Abstract
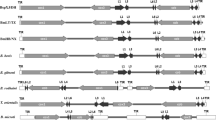
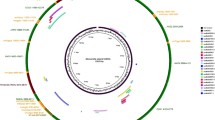
Babesia gibsoni (B. gibsoni), an intracellular apicomplexan protozoan, poses great threat to canine health. Currently, little information is available about the B. gibsoni (WH58) endemic to Wuhan, China. Here, the mitochondrial (mt) genome of B. gibsoni (WH58) was amplified by five pairs of primers and sequenced and annotated by alignment with the reported mt genome sequences of Babesia canis (B. canis, KC207822), Babesia orientalis (KF218819), Babesia bovis (AB499088), and Theileria equi (AB499091). The evolutionary relationships were analyzed with the amino acid sequences of cytochrome c oxidase I (cox1) and cytochrome b (cob) genes in apicomplexan parasite species. Additionally, the mt genomes of Babesia, Theileria, and Plasmodium spp. were compared in size, host infection, form, distribution, and direction of the protein-coding genes. The full size of the mt genome of B. gibsoni (WH58) was 5865 bp with a linear form, containing terminal-inverted repeats on both ends, six large subunit ribosomal RNA fragments, and three protein-coding genes: cox1, cob, and cytochrome c oxidase III (cox3). Babesia, Theileria, and Plasmodium spp. had a similar mt genome size of about 6000 bp. The mt genomes of parasites that cause canine babesiosis showed a slightly smaller size than the other species. Moreover, Babesia microti (R1 strain) was about 11,100 bp in size, which was twice larger than that of the other species. The mt form was linear for Babesia and Theileria spp. but circular for Plasmodium falciparum and Plasmodium knowlesi. Additionally, all the species contained the three protein-coding genes of cox1, cox3, and cob except Toxoplasma gondii (RH strain) which only contained the cox1 and cob genes. The phylogenetic analysis indicated that B. gibsoni (WH58) was more identical to B. gibsoni (AB499087), B. canis (KC207822), and Babesia rossi (KC207823) and most divergent from Babesia conradae in Babesia spp. Despite the highest similarity to B. gibsoni (AB499087) reported in Japan, B. gibsoni (WH58) showed notable differences in the sequence of nucleotides and amino acids and the property in virulence to host and in vitro cultivation. This study compared the mt genomes of the two B. gibsoni isolates and other parasites in the phylum Apicomplexa and provided new insights into their differences and evolutionary relationships.


Similar content being viewed by others
Abbreviations
- B. gibsoni :
-
Babesia gibsoni
- P. falciparum :
-
Plasmodium falciparum
- B. conradae :
-
Babesia conradae
- B. canis :
-
Babesia canis
- B. rossi :
-
Babesia rossi
- B. microti :
-
Babesia microti
- B. rodhaini :
-
Babesia rodhaini
- T. equi :
-
Theileria equi
- Mt.:
-
mitochondrial
- Cob :
-
cytochrome b
- Cox1 :
-
cytochrome c oxidase I
- Cox3 :
-
cytochrome c oxidase III
- LSU:
-
large subunit
- SSU:
-
small subunit
- rRNAs:
-
ribosomal RNAs
- TIRs:
-
terminal inverted repeats
- PPE:
-
parasitized erythrocytes
- gDNA:
-
genomic DNA
- PCR:
-
polymerase chain reaction
- ORF:
-
open reading frame
- tRNA:
-
transfer RNA
References
Burland TG (2000) DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol 132:71–91
Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, Peterson JD, Pop M, Kosack DS, Shumway MF, Bidwell SL, Shallom SJ, van Aken SE, Riedmuller SB, Feldblyum TV, Cho JK, Quackenbush J, Sedegah M, Shoaibi A, Cummings LM, Florens L, Yates JR, Raine JD, Sinden RE, Harris MA, Cunningham DA, Preiser PR, Bergman LW, Vaidya AB, van Lin LH, Janse CJ, Waters AP, Smith HO, White OR, Salzberg SL, Venter JC, Fraser CM, Hoffman SL, Gardner MJ, Carucci DJ (2002) Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512–519. https://doi.org/10.1038/nature01099
Cornillot E, Hadj-Kaddour K, Dassouli A, Noel B, Ranwez V, Vacherie B, Augagneur Y, Brès V, Duclos A, Randazzo S, Carcy B, Debierre-Grockiego F, Delbecq S, Moubri-Ménage K, Shams-Eldin H, Usmani-Brown S, Bringaud F, Wincker P, Vivarès CP, Schwarz RT, Schetters TP, Krause PJ, Gorenflot A, Berry V, Barbe V, Ben Mamoun C (2012) Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti. Nucleic Acids Res 40:9102–9114. https://doi.org/10.1093/nar/gks700
Cornillot E, Dassouli A, Garg A, Pachikara N, Randazzo S, Depoix D, Carcy B, Delbecq S, Frutos R, Silva JC, Sutton R, Krause PJ, Mamoun CB (2013) Whole genome mapping and re-organization of the nuclear and mitochondrial genomes of Babesia microti isolates. PLoS One 8:e72657. https://doi.org/10.1371/journal.pone.0072657
El-Dakhly KM, Goto M, Noishiki K, El-Nahass el S, Sakai H, Yanai T, Takashima Y (2015) Distribution patterns of Babesia gibsoni infection in hunting dogs from nine Japanese islands. J Parasitol 101:160–166. https://doi.org/10.1645/14-564.1
Frederick RL, Shaw JM (2007) Moving mitochondria: establishing distribution of an essential organelle. Traffic 8:1668–1675. https://doi.org/10.1111/j.1600-0854.2007.00644.x
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. https://doi.org/10.1038/nature01097
Gjerde B (2013) Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of Toxoplasma gondii, Neospora caninum, Hammondia heydorni and Hammondia triffittae (Apicomplexa: Sarcocystidae). Parasitol Res 112:1493–1511. https://doi.org/10.1007/s00436-013-3296-4
Goo YK, Xuan X (2014) New molecules in Babesia gibsoni and their application for diagnosis, vaccine development, and drug discovery. Korean J Parasitol 52:345–353. https://doi.org/10.3347/kjp.2014.52.4.345
Hall TA (1999) BioEdit : a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
He L, Miao X, Hu J, Huang Y, He P, He J, Yu L, Malobi N, Shi L, Zhao J (2017) First molecular detection of Babesia gibsoni in dogs from Wuhan, China. Front Microbiol 8:1577. https://doi.org/10.3389/fmicb.2017.01577
Hikosaka K, Watanabe Y, Tsuji N, Kita K, Kishine H, Arisue N, Palacpac NMQ, Kawazu S, Sawai H, Horii T, Igarashi I, Tanabe K (2010) Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol Biol Evol 27:1107–1116. https://doi.org/10.1093/molbev/msp320
Hikosaka K, Watanabe Y, Kobayashi F, Waki S, Kita K, Tanabe K (2011) Highly conserved gene arrangement of the mitochondrial genomes of 23 Plasmodium species. Parasitol Int 60:175–180. https://doi.org/10.1016/j.parint.2011.02.001
Katoh K, Rozewicki J, Yamada KD (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. https://doi.org/10.1093/bib/bbx108
Ke H, Dass S, Morrisey JM, Mather MW, Vaidya AB (2018) The mitochondrial ribosomal protein L13 is critical for the structural and functional integrity of the mitochondrion in Plasmodium falciparum. J Biol Chem 293:8128–8137. https://doi.org/10.1074/jbc.RA118.002552
Lau AO (2009) An overview of the Babesia, Plasmodium and Theileria genomes: a comparative perspective. Mol Biochem Parasitol 164:1–8. https://doi.org/10.1016/j.molbiopara.2008.11.013
