector-borne diseases: ticks are different. Trends Parasitol 32:646–656. https://doi.org/10.1016/j.pt.2016.04.015
Orkun Ö, Karaer Z, Çakmak A, Nalbantoğlu S (2014) Spotted fever group rickettsiae in ticks in Turkey. Ticks Tick Borne Dis 5:213–218. https://doi.org/10.1016/j.ttbdis.2012.11.018
Otranto D (2018) Arthropod-borne pathogens of dogs and cats: from pathways and times of transmission to disease control. Vet Parasitol 251:68–77. https://doi.org/10.1016/j.vetpar.2017.12.021
Otranto D, Dantas-Torres F, Breitschwerdt EB (2009a) Managing canine vector-borne diseases of zoonotic concern: part one. Trends Parasitol 25:157–163. https://doi.org/10.1016/j.pt.2009.01.003
Otranto D, Dantas-Torres F, Breitschwerdt EB (2009b) Managing canine vector-borne diseases of zoonotic concern: part two. Trends Parasitol 25:228–235. https://doi.org/10.1016/j.pt.2009.02.005
Otranto D, Paradies P, De Caprariis D et al (2009c) Toward diagnosing Leishmania infantum infection in asymptomatic dogs in an area where leishmaniasis is endemic. Clin Vaccine Immunol 16:337–343. https://doi.org/10.1128/CVI.00268-08
Otranto D, Dantas-Torres F, Mihalca AD et al (2017) Zoonotic parasites of sheltered and stray dogs in the era of the global economic and political crisis. Trends Parasitol 33:813–825. https://doi.org/10.1016/j.pt.2017.05.013
Otranto D, Iatta R, Baneth G et al (2019) High prevalence of vector-borne pathogens in domestic and wild carnivores in Iraq. Acta Trop 197:105058. https://doi.org/10.1016/j.actatropica.2019.105058
Panarese R, Iatta R, Latrofa MS et al (2020) Hyperendemic Dirofilaria immitis infection in a sheltered dog population: an expanding threat in the Mediterranean region. Int J Parasitol 50:555–559. https://doi.org/10.1016/j.ijpara.2020.04.002
Parola P, Paddock CD, Socolovschi C et al (2013) Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev 26:657–702. https://doi.org/10.1128/CMR.00032-13
Regnery RL, Spruill CL, Plikaytis BD (1991) Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol 173:1576–1589. https://doi.org/10.1128/jb.173.5.1576-1589.1991
Rezaei A, Gharibi D, Pourmahdi Borujeni M, Mosallanejad B (2016) Seroprevalence of Lyme disease and Q fever in referred dogs to veterinary hospital of Ahvaz. Iran Vet J 11:34–41. https://doi.org/10.22055/IVJ.2016.13016
Rubini AS, Paduan KS, Martins TF et al (2009) Acquisition and transmission of Hepatozoon canis (Apicomplexa: Hepatozoidae) by the tick Amblyomma ovale (Acari: Ixodidae). Vet Parasitol 164:324–327. https://doi.org/10.1016/j.vetpar.2009.05.009
Shokri A, Fakhar M, Teshnizi SH (2017) Canine visceral leishmaniasis in Iran: a systematic review and meta-analysis. Acta Trop 165:76–89. https://doi.org/10.1016/j.actatropica.2016.08.020
Soltani R, Dalimi A (2018) A molecular study on Hepatozoon canis infection in dogs in Tehran (Iran). Arch Razi Inst 73:257–263. https://doi.org/10.22092/ARI.2017.110293.1125
Tabar M-D, Altet L, Francino O et al (2008) Vector-borne infections in cats: molecular study in Barcelona area (Spain). Vet Parasitol 151:332–336. https://doi.org/10.1016/j.vetpar.2007.10.019
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9:678–687. https://doi.org/10.1093/oxfordjournals.molbev.a040752
Travi BL, Cordeiro-da-Silva A, Dantas-Torres F, Miró G (2018) Canine visceral leishmaniasis: diagnosis and management of the reservoir living among us. PLoS Negl Trop Dis 12:1–13. https://doi.org/10.1371/journal.pntd.0006082
Ullah Q, El-Adawy H, Jamil T et al (2019) Serological and molecular investigation of Coxiella burnetii in small ruminants and ticks in Punjab, Pakistan. Int J Environ Res Public Health 16:4271. https://doi.org/10.3390/ijerph16214271
Yaghoobi-Ershadi MR (2016) Control of phlebotomine sand flies in Iran: a review article. J Arthropod Borne Dis 10:429–444
Acknowledgments
This study was planned under the academic agreement between the Bu-Ali Sina University Hamedan (Iran) and the University of Bari (Italy). We thank all who assisted us in sampling especially Dr. Seyedmasoud Zolhavarieh (Bu-Ali Sina University), Dr. Omid Hosseininejad and Dr. Fatemeh Arabifar (in Ahvaz), Mrs. Shilan Lorestani and Dr. Saman Salmani (in Kermanshah), Mrs. Faranak Rahmanpour (in Amol), and Mr. Seyed-Ali Vaziri (in Yazd).
Funding
Open Access funding provided by Università degli Studi di Bari Aldo Moro. This work was partially supported by the Bu-Ali Sina University, Hamedan, Iran (Grant Number 1/1/28864).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Guest Editor: Christina Strube
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Iatta, R., Sazmand, A., Nguyen, VL. et al. Vector-borne pathogens in dogs of different regions of Iran and Pakistan. Parasitol Res 120, 4219–4228 (2021). https://doi.org/10.1007/s00436-020-06992-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-020-06992-x