Abstract
Purpose
Gastric cancer (GC) is prevalent as one of the most common malignant tumors globally, with a particularly high incidence in China. The role of UBE2L3 in the initiation and progression of various cancers has been well documented, but its specific significance in GC is not yet fully elucidated. The objective of this study is to examine the expression and importance of UBE2L3 in human gastric cancer tissues.
Methods
Immunohistochemical staining and survival analysis were conducted on 125 cases of GC. Western blot and quantitative real-time polymerase chain reaction (qRT-PCR) were employed to assess the expression of UBE2L3 in GC cell lines. Cell lines with UBE2L3 knockdown and overexpression were cultured through lentivirus transfection and subsequently assessed using Western blot analysis. The involvement of UBE2L3 in the proliferation, invasion, and apoptosis of GC cells was confirmed through in vitro experiments, and its capacity to facilitate tumor growth was also validated in in vivo studies.
Results
The up-regulation of UBE2L3 expression was observed in GC, and its high expression was found to be significantly associated with the degree of differentiation (χ2 = 6.153, P = 0.0131), TNM stage (χ2 = 6.216, P = 0.0447), and poor overall survival. In vitro, UBE2L3 has been shown to enhance functions in GC cell lines, such as promoting proliferation and invasion, and inhibiting apoptosis. In vivo experiments have validated the role of UBE2L3 in promoting tumor growth.
Conclusions
The findings of our study demonstrate the significant involvement of UBE2L3 in the pathogenesis and advancement of gastric cancer, suggesting its potential as a therapeutic target.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is a widespread disease and a significant threat to human health (Bray et al. 2018; Smyth et al. 2020). According to the GLOBOCAN 2020 report, the incidence of GC ranked fifth and its mortality ranked fourth among all types of tumors (Smyth et al. 2020; Ferlay 2020. Accessed on September 30, 2021). In 2020, the global incidence of new cases exceeded one million (1,089,103), leading to 768,793 fatalities (Ferlay 2020. Accessed September 30, 2021). In recent years, the overall incidence and mortality of GC have shown a stable decrease, attributed to advancements in early diagnosis, standardization of treatment methods including surgery, neoadjuvant therapy, and targeted therapy (Yonemura et al. 2016; Tan 2019; Kawazoe et al. 2020; Sexton et al. 2020). Despite significant progress being achieved in the last decade, the long-term survival of patients with advanced GC still remains unsatisfactory. The 5-year survival rate following radical gastrectomy and chemotherapy for patients with GC varies from 30 to 50% (Cats et al. 2018; Al-Batran et al. 2019). Consequently, there is an urgent need to identify more precise and efficacious therapeutic targets.
The ubiquitin-conjugating enzyme E2 (E2) plays a crucial role in the ubiquitin–proteasome pathway by facilitating the transfer of ubiquitin to the substrate protein. As an E2 enzyme, UBE2L3 facilitates the ubiquitination process by catalyzing a variety of substrate proteins in conjunction with E1s and E3s. UBE2L3 is situated on chromosome 22 q11.2-13.1 and codes for 153 amino acid residues (Clague et al. 2015). An increasing body of research has demonstrated the involvement of UBE2L3 in the pathogenesis of various diseases through the regulation of protein stability. These diseases include systemic lupus erythematosus (SLE) (Kim et al. 2020; Mauro et al. 2023), hepatitis B virus (HBV) infection (Zhou et al. 2019), Alzheimer’s disease (AD) (Wei et al. 2019), ischemic stroke (IS) (Wei et al. 2019), celiac disease (CeD) (Fernandez-Jimenez and Bilbao 2019), and rheumatoid arthritis (RA) (Zeng et al. 2021). Furthermore, it has been observed that UBE2L3 exhibits abnormally high expression in various human tumor types, indicating a potential role for UBE2L3 as an oncogene. UBE2L3 has been demonstrated to facilitate the migration of cervical cancer cells (Weinberg et al. 2020; Yi et al. 2020a, b). Furthermore, several studies have suggested that the depletion of UBE2L3 inhibits the proliferation and induces apoptosis of hepatocellular carcinoma cells (Tao et al. 2020). Additionally, UBE2L3 in promoting the migration and invasion of lung cancer cells (Ma et al. 2022), and can be used as a potential response biomarker to enhance the efficacy of HSP90 inhibitors against lung adenocarcinoma (Marrugal et al. 2023). Another investigation demonstrated that UBE2L3 facilitated the cellular malignant characteristics of oral squamous cell carcinoma, such as proliferation, invasion, migration, and in vivo tumor growth (Cui et al. 2022). Nevertheless, the specific function of UBE2L3 in gastric cancer remains unclear.
This study aimed to examine the clinical significance of UBE2L3 expression in human GC tissues, explore the relationship between UBE2L3 expression and clinicopathological features, and assess its prognostic value using a multi-experimental approach.
Materials and methods
The analysis of gene expression data
The UBE2L3 gene expression analysis of various tumors can be accessed from the GEPIA database (http://gepia.cancer-pku.cn/index.html).
