Abstract
Sarcocysts from four Polish roe deer were collected and examined by light microscopy, small subunit ribosomal RNA (ssu rRNA), and the subunit I of cytochrome oxidase (cox1) sequence analysis. This resulted in identification of Sarcocystis gracilis, Sarcocystis oviformis, and Sarcocystis silva. However, we were unable to detect Sarcocystis capreolicanis, the fourth Sarcocystis species found previously in Norwegian roe deer. Polish sarcocysts isolated from various tissues differed in terms of their shape and size and were larger than the respective Norwegian isolates. Analysis of ssu rRNA gene revealed the lack of differences between Sarcocystis isolates belonging to one species and a very low degree of genetic diversity between Polish and Norwegian sarcocysts, ranging from 0.1 % for Sarcocystis gracilis and Sarcocystis oviformis to 0.44 % for Sarcocystis silva. Contrary to the results of the ssu rRNA analysis, small intraspecies differences in cox1 sequences were found among Polish Sarcocystis gracilis and Sarcocystis silva isolates. The comparison of Polish and Norwegian cox1 sequences representing the same Sarcocystis species revealed similar degree of sequence identity, namely 99.72 % for Sarcocystis gracilis, 98.76 % for Sarcocystis silva, and 99.85 % for Sarcocystis oviformis. Phylogenetic reconstruction and genetic population analyses showed an unexpected high degree of identity between Polish and Norwegian isolates. Moreover, cox1 gene sequences turned out to be more accurate than ssu rRNA when used to reveal phylogenetic relationships among closely related species. The results of our study revealed that the same Sarcocystis species isolated from the same hosts living in different geographic regions show a very high level of genetic similarity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sarcocystis spp. are obligatory intracellular protozoa, which parasitize many different animals, including mammals, birds, and fish (Tenter 1995; Fayer 2004). They have an obligatory two-host life cycle. The parasites reproduce sexually in the intestine of the carnivorous definitive host, and the asexual phase of life cycle takes place in the muscles or nervous tissue of the herbivore or omnivore intermediate hosts (Dubey et al. 1988). Pathogenic species of Sarcocystis cause acute disease only in intermediate hosts. Most pathogenic species of Sarcocystis are those infecting ruminants; in the case of massive infections, they cause hemorrhagic diathesis, encephalitis, encephalomyelitis, and, eventually, death of the animal. Acute infections of pregnant females with Sarcocystis spp. are associated with fetal death, abortion, and premature birth (Dubey et al. 1988; Dubey and Rommel 1992; Jeffrey 1993). The chronic infections cause economic losses due to reduced quality and quantity of animal meat (Fayer and Elsasser 1991). Apart from domestic ruminants (Tenter 1995), Sarcocystis spp. were found in several species of cervids, such as reindeer (Dahlgren and Gjerde 2007, Dahlgren et al. 2007, 2008), red deer (Dahlgren and Gjerde 2010), and roe deer (Gjerde 2012).
Three different Sarcocystis species, namely Sarcocystis gracilis, Sarcocystis capreolicanis, and Sarcocystis cf. hofmanni, were identified in European roe deer using light microscopy (LM), transmission electron microscopy (TEM), and combination thereof (Erber et al. 1978; Entzeroth 1981, 1982; Enzeroth 1985; Sedlaczek and Wesemeier 1995; Santini et al. 1997; Stolte et al. 1996, 1998). The fourth unnamed Sarcocystis spp. that was discovered in roe deer with an aid of TEM (Schramlová and Blažek 1978; Entzeroth 1982; Santini et al. 1997) resembled sarcocysts of Sarcocystis grueneri found in reindeer (Gjerde 1985), Sarcocystis cevicanis in red deer, and Sarcocystis wapiti in wapiti (Hernandez et al. 1981; Speer and Dubey 1982). The last Sarcocystis spp. identified in roe deer with an aid of morphological methods (Stolte et al. 1997) has similar morphology as Sarcocystis ovalis (Dahlgren and Gjerde 2010).
The accuracy of Sarcocystis spp. identification and appropriate characterization of phylogenetic relationships, inferred solely from phenotypic characters, is often uncertain and can be highly subjective. To overcome such problems, the morphological and biological observations were complemented with molecular data obtained with an aid of such techniques as isozyme analysis (O’Donoghue et al. 1986) and, more recently, small subunit ribosomal RNA (ssu rRNA) sequencing (Ellis et al. 1995; Fenger et al. 1994; Johnson et al. 1988; Tenter et al. 1992). Species-specific sequences can be detected with the latter method and further amplified by PCR for identification of Sarcocystis spp. (Tenter et al. 1994; Stojecki et al. 2012). Amplification of DNA and sequencing of the ssu rRNA gene from sarcocyst found in roe deer from Norway confirmed the presence of two Sarcocystis spp., the already described S. gracilis and S. capreolicanis, and revealed the existence of two new species, Sarcocystis oviformis (Dahlgren and Gjerde 2009) and Sarcocystis silva (Gjerde 2012).
