Abstract
The invasive and motile life stages of malaria parasites (merozoite, ookinete and sporozoite) possess a distinctive cortical structure termed the pellicle. The pellicle is characterised by a double-layered ‘inner membrane complex’ (IMC) located underneath the plasma membrane, which is supported by a cytoskeletal structure termed the subpellicular network (SPN). The SPN consists of intermediate filaments, whose major constituents include a family of proteins called alveolins. Here, we re-appraise the alveolins in the genus Plasmodium with respect to their repertoire, structure and interrelatedness. Amongst 13 family members identified, we distinguish two domain types that, albeit distinct at the primary structure level, are structurally related and contain tandem repeats with a consensus 12-amino acid periodicity. Analysis in Plasmodium berghei of the most divergent alveolin, PbIMC1d, reveals a zoite-specific expression in ookinetes and a subcellular localisation in the pellicle, consistent with its predicted role as a SPN component. Knockout of PbIMC1d gives rise to a wild-type phenotype with respect to ookinete morphogenesis, tensile strength, gliding motility and infectivity, presenting the first example of apparent functional redundancy amongst alveolin family members.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malaria parasite transmission begins when gametocytaemic blood is ingested by a vector mosquito. This initiates rapid gametogenesis followed by fertilisation. Zygotes transform into motile ookinetes that cross the midgut wall of the insect and transform into oocysts (Meis and Ponnudurai 1987; Meis et al. 1989). An approximately 2-week period of growth and replication concludes in hundreds of motile sporozoites being released from each oocyst and invading the salivary glands. Blood feeding of the insect transmits the sporozoites to the vertebrate host, where they replicate to each produce thousands of merozoites. The motile merozoites are released into the bloodstream, where they infect red blood cells and either replicate to form more merozoites or differentiate into sexual stage male and female gametocytes to complete the life cycle.
The three zoite stages of Plasmodium species (i.e. ookinetes, sporozoites and merozoites) possess a characteristic peripheral cytoskeletal structure known as the pellicle. The pellicle is defined by a double-membrane structure termed the inner membrane complex (IMC) (Bannister et al. 2000; Morrissette and Sibley 2002; Santos et al. 2009). The IMC is equivalent to a system of flattened membranous sacs that underlie the plasma membrane, the so-called ‘alveoli’, which are a defining feature of unicellular microorganisms belonging to the phyla Apicomplexa, Ciliophora (ciliates) and Dinoflagellata (dinoflagellates) within the protist Alveolata superphylum. Tightly associated with the IMC on its cytoplasmic side lies a network of intermediate filaments termed the subpellicular network (SPN), which supports the pellicular membranes and provides mechanical strength to the cell (Mann and Beckers 2001). Members of an Apicomplexa-specific family of proteins, termed IMC1 proteins, were identified as building blocks of the SPN (Khater et al. 2004; Mann and Beckers 2001). Subsequently, structurally related proteins from ciliates and dinoflagellates were identified and added to this protein family renamed ‘alveolins’ (Gould et al. 2008). In the genus Plasmodium, the alveolin family members display differential expression between different zoite stages of the parasite. In the rodent malaria species Plasmodium berghei, it was shown that disruption of individual alveolin family members expressed in sporozoites (PbIMC1a), in ookinetes (PbIMC1b) or in both these zoites (PbIMC1h) results in morphological abnormalities that are accompanied by reduced tensile strength of the zoite stages in which they are expressed (Khater et al. 2004; Tremp and Dessens 2011; Tremp et al. 2008; Volkmann et al. 2012). Besides their roles in morphogenesis and tensile strength, the Plasmodium alveolins are also involved in gliding motility, most likely through interactions with components of the glideosome that are situated within the pellicular cytoplasm (Khater et al. 2004; Tremp and Dessens 2011; Tremp et al. 2008; Volkmann et al. 2012). Apart from their expression throughout the Plasmodium life cycle, alveolins are essential for parasite development both in the vertebrate and insect hosts (Khater et al. 2004; Tremp et al. 2014; Tremp and Dessens 2011), which makes them potentially attractive targets for malaria treatment, prophylaxis and transmission control. For this reason, it is important to better understand their core architecture, as well as the underlying mechanisms for their assembly into the supramolecular structures that make up the cortical cytoskeleton of the zoite stages.
In this study, we carried out a critical re-evaluation of the Plasmodium alveolins with respect to their repertoire, structure and interrelatedness. Our analyses identify two distinct domain types that are structurally and functionally related without possessing significant homology at the primary structure level.