Lin RQ, Qiu LL, Liu GH, Wu XY, Weng YB, Xie WQ, Hou J, Pan H, Yuan ZG, Zou FC, Hu M, Zhu XQ (2011) Characterization of the complete mitochondrial genomes of five Eimeria species from domestic chickens. Gene 480:28–33. https://doi.org/10.1016/j.gene.2011.03.004
Lloyd YM, Esemu LF, Antallan J, Thomas B, Tassi Yunga S, Obase B, Christine N, Leke RGF, Culleton R, Mfuh KO, Nerurkar VR, Taylor DW (2018) PCR-based detection of Plasmodium falciparum in saliva using mitochondrial cox3 and varATS primers. Trop Med Health 46:22. https://doi.org/10.1186/s41182-018-0100-2
Mogi T, Kita K (2010) Diversity in mitochondrial metabolic pathways in parasitic protists Plasmodium and Cryptosporidium. Parasitol Int 59:305–312. https://doi.org/10.1016/j.parint.2010.04.005
Preiser PR, Wilson RJ, Moore PW, McCready S, Hajibagheri MA, Blight KJ, Strath M, Williamson DH (1996) Recombination associated with replication of malarial mitochondrial DNA. EMBO J 15:684–693
Schnittger L, Rodriguez AE, Florin-Christensen M, Morrison DA (2012) Babesia: a world emerging. Infect Genet Evol 12:1788–1809. https://doi.org/10.1016/j.meegid.2012.07.004
Schreeg ME, Marr HS, Tarigo JL, Cohn LA, Bird DM, Scholl EH, Levy MG, Wiegmann BM, Birkenheuer AJ (2016) Mitochondrial genome sequences and structures aid in the resolution of Piroplasmida phylogeny. PLoS One 11:e0165702. https://doi.org/10.1371/journal.pone.0165702
Solano-Gallego L, Sainz A, Roura X, Estrada-Pena A, Miro G (2016) A review of canine babesiosis: the European perspective. Parasit Vectors 9:336. https://doi.org/10.1186/s13071-016-1596-0
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Uilenberg G (2006) Babesia—a historical overview. Vet Parasitol 138:3–10. https://doi.org/10.1016/j.vetpar.2006.01.035
Wickramasekara Rajapakshage BK et al (2012) Involvement of mitochondrial genes of Babesia gibsoni in resistance to diminazene aceturate. J Vet Med Sci 74:1139–1148. https://doi.org/10.1292/jvms.12-0056
Yang X, Zhao Y, Wang L, Feng H, Tan L, Lei W, Zhao P, Hu M, Fang R (2015) Analysis of the complete Fischoederius elongatus (Paramphistomidae, Trematoda) mitochondrial genome. Parasit Vectors 8:279. https://doi.org/10.1186/s13071-015-0893-3
Funding
This study was financially supported by the National Key Research and Development Program of China (Grant No. 2017YFD0500402), the National Basic Science Research Program (973 program) of China (Grant No. 2015CB150300), the National Natural Science Foundation of China (Grant No. 31772729), and the Natural Science Foundation of Hubei Province (Grant No. 2017CFA020).
Author information
Authors and Affiliations
Contributions
Performed the experiments: JG, PH, and XM. Participated in the data analysis: JG, LH, XM, PH, JC, SW, and ML. Helped with the diagnostic assays: XM and PH. Edited the manuscript: LH, JG, CH, and JZ. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All experiments were performed under the approval of Laboratory Animals Research Centre of Hubei Province and the Ethics Committee of Huazhong Agricultural University (Permit number: HZAUCA-2016-007).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Section Editor: Dana Mordue
Rights and permissions
About this article
Cite this article
Guo, J., Miao, X., He, P. et al. Babesia gibsoni endemic to Wuhan, China: mitochondrial genome sequencing, annotation, and comparison with apicomplexan parasites. Parasitol Res 118, 235–243 (2019). https://doi.org/10.1007/s00436-018-6158-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-018-6158-2