The mitochondrial gene encoding the subunit I of cytochrome oxidase (cox1) has proven to be particularly suitable as a molecular marker for taxonomic differentiation and evolutionary studies of different eukaryotic groups. Indeed, it was used recently as a target gene to analyze phylogenetic relationships among 22 Sarcocystis spp. of cervids, cattle, and sheep (Gjerde 2013). The analysis of cox1 sequences allowed a better distinction between the closely related species than the examination of ssu rRNA sequences and confirmed the presence of four Sarcocystis spp. (S. gracilis, S. capreolicanis, S. oviformis, and S. silva) in roe deer.
To date, data on phylogenetic relationships between different Sarcocystis spp. found in roe deer were based solely on the analysis of specimens collected in Norway. Therefore, the objectives of the present study included molecular identification of Sarcocystis spp. collected from Polish roe deer, their genetic comparison with the same species described by Gjerde et al. in Norway, and analysis of phylogenetic relationships between Sarcocystis spp. from Polish and Norwegian roe deer (Dahlgren and Gjerde 2009; Gjerde 2012).
Material and methods
Isolation and morphological identification of sarcocysts
The study included 14 sarcocysts isolated from four adult (4- and 5-year-old) roe deer found dead in the fall and winter of 2009 in Greater Poland, Forest District Piaski (animal Rd1 and Rd2) and Forest District Przedborów (animal Rd3 and Rd4). Massive invasion of Sarcocystis spp. was diagnosed during necropsy in all animals. Sarcocysts were isolated from skeletal muscles (musculus latissimus dorsi), esophagus, heart, and tongue (Table 1). Individual sarcocysts were excised under stereomicroscope, using fine needles. A new needle was used for each specimen. Excised cysts were washed three times in PBS and examined under LM. Subsequently, the material was stored up to 3 months at −20 °C until DNA isolation.
DNA isolation and amplification of ssu rRNA and cox1 genes
Extraction of genomic and mitochondrial DNA was performed immediately upon thawing, with DNeasy Blood & Tissue Kit (Qiagen GmbH, Germany). Isolation procedure was performed according to the manufacturer’s protocol. Briefly, frozen sarcocysts were thawed on ice, and ATL buffer (180 μl) and proteinase K (20 μl) were added. Samples were mixed by vortexing and incubated at 56 °C, until the cysts were completely lysed. Then, 200 μl of AL buffer and 200 μl of 96 % ethanol were added, and samples were again mixed by vortexing. Subsequently, the DNA-containing mixture was transferred on a column supplied by the manufacturer. The column was washed with AW1 buffer (500 μl) and AW2 buffer (500 μl), and the DNA was eluted with 100 μl of AE buffer.
The ssu rRNA gene was amplified by PCR, according to slightly modified procedure described by Dahlgren and Gjerde (2007). PCR was carried with primers shown in Table 2, synthesized at the Genomed SA (Poland). Reaction mix (50 μl) contained template DNA, 10× DreamTaq Green Buffer [KCl, (NH4)2SO4, 20 mM MgCl2], 0.2 mM dNTP mix, 0.2 μM of each primer, and 1 U of DreamTaq polymerase (Fermentas). PCR was started with initial denaturation (95 °C, 5 min), followed by 35 cycles of denaturation (95 °C, 30 s), annealing (55 °C, 30 s), elongation (72 °C, 1 kb/min), and final elongation (72 °C, 10 min). The PCR amplification of cox1 gene was carried out with primers showed in Table 2, synthesized at the BioTeZ GmbH (Germany). Reaction mix (50 μl) contained DNA (1 μl), Phusion HF Buffer (Thermo Scientific), MgCl2 at final concentration of 4 mM, 0.5 U of Phusion High-Fidelity DNA Polymerase (Thermo Scientific), 0.2 mM dTNPs mix, and 0.2 μM of each primer. All amplification reactions were started with initial denaturation (98 °C, 30 s), followed by 40 cycles of denaturation (98 °C, 5 s), annealing (52 °C, 30 s), elongation (72 °C, 30 s), and final elongation (72 °C, 10 min). PCR products were analyzed by electrophoresis in 1.5 % agarose gel and stained with ethidium bromide.
Sequencing
The nucleotide sequences of the ssu rRNA and cox1 genes were determined by the BigDye® Terminator v3.1 cycle sequencing (Life Technologies) and analyzed on an ABI fluorescence automated DNA sequencer, at the Genomed SA (Poland) and the LGC Genomics (Germany), respectively. CAP3 software (Huang and Madan 1999) was used to align the nucleotide sequences of ssu rRNA and cox1 genes.
Sequence alignment, comparison, and phylogenetic analyses
Obtained nucleotide sequences were submitted to BLAST (Altschul et al. 1990), and GenBank database was searched for similar sequences with the use of “blastn” and “megablast” algorithms.