Materials and methods
Animal use
All laboratory animal work undergoes regular ethical review by the London School of Hygiene & Tropical Medicine and has been approved by the UK Home Office. Work was carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 implementing European Directive 2010/63 for the protection of animals used for experimental purposes. Experiments were conducted in 6–8-week-old female CD1 mice, specific pathogen free and maintained in filter cages. Animal welfare was assessed daily, and animals were humanely killed upon reaching experimental or humane endpoints. Mice were infected with parasites suspended in RPMI or phosphate-buffered saline (PBS) by intraperitoneal injection or by infected mosquito bite on anaesthetised animals. Parasitaemia was monitored regularly by collecting of a small drop of blood from a superficial tail vein. Drugs were administered by intraperitoneal injection or where possible were supplied in drinking water. Parasitised blood was harvested by cardiac bleed under general anaesthesia without recovery.
Parasite maintenance, transmission, culture and purification
P. berghei ANKA clone 234 parasites were maintained as cryopreserved stabilates or by mechanical blood passage and regular mosquito transmission. Ookinete cultures were set up overnight from gametocytaemic blood as previously described (Arai et al. 2001). After 20–24 h, ookinetes were purified via ice-cold 0.17 M ammonium chloride lysis and centrifugation at 800×g for 10 min, followed by PBS washes. Mosquito infection and transmission assays were as previously described using Anopheles stephensi (Dessens et al. 1999; Khater et al. 2004), and infected insects were maintained at 20 °C at approximately 70 % relative humidity.
Gene targeting constructs
The entire pbimc1d coding sequence plus ca. 0.55 kb of upstream sequence was PCR amplified from genomic DNA with primers pDNR-IMC1d-F (ACGAAGTTATCAGTCGAGGTACCAGCCAAAATCACCGAAAAG) and pDNR-IMC1d-R (ATGAGGGCCCCTAAGCTTTCAGATATTAAAGGAGCATTATCAATG) and cloned into SalI/HindIII-digested pDNR-EGFP by in-fusion cloning to give plasmid pDNR-IMC1d/GFP. The 3′ untranslated region of pbimc1d was amplified with primers pLP-IMC1d-F (ATATGCTAGAGCGGCCTAGTAAGTCTTTTGCATTTTATCAATGC) and pLP-IMC1d-R (CACCGCGGTGGCGGCCAAAATATGAAGAAATGACAAAACAGAAG) and the resulting ca. 0.62-kb fragment cloned into NotI-digested pLP-hDHFR by in-fusion cloning to give plasmid pLP-hDHFR/IMC1d. The pbimc1d/gfp-specific sequence from pDNR-IMC1d/GFP was transferred to pLP-hDHFR/IMC1d by Cre/loxP recombination to give the final construct pLP-IMC1d/GFP. This plasmid served as template in PCR-based site-directed mutagenesis using primers IMC1d-KO-F (AGCCAGTGATGAGTAAAGGAGAAGAACTTTTCAC) and IMC1d-KO-R (TTACTCATCACTGGCTTATAAAATGCATTTATT). The resulting PCR product was circularised using in-fusion to give plasmid pLP-IMC1d-KO.
Generation and genotyping of genetically modified parasites
Parasite transfection, pyrimethamine selection and dilution cloning were performed as previously described (Waters et al. 1997). Prior to performing transfections, plasmid DNA was digested with KpnI and SacII to remove the vector backbone. Genomic DNA extraction was performed as previously described (Dessens et al. 1999). Integration into the pbimc1d locus was confirmed with primers IMC1d-5′F (TACCCGCATATTTATCATTG) and LAP-GFP-R (GTGCCCATTAACATCACC), and the absence of the wild-type allele was confirmed using primers IMC1d-5′F and IMC1d-3′R (GGTTACATGTATTTTTATTTCCGC).
RT-PCR analysis
Reverse transcription PCR (RT-PCR) analysis was carried out as described (Claudianos et al. 2002) using primers IMC1d-ORF-F (TTGAAAATGGAGATGCTATTACAAG) and pDNR-IMC1d-R (for pbimc1d) and primers tub1-F (GAAGTAATAAGTATACATGTAGG) and tub1-R (ACACATCAATGACTTCTTTACC) (for pbtubulin1).