Tissue specimens
Human tissue specimens were obtained from GC patients who underwent surgical excision at the Second Hospital of Lanzhou University between December 2016 and June 2017. The informed consent was signed by all patients, and the study protocol received approval from the Ethics Committee of the Second Hospital of Lanzhou University. The study included 125 patients with primary gastric cancer, with 11 patients excluded due to less than 5 years of follow-up. The cohort consisted of 114 patients, comprising 87 men and 27 women, all of whom did not undergo chemotherapy or radiotherapy before the surgical procedure, and all completed a 5-year follow-up period. All 114 specimens underwent confirmation through pathological examination and TNM staging.
Cell culture
The six cell lines, namely GES-1, MKN45, MKN28, N87, AGS, and HGC27, were acquired from the Digestive Tumor Laboratory of Gansu Province. The six cell lines were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA), and maintained in a humidified incubator at 37 °C with a 5% CO2 atmosphere.
Lentiviral transfection
In the MKN45, MKN28, and AGS cell lines, the experimental group consisted of cells transfected with lentivirus obtained from GenePharma, China, while the negative control group comprised cells transfected with an empty lentiviral vector. Cell transfection was conducted using Invitrogen following the manufacturer’s protocol. Cells were cultured in six-well plates (Corning, USA) until reaching a cell density of 20–30%. Subsequently, the cells were harvested 18 h after transfection with lentivirus. After 72 h of continuous culture, the cells were subjected to puromycin screening for an additional 72 h. The transfection efficiency was assessed by observing green fluorescent protein expression under a microscope, and it was found that the fluorescence efficiency exceeded 80%, indicating a successful transfection.
Cell viability assay
In this study, the cell proliferation potential of GC cells was assessed using the Cell Counting Kit-8 (CCK-8) assay kit (Solarbio, China). The cells (MKN45, MKN28, AGS) were seeded at a density of 1000 cells per well in 96-well plates (Corning, USA). The medium in each well was removed at different time intervals (0, 24, 48, 72, and 96 h). Subsequently, 10 μL of the prepared CCK-8 reagent was introduced into each well and incubated for 1 h at 37 °C in a light-sheltered incubator. Following incubation, the absorbance value (OD) was assessed at a wavelength of 450 nm.
Colony-forming assay
The cells were cultured at a density of 500 cells per well in 6-well plates. The cells were cultured without disturbance for a period of two weeks to facilitate the formation of proliferating colonies. Subsequently, the proliferating cells underwent two gentle washes with phosphate-buffered saline (PBS) to eliminate any non-adherent cells. Subsequently, the cells were treated with paraformaldehyde for 15 min and then subjected to staining with 0.1% crystal violet (Solarbio, China) for an additional 15 min. The number of colonies was determined using Image J software.
Cell invasion assay
The cells that were treated with trypsin were subsequently placed in the upper chamber of the transwell system (Corning, USA) that was coated with Matrigel (BD Bioscience). The cell count was 1 × 105 cells per upper chamber. The cells underwent two gentle washes with PBS following 18–72 h of incubation. Subsequently, the cells underwent fixation and staining using the identical procedure employed in the colony formation assay. The number of invasive cells was visualized and quantified.
Apoptosis assay
Apoptosis in GC cells was evaluated using flow cytometry. To analyze apoptosis, the apoptosis rate of adherent cells was assessed using the AnnexinV-APC/7-ADD apoptosis kit (Multisciences, China). The GC cell samples were suspended again in a binding buffer. 5 μl of Annexin V-APC and 10 μl of 7-AAD were added to each tube, followed by a 5-min incubation at room temperature in the dark. At the conclusion of the incubation period, the samples underwent processing using flow cytometry (Canto BD).
Western blot analysis
Proteins were initially extracted using RIPA buffer containing a protease inhibitor cocktail (Beyotime), and the quantification of protein levels was determined using the BCA assay (Beyotime). Following the isolation of proteins, the entire protein content was separated using 10% SDS-PAGE and subsequently transferred to PVDF membranes for Western blot analysis. Subsequently, the membranes were blocked with 5% skim milk for 1 h, followed by sequential incubation with primary antibodies overnight and secondary antibodies for 1 h. To analyze and document the protein bands, the membrane underwent imaging using the enhancement chemiluminescence reagent (Biosharp, China) and a chemiluminescence imager (Tanon 4600, China). The antibodies used for Western blot analysis were as follows: anti-UBE2L3 (dilution 1:10,000, Abcam, USA) and anti-GAPDH (dilution 1:5000, ImmunoWay Biotechnology, USA).
Quantitative real-time PCR (qRT-PCR)
The total RNA was isolated using AG RNAex Pro Reagent (Accurate Biology, China), followed by RNA conversion to cDNA, and subsequently subjected to quantitative real-time PCR using a CFX96 Touch qRT-PCR system (Bio-Rad). GAPDH was employed as an internal control for mRNA analysis. The values of the target genes were determined using the 2−ΔΔCt method. The primer sequences have been documented in Table 1.