The ssu rRNA and cox1 gene sequences were aligned with ClustalW in default settings (Larkin et al. 2007). MEGA 5.0 (Tamura et al. 2011) was used to find the best nucleotide substitution model and generate maximum-likelihood (ML) phylograms of ssu rRNA and cox1 gene sequences. The Kimura 2-parameter (K2) (Kimura 1980), with gamma distribution and invariable sites (K2 + G + I), was found to be the optimal nucleotide substitution model for construction of the ML tree. Subtree-pruning-regrafting (SPR) algorithm was used in order to construct maximum parsimony (MP) trees for both genes. For ssu rRNA gene, phylogenetic reconstruction consisted of 147 sequences, and for cox1 gene, 327 sequences. Bootstrap method was used to check the reliability of formed phylograms. All sequences used in phylogenetic reconstruction are listed in Table S1.
Population genetic analysis
Cox1 gene sequences were analyzed with Arlequin 3.5 Software (Excoffier and Lischer 2010). Genetic diversity values, including haplotype numbers and diversity, substitution numbers, polymorphic sites, and nucleotide diversity, were compared between Polish and Norwegian Sarcocystis isolates. Tajima’s D and Fu’s F s tests were used for the evaluation of genetic structure of the Sarcocystis under the population expansion. Population pairwise F st estimates were also obtained with Arlequin 3.5 software. Mean distance between groups was calculated with MEGA 5.0 software.
Results
Morphological characterization of sarcocysts
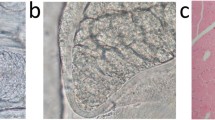
Several types of sarcocysts were identified in roe deer by light microscopy. Four spindle-shaped sarcocysts (isolates no. 4, 9, 10, and 13; Fig. 1a) and three long cysts with rounded ends (isolates no. 11, 12, and 14; Fig. 1b) were found in skeletal muscles. While the spindle-shaped cysts were similar in size (11.1 to 12.2 mm in length), the long cysts with rounded ends differed significantly in terms of their sizes (7.4 to 10.9 mm in length). All the three cysts isolated from the esophagus were spindle-shaped (isolates no. 1–3) and had similar sizes as those isolated from the muscles (11.1–12.7 mm). In turn, the two Sarcocystis residing in the tongue were long cysts with rounded ends (isolates no. 5 and 6) and 7.7 to 9.2 mm in length. The third type of cysts (n = 2) with different morphology, described as sac-like with rounded ends (isolates no. 7–8; Fig. 1c), was isolated from the heart. These cysts were markedly smaller (1.5 mm in length) than the other specimens.
Amplification and sequencing of ssu rRNA gene
The samples of genomic DNA from 14 Sarcocystis specimens collected from Polish roe deer were used as template to amplify ssu rRNA gene by PCR. The sequencing of PCR amplicons revealed the presence of three different nucleotide sequences, designated as ssurRNA1 (1,805 bp), ssurRNA2 (1,833 bp), and ssurRNA3 (1,752 bp). The obtained sequences were subjected to BLAST analysis and then aligned with published sequences of S. gracilis (FJ196261, JN226126, JN226127), S. oviformis (FJ196262, KC209745, KC209746), and S. silva (JN226124, JN226122, JN226123, JN226125 and EU282016) with an aid of ClustalW software (Dahlgren and Gjerde 2009; Gjerde 2012, 2013). The seven sequences (isolates no. 1, 2, 3, 7, 9, 10, and 13) designated as ssurRNA1 turned out to be almost identical (99.9 %) with the sequence of ssu rRNA gene of Norwegian S. gracilis. Another three sequences (isolates no. 4, 5, and 6) referred to as ssurRNA2 were almost identical (99.9 %) with the sequence of ssu rRNA gene of S. oviformis, and four ssurRNA3 sequences (isolates no. 8, 11, 12, and 14) closely resembled (99.37–99.66 %, average identity 99.56 %) previously reported sequences of ssu rRNA gene from S. silva (Table 3). All three newly identified sequences, ssurRNA1 (ssu rRNA gene of S. gracilis), ssurRNA2 (ssu rRNA gene of S. oviformis), and ssurRNA3 (ssu rRNA gene of S. silva), were submitted to the GenBank and given the accession numbers KF880741, KF880742, and KF880743, respectively.
The sequences of Polish S. gracilis and S. oviformis ssu rRNA genes differed from their Norwegian counterparts only by one nucleotide. The ssu rRNA gene from Polish S. gracilis isolates had an insertion of T in position 682, and ssu rRNA gene from Polish S. oviformis had a substitution of T to A in position 187. Five nucleotide differences, unique for Polish sequence, were identified in the case of ssu rRNA gene from Polish S. silva sarcocysts. A transversion of C for G was found in position 3 when the Polish sequence was compared with Norwegian sequences designated as JN226122, JN226123, JN226124, and EU282016, and another transversion of A for C was identified in position 5 as compared to Norwegian sequences designated as JN226122, JN226123, JN226124, and EU282016. Moreover, an insertion of G in position 885, deletion of C in position 1468, and transition of G for A in position 1736 were found in the Polish sequence when compared to all the Norwegian sequences (Table S2).