Western blot analysis
Parasite samples were heated directly in SDS-PAGE loading buffer at 70 °C for 10 min. Proteins were fractionated by electrophoresis through NuPage 4–12 % Bis-Tris precast gels (Invitrogen) and transferred to PVDF membrane (Invitrogen) according to the manufacturer’s instructions. Membranes were blocked for non-specific binding in PBS supplemented with 0.1 % Tween 20 and 5 % skimmed milk for 1 h at room temperature. Goat polyclonal antibody to GFP conjugated to horseradish peroxidase (Abcam ab6663) diluted 1:5000 was applied to the membrane for 1 h at room temperature. After washing, signal was detected by chemiluminescence (Pierce ECL western blotting substrate) according to the manufacturer’s instructions.
Tensile strength and viability assays
Unpurified ookinetes present in ookinete cultures were subjected to hypo-osmotic shock of 0.5× normal osmotic strength by adding an equal volume of water. After 5 min, normal osmotic conditions were restored by adding an appropriate amount of 10× PBS. Cell viability was scored by fluorescence microscopy in the presence of 5 mL/L propidium iodide and 1 % Hoechst 33258. Ookinetes whose nucleus stained positive for both propidium iodide and Hoechst were scored as non-viable, whereas ookinetes whose nucleus only stained positive for Hoechst were scored as viable.
Assessment of ookinete shape and motility
Images of Giemsa-stained ookinetes were captured by microscopy and their length and width measured. The ookinete motility assay was performed as previously described (Moon et al. 2009). Ookinete cultures were added to an equal volume of Matrigel (BD Biosciences) on ice, mixed thoroughly, spotted onto a microscope slide and covered with a Vaseline-rimmed cover slip. The Matrigel was allowed to set at room temperature for at 30 min. Time-lapse videos (one frame every 10 s for 10 min) were taken on a Zeiss Axioplan II microscope. Movies were analysed with ImageJ using the Manual Tracking plugin (http://fiji.sc/wiki/index.php/Manual_Tracking).
Microscopy
For assessment of fluorescence, live parasite samples were assessed and images captured on a Zeiss LSM510 inverted laser scanning confocal microscope.
Bioinformatics
Conserved domains were identified by multiple alignments of orthologous proteins from P. berghei, P. falciparum, P. vivax and P. knowlesi. Multiple alignments were obtained using Clustal Omega, and phylogenetic analyses were carried out using ClustalW2 Phylogeny, accessed through the EMBL-EBI website. Amino acid sequence similarity searches were carried out by protein BLAST, accessed through the National Centre for Biotechnology Information (NCBI), PlasmoDB or ToxoDB. Trees were drawn using TreeDraw. The program HHrepID was accessed through the Bioinformatics Toolkit, Max-Planck Institute for Developmental Biology.
Results
Repertoire and interrelatedness of Plasmodium alveolins
The existence of an alveolin protein family in Plasmodium was first reported in 2004, identifying eight putative members named IMC1a through to IMC1h (Khater et al. 2004). More recent studies identified several additional alveolins (Gould et al. 2008; Kono et al. 2012; Tremp et al. 2013), here named IMC1i to IMC1l in keeping with original nomenclature (Table 1). To evaluate the structural interrelatedness of these alveolins, we carried out a systematic analysis of the Plasmodium genome using BLAST similarity searches with each of the family members. In the process, we identified a 13th family member, named IMC1m (Table 1). Alveolin hits from each of the BLAST searches were given an arbitrary integer score (relating to the scores of the BLAST hits, lowest score receives 1) to generate a similarity matrix (Table 2). For each alveolin, a total score was then calculated to reflect its structural similarity to the Plasmodium alveolin family as a whole. This, in turn, allowed a ranking of the 13 alveolins with respect to their interrelatedness. Accordingly, IMC1e (ranked 1) was identified as being structurally most similar to the alveolin population: it both detects and is detected by the highest number of family members (Table 2). In contrast, IMC1d (ranked 13) has the most divergent structure (Table 2). This suggests that IMC1e represents the most recent common ancestor. Indeed, PbIMC1e was much more successful than PbIMC1d at detecting alveolins in other genera within the Apicomplexa phylum. For example, in Toxoplasma gondii, which encodes 14 alveolins (named TgIMC1 and TgIMC3–TgIMC15) (Anderson-White et al. 2011), PbIMC1e detected 13 family members in protein BLAST, whilst PbIMC1d detected five.