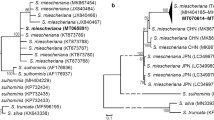
The phylogenetic relationships among Polish and Norwegian Sarcocystis isolates were established by constructing phylogenetic trees obtained with the ML and MP methods, as reported previously by Gjerde (2013) for Sarcocystis species from cervids, cattle, and sheep. Both methods produced similar results: Only minor differences in branching of aligned sequences were observed, mostly in the case of branches with low bootstrap values. Phylogenetic tree based on ssu rRNA gene sequences, obtained by the MP method, is shown in Fig. 2a. Polish isolates of S. gracilis represented a sister leaf to Norwegian isolates with high bootstrap value (99 %), dividing these sequences. Polish S. oviformis isolates were nested in three Norwegian isolates. All S. silva isolates formed clade with one isolate of Sarcocystis truncata. Thus, phylogenetic analysis confirmed close relationships between all the Polish and Norwegian isolates of Sarcocystis species, namely S. gracilis, S. oviformis, and S. silva.
Maximum parsimony of DNA phylograms of selected Sarcocystidae and Coccidia. a The ssu rRNA tree was constructed based on the alignment of nearly complete ssu rRNA gene sequences of 14 Polish Sarcocystis spp. isolates and available ssu rRNA gene sequences of related species deposited in GenBank. b The cox1 tree was constructed based on the alignment of nearly complete ssu rRNA gene sequences of 14 Polish Sarcocystis spp. isolates and available ssu rRNA gene sequences of related species deposited in GenBank. Trees were rooted with Eimeria spp. The italicized values among the branches indicate percent bootstrap value per 1,000 replicates. Bootstrap values below 50 % are not shown. Polish isolates are marked in red. The GenBank accession numbers of all sequences used for construction of the trees are given in Table S1 (color figure online)
Amplification and sequencing of cox1 gene
Mitochondrial DNA isolated from the abovementioned Sarcocystis specimens was used as a template to amplify the sequences of cox1 gene by PCR. All the obtained amplicons were sequenced, and the sequences were submitted to GenBank and given the accession numbers shown in Table 1. The sequencing of PCR amplicons produced three sets of closely related sequences, designated as cox1.I, cox1.II, and cox1.III.
As for ssu rRNA gene, the cox1 sequences of Polish roe deer isolates were first subjected to BLAST analysis, and then ClustalW software was used to align them with the published sequences of cox1 gene from Norwegian isolates of S. gracilis (KC209605–KC209614, KF241330–KF241341), S. oviformis (KC209656–KC209661), and S. silva (KC209686–KC209689, KF241410) (Gjerde 2013, 2014). Based on these analyses, cox1.I sequences (isolates no. 1, 2, 3, 7, 9, 10, and 13) were identified as S. gracilis, cox1.II sequences (isolates no. 4, 5, and 6) as S. oviformis, and cox1.III sequences (isolates no. 8, 11, 12, and 14) as S. silva.
Clustal alignment was further analyzed with Arlequin 3.5 software for the purpose of population genetic analysis. Generally, five haplotypes were found when Polish S. gracilis isolates were compared with each other. The comparison of cox1 gene sequences identified in our study with Norwegian sequences revealed that both groups were characterized by similar haplotype frequency and nucleotide diversity (Table 4). The average number of pairwise differences for Polish isolates was 1.9 (±1.4) with 6 variable sites, and for Norwegian isolates 2.6 (±1.6) with 16 variable sites. Figure 3a presents interhaplotypic distance matrix for all S. gracilis isolates. Sequence identity between Polish isolates of S. gracilis was 99.33–100 % (average identity 99.70 %), and the identity between Polish and Norwegian amounted to 99.11–100 % (average identity 99.72 %) (Table S3). Between-group mean distance value for S. gracilis was 0.0024.
Interhaplotypic distance matrix for S. gracilis, S. oviformis, and S. silva haplotypes. Heat map for each species shows differences between haplotypes. GenBank accession numbers of one representative strain from each haplotype are displayed on the x- and y-axes. Full list of isolates belonging to presented haplotypes is shown in Tables S2 and S3. A color key containing histogram is added to each chart. “Value” on the x-axes and the color gradient correspond to the number of nucleotide differences between two haplotypes. “Count” on the y-axes describes the number of squares with occurring value
Each of the three Polish isolates of S. oviformis had the same nucleotide sequence of cox1 gene. The average number of pairwise differences for Polish isolates was 0 (±0) with no variable sites, and for Norwegian isolates 0.6 (±0.6) with one variable site. Figure 3b presents interhaplotypic distance matrix for all S. oviformis isolates. Sequence identity between Polish and Norwegian isolates was 99.85 % (Table S4). Between-group mean distance value for S. oviformis was 0.0015.
S. silva showed the highest genetic variability. Each of the four Polish and five Norwegian isolates had unique sequences. The average number of pairwise differences for Polish isolates was 7.6 (±5.4) with 15 variable sites, and for Norwegian isolates 11.0 (±7.6) with 20 variable sites. Figure 3c presents interhaplotypic distance matrix for all S. silva isolates. The identity of sequences between Polish isolates ranged from 98.48 to 99.78 % (average identity 99.17 %) and between Polish and Norwegian from 98.48 to 99.57 % (average identity 98.76 %) (Table S5). Between-group mean distance value for S. silva was 0.016. Tajima’s D and Fu’s F s tests showed no significant differences between any of the species, (p > 0.05). Pairwise F st values for Polish and Norwegian populations showed no significant differences (data not shown).
As for ssu rRNA gene, phylogenetic analyses based on cox1 gene gave similar results for the MP and ML methods. Minor differences in branching of aligned sequences were found in branches with low bootstrap values. The analysis of phylogenetic tree based on MP (Fig. 2b) revealed that the Polish sequences of cox1 gene of all the three identified Sarcocystis species co-localized with other cox1 sequences representing the same species. The Polish isolates of S. gracilis were distributed among the Norwegian isolates and did not form a separate group. Similarly, Polish S. oviformis isolates were nested in a clade of all Norwegian isolates. In turn, S. silva sequences were divided in three clades. The first of them consisted of three Polish isolates and one Norwegian isolate. The second clade consisted of one Polish and three Norwegian isolates. The first and the second clades turned out to be sister clades. The third clade comprised one Norwegian sequence and represented a sister clade for the remaining two clades.
Discussion
The molecular studies on Sarcocystis species, which parasatize cervids, are rare, (Gjerde 2013) and such studies were not performed so far in Poland. Therefore, the present study was undertaken to identify the Sarcocystis spp. parasitizing Polish roe deer and to compare their characteristics with those of Norwegian isolates, as available data on the specimens collected in various geographical locations are sparse.
Polish sarcocysts isolated from various tissues differed in terms of their shape and size. S. gracilis and S. silva found in the skeletal muscles, esophagus, and tongue had morphology typical for the species but were larger than the respective Norwegian isolates (Dahlgren and Gjerde 2009; Gjerde 2012). In contrast, the myocardial cysts of both species were generally smaller and sac-like shaped. Generally, the morphology of cysts found in Polish roe deer was similar to that of Norwegian isolates representing the same Sarcocystis species (Dahlgren and Gjerde 2009; Gjerde 2012). However, Polish and Norwegian isolates turned out to differ considerably in terms of their size distribution. The length of S. gracilis cysts isolated from the skeletal muscles, esophagus, and myocardium of Polish roe deer was 6.7, 12.2, and 1.5 mm, respectively. In turn, the size of typical S. gracilis cysts from Norwegian roe deer ranges from 1.0 to 2.5 mm for diaphragm isolates and between 0.3 and 0.9 mm for myocardial isolates (Dahlgren and Gjerde 2009). Cysts of Polish S. silva found in skeletal muscles and heart measured 7.4–11.5 and 1.5 mm, respectively. Similar to other Polish species, the cysts were larger than those isolated and described by Gjerde (2012) in Norway (0.6 and 0.55 mm, respectively). It should be noted, however, that the Norwegian cysts were isolated from the esophagus. The Polish cysts of S. oviformis isolated from the tongue and skeletal muscles measured about 7.7–9.2 and 10.9 mm, respectively. Cysts of the same species described by Dahlgren and Gjerde (2009) were much smaller (1.6–2.0 mm), but again they were isolated from other tissues, namely the esophagus and diaphragm. Also, the study by Pérez-Creo et al. (2013), who analyzed the prevalence and morphology of Sarcocystis spp. in Spanish roe deer, has shown that cysts of different species isolated from the diaphragm and myocardium were smaller than 1.5 mm. Larger cysts (mean size of 2.055 mm) were observed solely in the esophagus. The abovementioned discrepancies in the size of Polish, Norwegian, and Spanish Sarcocystis isolates may result from their different localization. The Polish sarcocysts were isolated from musculus latissimus dorsi, whereas the Norwegian and Spanish isolates originated from the diaphragm (Dahlgren and Gjerde 2009; Gjerde 2012; Pérez-Creo et al. 2013). Moreover, it should be remembered that only the largest specimens were included in our study.