Domain structure of Plasmodium alveolins
Plasmodium alveolins are typified by possessing, within their primary amino acid sequences, highly conserved regions that are flanked by sequences more variable in length and amino acid composition (Khater et al. 2004; Tremp et al. 2008, 2014; Tremp and Dessens 2011). The functional properties of the alveolins are likely to be defined by these conserved domains. Closer examination showed that the sequence similarities between the alveolins identified from the BLAST searches (Table 2) were largely confined to two conserved domains, here named type 1 and type 2, which by phylogenetic analysis split into distinct clades and which are variably distributed amongst the family members (Fig. 1). The alveolins PbIMC1a and PbIMC1b are the only family members that possess interspersed type 1 and type 2 domains (Fig. 1b) (Khater et al. 2004; Tremp et al. 2008). Whilst these different domain types share little sequence homology, both have a strong compositional bias for the amino acids P, I, V, D, E and K (e.g. 66 % for PbIMC1a type 1, 62 % for PbIMC1a type 2, 64 % for PbIMC1b type 1, 66 % for PbIMC1b type 2). These observations suggested that the type 1 and type 2 domains could be structurally related, despite a lack of discernible primary amino acid sequence homology.
Repertoire and domain structure of Plasmodium alveolins. a Phylogeny of conserved domains within the alveolin family members PbIMC1a to PbIMC1m (a–m). Numbers give amino acid coordinates of the conserved domains in the corresponding PbIMC1 protein. Type 1 (red) and type 2 (green) domains separate into distinct clades. b Schematic diagram depicting the 13 alveolin family members (a–m), showing relative positions of the type 1 (red) and type 2 (green) domains
PbIMC1d is expressed in ookinetes and localises to the pellicle/SPN
PbIMC1d is not only the most divergent alveolin family member (Table 2) but is the only alveolin that possesses only a type 2 domain (Fig. 1). We used PbIMC1d to assess the functional relationship between the type 1 and type 2 domains by determining its life stage expression, subcellular distribution and contribution to parasite development. PbIMC1d is encoded by a two-exon gene, separated by a 170-bp intron. The gene is annotated as a putative heat shock protein 90 (hsp90) in P. berghei (hsp86 in P. falciparum), but it has no actual sequence similarity to heat shock proteins. The full-length protein is composed of 249 amino acids with a calculated M r of 29,361. The type 2 domain in PbIMC1d is highly conserved and has a 68 % amino acid content composed of P, I, V, D, E and K.
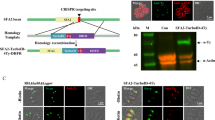
PbIMC1d expression and localisation was studied by tagging the gene with enhanced green fluorescent protein (GFP) in genetically modified parasites. To achieve this, we used a strategy of double crossover homologous recombination in which the wild-type allele was replaced with a recombinant full-length wild-type allele fused to enhanced GFP at its carboxy terminus (Fig. 2a), giving rise to stably transfected parasites. To study the function of PbIMC1d, we generated a null mutant using a similar gene targeting strategy, but removing most of the pbimc1d coding sequence whilst leaving the gfp gene under control of the native pbimc1d promoter to act as a reporter (Fig. 2a). After transfection of purified schizonts, pyrimethamine-resistant parasites were selected and cloned by limiting dilution as described (Tremp and Dessens 2011; Tremp et al. 2008) to give parasite lines IMC1d/GFP and IMC1d-KO, respectively. PCR diagnostic for integration into the pbimc1d locus produced a specific band of approximately 1.1 kb in the IMC1d-KO parasite, whilst this product was approximately 1.7 kb in parasite line IMC1d/GFP (Fig. 2b). The size difference between these PCR products reflects the removal of the pbimc1d coding sequence in the null mutant. PCR diagnostic for the presence of the wild-type pbimc1d allele gave a specific band of approximately 2.4 kb only in wild-type parasites (Fig. 2b). Parasite line IMC1d/GFP developed normally in mouse and mosquito. GFP fluorescence was only observed in zygotes and ookinetes and in the latter was predominantly distributed at the cell cortex (Fig. 2c). Immunoblot analysis of purified, cultured ookinetes with anti-GFP antibodies detected a specific band corresponding to the PbIMC1d::GFP fusion protein (Fig. 2d). These data show that PbIMC1d is specifically expressed in only one of the zoite stages, the ookinete, and is targeted to the pellicle structure/SPN. In T. gondii, the ‘alveolin domain’ was shown to be the main determinant in targeting the protein to the cell cortex (Anderson-White et al. 2011). Thus, the pellicular localisation of PbIMC1d in ookinetes suggests that the type 2 domain in its own right is able to target proteins to this cellular compartment, as does the type 1 domain in other Plasmodium alveolins (Tremp et al. 2014; Tremp and Dessens 2011).