The hereby described morphological characteristics of cysts isolated from Polish roe deer and their comparison with the respective data of Norwegian specimens confirm that the identification of Sarcocystis spp. solely on the basis of phenotypic characterization is inaccurate. In order to overcome this issue, the results of morphological analyses should be supported by DNA-based molecular techniques based on sequencing of ssu rRNA and cox1 genes (Gjerde 2013).
The analysis of ssu rRNA and cox1 genes from sarcocysts isolated from Polish roe deer revealed the presence of the same three species: S. gracilis, S. silva, and S. oviformis, as in Norwegian roe deer (Dahlgren and Gjerde 2009; Gjerde 2012). However, we were unable to isolate the fourth Sarcocystis species found in roe deer, S. capreolicanis (Gjerde 2012), probably due to the limited number of available samples. The sequence analysis of ssu rRNA gene revealed the lack of differences between Sarcocystis isolates belonging to one species, and a very low degree of genetic diversity, close to zero, between Polish and Norwegian sarcocysts (the intraspecific variation rate was 99.9 % for S. gracilis and S. oviformis and 99.56 % for S. silva). Thus, the Sarcocystis specimens representing three species isolated from roe deer living in different geographical areas displayed a very low level of genetic diversity.
Contrary to the results of ssu rRNA analysis, small intraspecies differences in cox1 sequences were found among the Polish Sarcocystis isolates. Five haplotypes were found when S. gracilis isolates were compared to each other, with average identity of 99.70 %. Similarly, four haplotypes were identified on comparison of S. silva isolates, which corresponded to a 99.17 % identity on average. In turn, only one haplotype was found in the case of S. ovifiormis.
The comparison of Polish and Norwegian isolates representing the same Sarcocystis species revealed similar degree of sequence identity, namely 99.72 % for S. gracilis, 98.76 % for S. silva, and 99.85 % for S. oviformis.
High degree of identity between Polish and Norwegian isolates was further confirmed by the results of genetic population analysis; the values of haplotype diversity (H d), average number of pairwise differences (theta-π), and nucleotide diversity (π) turned out to be similar for sequences obtained in Norway and Poland. On phylogenetic reconstruction, Polish isolates were not separated from their Norwegian counterparts. This conclusion was further supported by spatially nonrestricted display of haplotypes on heat maps (Fig. 3).
Gjerde (2013, 2014) put into question the usefulness of ssu rRNA gene in phylogenetic studies of Sarcocystis; this author found that discriminatory power of analyses based on sequences of cox1 gene is higher than the analyses based on the sequencing of ssu rRNA gene. This observation is consistent with data published by Ogedengbe et al. (2011), who showed that partial cox1 sequences are more useful for identifying Eimeria species than nearly complete ssu rRNA gene sequences. Moreover, cox1 gene sequences turned out to be more accurate when used to reveal phylogenetic relationships among closely related taxa. This is confirmed by our findings presented in Table 5, illustrating pairwise distances between species for S. gracilis, S. oviformis, and S. silva determined on the basis of ssu rRNA and cox1 sequences. The genetic matrix based on cox1 sequences points to a higher resolution of interspecies genetic differences than in the case of ssu rRNA sequences. Similar results were obtained in the case of other species subjected to phylogenetic analyses (data not shown).
To summarize, our study revealed that the same Sarcocystis species isolated from the same hosts living in different geographic regions show a considerable level of genetic similarity. These results are consistent with previously published findings by Gjerde (2014), who observed that Sarcocystis spp. from cervids, namely S. elongata and S. truncata, show a low degree of intraspecies variation in the ssu rRNA gene. Furthermore, the other species (e.g., S. gracilis, S. oviformis) found in cervids showed little or no variation in this gene, which suggests that (nearly) identical sequences can be obtained from different sarcocysts isolated from different animals. The limited genetic diversity of Sarcocystis isolates belonging to the same species was also observed in the case of Sarcocystis neurona; in the case of this species, analysis of multiple genetic markers turned out to be insufficient to distinguish between terrestrial isolates and strains infecting marine mammals (Wendte et al. 2010). The results of a study of closely related species Toxoplasma gondii (Grigg and Sundar 2009) suggest that the phenomenon of limited genetic diversity among Sarcocystis isolates can result from the ability of few haplotypes to clonal propagation. However, further research, including more isolates from various countries and continents, is required in order to confirm this hypothesis.