Generation and molecular analyses of genetically modified parasite lines. a General targeting strategy for the GFP tagging and gene disruption of pbimc1d via double crossover homologous recombination. Both wild-type (WT) GFP-tagged (IMC1d/GFP) and disrupted (IMC1d-KO) alleles are shown. The pbimc1d gene is indicated with coding sequence (wide bars) and non-coding sequence (narrow bars). Also indicated are the enhanced GFP module (gfp), the hDHFR selectable marker gene cassette (hdhfr) and primers used for diagnostic PCR amplification (P1–P3). b PCR diagnostic for the presence of the modified GFP-tagged pbimc1d alleles using primers P2 and P3 and the absence of the wild-type pbimc1d allele using primers P1 and P2, from clonal parasite populations of IMC1d/GFP and IMC1d-KO. WT parasites are included as positive controls for the wild-type alleles. c Confocal brightfield and GFP fluorescence image of a cultured, mature ookinete of parasite line IMC1d/GFP, showing cortical fluorescence. d Western blot analysis of purified, cultured ookinetes of parasite lines IMC1d/GFP using anti-GFP antibodies, showing the PbIMC1d::GFP fusion protein. e RT-PCR analysis of wild-type parasite samples enriched for asexual stages (ASX), gametocytes (GCT) and ookinetes (OOK) using primers specific for pbimc1d and pbtubulin1. Due to the primers flanking introns, for each gene, the larger PCR products are amplified from gDNA and the smaller from cDNA. f Confocal brightfield and GFP fluorescence image of a cultured, mature ookinete of parasite line IMC1d-KO, showing cytoplasmic fluorescence
Expression profiling of pbimc1d messenger RNA by RT-PCR with pbimc1d-specific primers flanking its intron amplified an approximately 0.75-kb messenger RNA (mRNA)-specific product in both gametocytes and ookinetes, but in asexual blood stages only amplified an approximately 0.9-kb product from genomic DNA (this product is larger in size due to the intron) (Fig. 2e). By contrast, mRNA of the control gene tubulin1 was present in all parasite samples (Fig. 2e). The discrepancy between pbimc1d mRNA and PbIMC1d protein expression in gametocytes strongly points to translational repression of the pbimc1d gene, as is predicted for the majority of alveolins in P. berghei (Mair et al. 2006). Translational repression is a female gametocyte-specific mechanism of translational silencing involved in the development of the parasite post-fertilisation (Mair et al. 2006).
PbIMC1d is functionally redundant
Ookinetes of the PbIMC1d-null mutant displayed cytoplasmic GFP fluorescence resulting from expression of the GFP reporter from the pbimc1d promoter (Fig. 2f), which is consistent with the expression pattern of PbIMC1d as determined by GFP tagging. The cell shape of PbIMC1d-null mutant ookinetes was not significantly different from that of control IMC1d/GFP ookinetes (length 11.49 ± 0.10 μm for IMC1d/GFP, 11.26 ± 0.12 μm for IMC1d-KO; width 2.15 ± 0.036 μm for IMC1d/GFP, 2.23 ± 0.034 μm for IMC1d-KO; n = 100), and the ookinetes were equally effective in producing oocysts in mosquitoes (140 ± 39 oocysts per mosquito for IMC1d/GFP, 140 ± 28 for IMC1d-KO; n = 20), indicating that they possess normal tensile strength and motility. Indeed, ookinetes of both parasite lines displayed similar resistance to hypo-osmotic shock (78 and 76 % survival for IMC1d/GFP and IMC1d-KO, respectively; n = 100), indicating that the knockout did not adversely affect tensile strength. Furthermore, ookinete motility through Matrigel was comparable between the two parasite lines (42.6 ± 1.7 μm per 10 min for IMC1d/GFP; 41.3 ± 2.6 μm for IMC1d-KO; n = 15). Finally, sporozoites of both parasite lines were readily transmitted by mosquito bite. Thus, PbIMC1d appears to be functionally redundant under our experimental conditions.
Alveolins possess tandem repeat sequences with a 12-amino acid periodicity
Alveolins have been reported to possess a core of repeated sequence motifs (Gould et al. 2008). We tested each of the family members for the presence of tandem repeat sequences using the program HHrepID. This method predicts structural repeats in protein sequences based on Hidden Markov Model (HMM)-HMM comparison, exploiting evolutionary information derived from multiple sequence alignment of homologues (Biegert and Soding 2008). These analyses revealed that all 13 Plasmodium alveolins possess predicted multi-repeat sequences within their conserved type 1 and type 2 domains, supporting the notion that they constitute genuine alveolins. Repeats within the large majority of type 1 domains revealed a clear minimum periodicity of 12 residues (e.g. PbIMC1e, Fig. 3a). Tandem 12-amino acid repeats were also identified in type 2 domains (e.g. PbIMC1b, Fig. 3b), albeit typically with lower probability scores. Articulins and plateins, cortical cytoskeleton proteins from Euglena and Euplotes spp., respectively, have also been reported to have 12-amino acid repetitive motifs rich in valine and proline residues and, in this respect, are similar to alveolins (Huttenlauch et al. 1995; Huttenlauch and Stick 2003; Kloetzel et al. 2003a, b; Marrs and Bouck 1992). This was confirmed by HHrepID analysis, which readily detected 12-amino acid tandem repeat structures in these proteins (Fig. 3c, d).
Discussion
This study brings the number of Plasmodium alveolin family members that have been identified to 13. Of these, seven have now experimentally been shown to localise to the pellicle structure. This includes the most divergent family member, IMC1d, characterised here, which gives strong support to the concept that all Plasmodium alveolins are fundamentally involved with the cortical cytoskeleton in malaria parasites. This is supported by several alveolin knockout studies that reveal key roles in morphogenesis, tensile strength and gliding motility. Studies involving double knockout of PbIMC1b and PbIMC1h showed that the functional contribution of these alveolins to tensile strength and motility is both cumulative and mutually independent (Tremp and Dessens 2011). In contrast, the apparent no-phenotype knockout of PbIMC1d reported here points to a functional redundancy amongst the alveolin family members. The highly conserved type 2 domain of IMC1d indicates that the gene is under selective pressure, and we cannot rule out that the phenotype of the PbIMC1d-null mutant is very subtle, or might only become apparent under conditions that are different from our experimental set-up. However, another possible explanation for the seemingly redundant role of PbIMC1d is that the formation and function of the SPN entail a system of co-operating proteins that are able to bypass, or substitute for, each other’s functions. This attractive hypothesis, if true, would add robustness to a complex biological system that serves roles in many key processes such as morphogenesis, motility and invasion.
The creation of internal repetitions forms an important mechanism for proteins to adapt their structure and function under evolutionary pressure (Marcotte et al. 1999). However, after fixation of duplications, sequence similarities amongst repeats can quickly erode whilst structure and function are preserved (Andrade et al. 2001). Although such mismatch or ‘fuzzy’ repeats are widespread, they are difficult to detect due to their low similarity, polymorphism and vast diversity. This is further confounded by the potential for non-canonical repeats, degenerate repeats and discontinuities (i.e. short insertions between repeats) to arise, which for example are known to frequently occur in coiled-coil domains, a well-known type of tandem repeat structure with a consensus 7-amino acid periodicity (Brown et al. 1996). The type 1 and type 2 domains identified here in the Plasmodium alveolins are likely to be the result of similar evolutionary forces. These alveolin domains lack discernible homology at the primary structure level and fall into distinct clades, but are clearly related with regard to their amino acid composition, tandem repeat structure and functional localisation. There is further indication that the type 2 repeats arose from type 1 repeats (or vice versa). First, in IMC1f, the type 1 and type 2 domains are located tandemly within a single uninterrupted block of conserved sequence (Fig. 1b). Second, four T. gondii alveolins share sequence similarity in protein BLAST with both PbIMC1e and PbIMC1d. In three of these (TgIMC6, TgIMC14 and TgIMC15), the regions of sequence similarity with PbIMC1e and PbIMC1d overlap, pointing to the presence of domains with ‘intermediate’ homology between type 1 and type 2. These combined observations suggest that the primary amino acid sequences of the type 1 and type 2 repeats have evolved in a way that has preserved their overall structure and function. This phenomenon of ‘constrained evolution’ has also been observed in other protist cytoskeletal proteins with repeat motifs (Gould et al. 2011).