References
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1016/S0022-2836(05)80360-2
Dahlgren SS, Gjerde B (2007) Genetic characterisation of six Sarcocystis species from reindeer (Rangifer tarandus tarandus) in Norway based on the small subunit rRNA gene. Vet Parasitol 146:204–213. doi:10.1016/j.vetpar.2007.02.023
Dahlgren SS, Gjerde B (2009) Sarcocystis in Norwegian roe deer (Capreolus capreolus): molecular and morphological identification of Sarcocystis oviformis n. sp. and Sarcocystis gracilis and their phylogenetic relationship with other Sarcocystis species. Parasitol Res 104:993–1003. doi:10.1007/s00436-008-1281-0
Dahlgren SS, Gjerde B (2010) Molecular characterization of five Sarcocystis species in red deer (Cervus elaphus), including Sarcocystis hjorti n. sp., reveals that these species are not intermediate host specific. Parasitology 137:815–840. doi:10.1017/S0031182009991569
Dahlgren SS, Gjerde B, Skirnisson K, Gudmundsdottir B (2007) Morphological and molecular identification of three species of Sarcocystis in reindeer (Rangifer tarandus tarandus) in Iceland. Vet Parasitol 149:191–198. doi:10.1016/j.vetpar.2007.08.015
Dahlgren SS, Gouveia-Oliveira R, Gjerde B (2008) Phylogenetic relationships between Sarcocystis species from reindeer and other Sarcocystidae deduced from ssu rRNA gene sequences. Vet Parasitol 151:27–35. doi:10.1016/j.vetpar.2007.09.029
Dubey J, Rommel M (1992) Durch Protozoen bedingte Aborte bei landwirtschaftlichen Nutztieren. Deut Tierarztl Wochenschr 99:355–362
Dubey J, Speer C, Fayer R (1988) Sarcocystosis of animals and man. CRC Press, Boca Raton, pp 1–125
Ellis TJ, Luton K, Baverstock PR et al (1995) Phylogenetic relationships between Toxoplasma and Sarcocystis deduced from a comparison of 18S rDNA sequences. Parasitology 110(Pt 5):521–528
Entzeroth R (1981) Untersuchungen an Sarkosporidien (Mieschersche Schläuche) des einheimischen Rehwildes (Capreolus capreolus L.). Z Jagdwiss 27:247–257. doi:10.1007/BF02243674
Entzeroth R (1982) A comparative light and electron microscope study of the cysts of Sarcocystis species of roe deer (Capreolus capreolus). Parasitol Res 66:281–292. doi:10.1007/BF00925345
Enzeroth R (1985) Light-, scanning-, and transmission electron microscope study of the cyst wall of Sarcocystis gracilis Ràtz, 1909 (Sporozoa, Coccidia) from the Roe Deer (Capreolus capreolus L.). Arch Protistenkd 129:183–186. doi:10.1016/S0003-9365(85)80020-8
Erber M, Boch J, Barth D (1978) Drei Sarkosporidienarten des Rehwildes. Berl Munch Tierärztl Wochenschr 91:482–486
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. doi:10.1111/j.1755-0998.2010.02847.x
Fayer R (2004) Sarcocystis spp. in Human Infections. Clin Microbiol Rev 17:894–902. doi:10.1128/CMR.17.4.894-902.2004
Fayer R, Elsasser TH (1991) Bovine sarcocystosis: how parasites negatively affect growth. Parasitol Today 7:250–255. doi:10.1016/0169-4758(91)90242-G
Fenger CK, Granstrom DE, Langemeier JL et al (1994) Phylogenetic relationship of Sarcocystis neurona to other members of the family Sarcocystidae based on small subunit ribosomal RNA gene sequence. J Parasitol 80:966–975
Gjerde B (1985) Ultrastructure of the cysts of Sarcocystis grueneri from cardiac muscle of reindeer (Rangifer tarandus tarandus). Parasitol Res 71:189–198
Gjerde B (2012) Morphological and molecular characterization and phylogenetic placement of Sarcocystis capreolicanis and Sarcocystis silva n. sp. from roe deer (Capreolus capreolus) in Norway. Parasitol Res 110:1225–1237. doi:10.1007/s00436-011-2619-6
Gjerde B (2013) Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 43:579–591. doi:10.1016/j.ijpara.2013.02.004
Gjerde B (2014) Sarcocystis species in red deer revisited: with a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology 141:441–452. doi:10.1017/S0031182013001819
Grigg ME, Sundar N (2009) Sexual recombination punctuated by outbreaks and clonal expansions predicts Toxoplasma gondii population genetics. Int J Parasitol 39:925–933. doi:10.1016/j.ijpara.2009.02.005
Hernandez S, Martinez-Gomez F, Navarrete I, Acosta-Garcia I (1981) Estudio al microscopio optico y electronico del quiste de Sarcocystis cervicanis. Rev Iber Parasitol 41:351–361
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Jeffrey M (1993) Sarcocystosis of sheep. Practice 15:2–8. doi:10.1136/inpract.15.1.2
Johnson AM, Illana S, Hakendorf P, Baverstock PR (1988) Phylogenetic relationships of the apicomplexan protist Sarcocystis as determined by small subunit ribosomal RNA comparison. J Parasitol 74:847–860
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
O’Donoghue PJ, Adams M, Dixon BR et al (1986) Morphological and biochemical correlates in the characterization of Sarcocystis spp. J Protozool 33:114–121
Ogedengbe JD, Hanner RH, Barta JR (2011) DNA barcoding identifies Eimeria species and contributes to the phylogenetics of coccidian parasites (Eimeriorina, Apicomplexa, Alveolata). Int J Parasitol 41:843–850. doi:10.1016/j.ijpara.2011.03.007
Pérez-Creo A, Panadero R, López C et al (2013) Prevalence and identity of Sarcocystis spp. in roe deer (Capreolus capreolus) in Spain: a morphological study. Res Vet Sci 95:1036–1040. doi:10.1016/j.rvsc.2013.08.003
Santini S, Mancianti F, Nigro M, Poli A (1997) Ultrastructure of the cyst wall of Sarcocystis sp. in roe deer. J Wildl Dis 33:853–859. doi:10.7589/0090-3558-33.4.853
Schramlová J, Blažek K (1978) Ultrastruktur der Cystenwand der Sarkosporidien des Rehes (Capreolus capreolus L.). Parasitol Res 55:43–48. doi:10.1007/BF00383473
Sedlaczek J, Wesemeier HH (1995) On the diagnostics and nomenclature of Sarcocystis species (Sporozoa) in roe deer (Capreolus capreolus). Appl Parasitol 36:73–82
Speer C, Dubey J (1982) Sarcocystis wapiti sp. nov. from the North American wapiti (Cervus elaphus). Can J Zool 60:881–888. doi:10.1139/z82-120
Stojecki K, Karamon J, Sroka J, Cencek T (2012) Molecular diagnostics of Sarcocystis spp. infections. Pol J Vet Sci 15:589–596
Stolte M, Bockhardt I, Odening K (1996) A comparative scanning electron microscopic study of the cyst wall in 11 Sarcocystis species of mammals. J Zool (Lond) 239:821–832. doi:10.1111/j.1469-7998.1996.tb05480.x
Stolte M, Bockhardt I, Odening K (1997) First report of Sarcocystis rangiferi and second Sarcocystis species with parasite-induced encapsulation in cervids from Central Europe. Acta Protozool 36:131–135
Stolte M, Bockhardt I, Odening K (1998) Scanning electron microscopic identification of Sarcocystis gracilis from roe deer and cattle. J Zool (Lond) 244:265–268
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Tenter AM (1995) Current research on Sarcocystis species of domestic animals. Int J Parasitol 25:1311–1330
Tenter AM, Baverstock PR, Johnson AM (1992) Phylogenetic relationships of Sarcocystis species from sheep, goats, cattle and mice based on ribosomal RNA sequences. Int J Parasitol 22:503–513
Tenter AM, Luton K, Johnson AM (1994) Species-specific identification of Sarcocystis and Toxoplasma by PCR amplification of small subunit ribosomal RNA gene fragments. Appl Parasitol 35:173–188
Wendte JM, Miller MA, Nandra AK et al (2010) Limited genetic diversity among Sarcocystis neurona strains infecting southern sea otters precludes distinction between marine and terrestrial isolates. Vet Parasitol 169:37–44. doi:10.1016/j.vetpar.2009.12.020
Acknowledgments
This work was supported by the BMBF (Federal Ministry of Education and Research, Germany) project InnoProfile-Transfer 03IP611X.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
GenBank accession numbers of ssu rRNA and cox1 gene sequences used for phylogenetic reconstructions. (DOCX 12 kb)
Table S2
Nucleotide variation in ssu rRNA gene sequences of Polish (KF880743) and Norwegian (JN226122-JN226125, EU282016) S. silva isolates. Nucleotides identical to the nucleotide sequence of KF880743 are marked by dots. Dashes indicate sites with no nucleotide aligned. The site numbers on the top of each gene sequence correspond to position of nucleotides in KF880743 sequence. (DOCX 13 kb)
Table S3
Intraspecific similarity (%) between partial cox1 gene sequences of S. gracilis isolates from different geographical areas. Isolates belonging to given haplotypes: F898100= KF898103= KF898104= KC209614= KC209613= KF241338= KF241337; KF898102= KF241333, KF898106= KC209612= KC209611= KC209605= KF241335= KF241334= KF241331= KF241330; KC209609= KF241340; KF241341= KF241336; KF241339= KF241332 (DOCX 14 kb)
Table S4
Intraspecific similarity (%) between partial cox1 gene sequences of S. oviformis isolates from different geographical areas. List of isolates belonging to a given haplotype: KF898107 = KF898108 = KF898109; KC209659 = KC209656 = KC209658; KC209661 = KC209657 = KC209660. (DOCX 12 kb)
Table S5
Intraspecific similarity (%) between partial cox1 gene sequences of S. silva isolates from different geographical areas. (DOCX 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Kolenda, R., Ugorski, M. & Bednarski, M. Molecular characterization of Sarcocystis species from Polish roe deer based on ssu rRNA and cox1 sequence analysis. Parasitol Res 113, 3029–3039 (2014). https://doi.org/10.1007/s00436-014-3966-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-014-3966-x