Many bioinformatics programs for internal repeat detection in proteins utilise algorithms based on local alignment and substitution matrices (Andrade et al. 2000; Heger and Holm 2000). These approaches have been modestly successful in predicting tandem repeats within the alveolins. By contrast, HHrepID is based on building and matching Hidden Markov Models to identify repeat sequences (Biegert and Soding 2008). Our findings demonstrate that HHrepID successfully predicts tandem repeats in the Plasmodium alveolins and show for the first time that these repeats have a consensus 12-amino acid length. Accordingly, the multi-repeat structures of alveolins are very similar, both in periodicity and amino acid composition, to those of cytoskeletal proteins in other protists: the articulins and plateins. Whilst Euplotes plateins possess canonical ER signal peptides and form plate-like structures inside alveoli (Kloetzel et al. 2003b), Euglena articulins form intermediate filaments with a more classic membrane skeleton role (Huttenlauch et al. 1995). Our findings give support to the concept that articulins, plateins and alveolins have a common structural scaffold and thus should be grouped within the same protein superfamily. Structural studies are being carried out to test this hypothesis. Interestingly, prokaryotic articulin homologues have been discovered in Caulobacter crescentus, the first bacterium described to rely on an intermediate filament-based cytoskeleton for its cell shape (Ausmees et al. 2003). Thus, articulin-like proteins like alveolins could be far more widespread than originally assumed.
References
Anderson-White BR, Ivey FD, Cheng K, Szatanek T, Lorestani A, Beckers CJ, Ferguson DJ, Sahoo N, Gubbels MJ (2011) A family of intermediate filament-like proteins is sequentially assembled into the cytoskeleton of Toxoplasma gondii. Cell Microbiol 13(1):18–31
Andrade MA, Ponting CP, Gibson TJ, Bork P (2000) Homology-based method for identification of protein repeats using statistical significance estimates. J Mol Biol 298(3):521–537
Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struc Biol 134(2–3):117–131
Arai M, Billker O, Morris HR, Panico M, Delcroix M, Dixon D, Ley SV, Sinden RE (2001) Both mosquito-derived xanthurenic acid and a host blood-derived factor regulate gametogenesis of Plasmodium in the midgut of the mosquito. Mol Biochem Parasitol 116(1):17–24
Ausmees N, Kuhn JR, Jacobs-Wagner C (2003) The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115(6):705–713
Bannister LH, Hopkins JM, Fowler RE, Krishna S, Mitchell GH (2000) A brief illustrated guide to the ultrastructure of Plasmodium falciparum asexual blood stages. Parasitol Today 16(10):427–433
Biegert A, Soding J (2008) De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics 24(6):807–814
Brown JH, Cohen C, Parry DA (1996) Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins 26(2):134–145
Claudianos C, Dessens JT, Trueman HE, Arai M, Mendoza J, Butcher GA, Crompton T, Sinden RE (2002) A malaria scavenger receptor-like protein essential for parasite development. Mol Microbiol 45(6):1473–1484
Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos FC, Sinden RE (1999) CTRP is essential for mosquito infection by malaria ookinetes. EMBO J 18(22):6221–6227
Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF (2008) Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Mol Biol Evol 25(6):1219–1230
Gould SB, Kraft LG, van Dooren GG, Goodman CD, Ford KL, Cassin AM, Bacic A, McFadden GI, Waller RF (2011) Ciliate pellicular proteome identifies novel protein families with characteristic repeat motifs that are common to alveolates. Mol Biol Evol 28(3):1319–1331
Heger A, Holm L (2000) Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41(2):224–237
Huttenlauch I, Stick R (2003) Occurrence of articulins and epiplasmins in protists. J Eukaryot Microbiol 50(1):15–18
Huttenlauch I, Geisler N, Plessmann U, Peck RK, Weber K, Stick R (1995) Major epiplasmic proteins of ciliates are articulins: cloning, recombinant expression, and structural characterization. J Cell Biol 130(6):1401–1412
Khater EI, Sinden RE, Dessens JT (2004) A malaria membrane skeletal protein is essential for normal morphogenesis, motility, and infectivity of sporozoites. J Cell Biol 167(3):425–432
Kloetzel JA, Baroin-Tourancheau A, Miceli C, Barchetta S, Farmar J, Banerjee D, Fleury-Aubusson A (2003a) Cytoskeletal proteins with N-terminal signal peptides: plateins in the ciliate Euplotes define a new family of articulins. J Cell Sci 116(7):1291–1303
Kloetzel JA, Baroin-Tourancheau A, Miceli C, Barchetta S, Farmar J, Banerjee D, Fleury-Aubusson A (2003b) Plateins: a novel family of signal peptide-containing articulins in euplotid ciliates. J Eukaryot Microbiol 50(1):19–33
Kono M, Herrmann S, Loughran NB, Cabrera A, Engelberg K, Lehmann C, Sinha D, Prinz B, Ruch U, Heussler V, Spielmann T, Parkinson J, Gilberger TW (2012) Evolution and architecture of the inner membrane complex in asexual and sexual stages of the malaria parasite. Mol Biol Evol 29(9):2113–2132
Mair GR, Braks JA, Garver LS, Wiegant JC, Hall N, Dirks RW, Khan SM, Dimopoulos G, Janse CJ, Waters AP (2006) Regulation of sexual development of plasmodium by translational repression. Science 313(5787):667–669
Mann T, Beckers C (2001) Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol 115(2):257–268
Marcotte EM, Pellegrini M, Yeates TO, Eisenberg D (1999) A census of protein repeats. J Mol Biol 293(1):151–160
Marrs JA, Bouck GB (1992) The two major membrane skeletal proteins (articulins) of Euglena gracilis define a novel class of cytoskeletal proteins. J Cell Biol 118(6):1465–1475
Meis JF, Ponnudurai T (1987) Ultrastructural studies on the interaction of Plasmodium falciparum ookinetes with the midgut epithelium of Anopheles stephensi mosquitoes. Parasitol Res 73(6):500–506
Meis JF, Pool G, van Gemert GJ, Lensen AH, Ponnudurai T, Meuwissen JH (1989) Plasmodium falciparum ookinetes migrate intercellularly through Anopheles stephensi midgut epithelium. Parasitol Res 76(1):13–19
Moon RW, Taylor CJ, Bex C, Schepers R, Goulding D, Janse CJ, Waters AP, Baker DA, Billker O (2009) A cyclic GMP signalling module that regulates gliding motility in a malaria parasite. PLoS Pathog 5(9):e1000599
Morrissette NS, Sibley LD (2002) Cytoskeleton of apicomplexan parasites. Microbiol Mol Biol Rev 66(1):21–38
Santos JM, Lebrun M, Daher W, Soldati D, Dubremetz JF (2009) Apicomplexan cytoskeleton and motors: key regulators in morphogenesis, cell division, transport and motility. Int J Parasitol 39(2):153–162
Tremp AZ, Dessens JT (2011) Malaria IMC1 membrane skeleton proteins operate autonomously and participate in motility independently of cell shape. J Biol Chem 286(7):5383–5391
Tremp AZ, Khater EI, Dessens JT (2008) IMC1b is a putative membrane skeleton protein involved in cell shape, mechanical strength, motility, and infectivity of malaria ookinetes. J Biol Chem 283(41):27604–27611
Tremp AZ, Carter V, Saeed S, Dessens JT (2013) Morphogenesis of Plasmodium zoites is uncoupled from tensile strength. Mol Microbiol 89(3):552–564
Tremp AZ, Al-Khattaf FS, Dessens JT (2014) Distinct temporal recruitment of Plasmodium alveolins to the subpellicular network. Parasitol Res 113(11):4177–4188
Volkmann K, Pfander C, Burstroem C, Ahras M, Goulding D, Rayner JC, Frischknecht F, Billker O, Brochet M (2012) The alveolin IMC1h is required for normal ookinete and sporozoite motility behaviour and host colonisation in Plasmodium berghei. PLoS One 7(7):e41409
Waters AP, Thomas AW, van Dijk MR, Janse CJ (1997) Transfection of malaria parasites. Methods 13(2):134–147
Acknowledgments
This work was supported by the Wellcome Trust, grant nos. 076648 and 088449, and a Studentship to FSA-K from the Cultural Bureau of the Royal Embassy of Saudi Arabia in London. We thank E McCarthy for assistance with microscopy. The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Fatimah S. Al-Khattaf and Annie Z. Tremp contributed equally to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Al-Khattaf, F.S., Tremp, A.Z. & Dessens, J.T. Plasmodium alveolins possess distinct but structurally and functionally related multi-repeat domains. Parasitol Res 114, 631–639 (2015). https://doi.org/10.1007/s00436-014-4226-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-014-4226-9