Subcutaneous xenograft model in nude mice
In this investigation, two cohorts of stably transfected MKN45 cells (1 × 106 cells) were subcutaneously administered to female Balb/c nude mice aged 5–6 weeks old, obtained from Chengdu Yaokang Biotechnology Co., LTD. Tumor dimensions were assessed at 3-day intervals over a period of 21 days. Subsequently, the mice were euthanized following inhalation of ether anesthesia, and the xenograft tumors were removed, weighed, and then stored in a refrigerator at − 80 ℃ after resection for future experiments.
Immunohistochemical and scoring method
The immunohistochemical method was used to detect the expression level of UBE2L3 protein in human tissues and tumor tissues of nude mice. Tissue microarrays were created using gastric cancer (GC) tissues and adjacent normal tissues from 125 GC patients. Each sample was represented by two cores with a diameter of 1mm. Subsequently, the tumor tissue samples in nude mice were fixed, embedded in paraffin, and sectioned into 5 µm slices. Subsequently, the sections underwent antigen retrieval, followed by overnight incubation at 4 °C with anti-UBE2L3 antibody (1:500, Abcam, USA) and anti-Ki67 antibody (1:500, Servicebio, China). Following incubation with secondary antibodies, the sections underwent staining with 3,3ʹ-diaminobenzidine, and subsequent incubation with hematoxylin. Two pathologists assessed UBE2L3 immunostaining based on the percentage of stained cells and the intensity of staining in each section. The scoring system was determined by the proportion of stained cells in the positive area (0 = 0–5%, 1 = 6–25%, 2 = 26–50%, 3 = 51–75%, 4 = 76–100%) and the intensity of staining (0 = no staining, 1 = weak staining, 2 = moderate staining, 3 = strong staining). The immunohistochemical score is calculated by multiplying the score of the staining area by the score of the staining intensity. The samples were subsequently categorized as negative (-, 0 points), weakly positive (+ , 1–4 points), moderately positive (+ + , 5–8 points), and strongly positive (+ + +, 9–12 points).
Statistical analysis
The study results were analyzed using GraphPad Prism (version 8.0, USA). The study utilized a Student’s t-test to compare two groups and a one-way analysis of variance (ANOVA) to compare multiple groups. The results of the study were reported as mean ± standard deviation (SD). P values less than 0.05 were deemed to be statistically significant. The chi-square test was employed to assess the correlation between UBE2L3 expression and clinicopathological data. Survival analysis was evaluated using the log-rank test.
Results
UBE2L3 gene expression in tumors
The GEPIA public database was utilized for the collection of UBE2L3 mRNA expression data in various tumor tissues, with the aim of further investigating the UBE2L3 mRNA expression in GC tissues. The various cancer tissues examined in this study comprised pancreatic cancer (n = 179) and normal pancreatic tissue (n = 171), gastric cancer (n = 408) and normal gastric tissue (n = 211), cholangiocarcinoma (n = 36) and normal bile duct tissue (n = 9), and normal pancreatic tissue (n = 10). Thymoma (n = 118) and normal thymic tissue (n = 339) were compared, as well as diffuse large B-cell tumor (n = 47) and normal tissue (n = 337). The data presented in this study provide insight into the high expression of UBE2L3 in various human cancers, suggesting that UBE2L3 may be involved in the pathogenesis of malignant tumors (Fig. 1A).
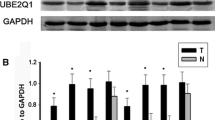
The expression of UBE2L3 in individuals diagnosed with gastric cancer and its association with prognosis. A UBE2L3 mRNA expression in different cancer tissues was assessed using the Gene Expression Profile Interaction Analysis (GEPIA) database. B Representative images illustrating UBE2L3 immunohistochemical staining in GC tissues and adjacent normal tissues. C The expression level of UBE2L3 in GC tissues was significantly higher compared to that in adjacent normal tissues. D High level of UBE2L3 was found to be correlated with a poorer overall survival (OS) in patients diagnosed with GC. The abbreviations STAD, THAM, PAAD, DLBC, and CHOL represent gastric adenocarcinoma, thymoma, pancreatic adenocarcinoma, diffuse large B-cell lymphoma, and cholangiocarcinoma, respectively
Upregulation of UBE2L3 in gastric cancer tissues has been found to be associated with a poor prognosis for patients
To clarify the potential clinical relevance of UBE2L3 in GC, we initially assessed the differential expression of UBE2L3 in GC tissues compared with adjacent normal tissues using immunohistochemical (IHC) staining on 125 specimens from 125 GC patients. As depicted in Fig. 1B, IHC data indicated that UBE2L3 is primarily expressed in the cytoplasm and nucleus of GC cells. The expression level of UBE2L3 in GC tissues was significantly higher than that in adjacent tissues (see Fig. 1C). Utilizing the median IHC score of UBE2L3, we segregated 114 GC patients into two categories: those with low UBE2L3 expression and those with high UBE2L3 expression. Table 2 illustrates the correlation between UBE2L3 expression and clinical pathological characteristics. The analysis of Table 2 reveals a strong correlation between the high expression of UBE2L3 in GC and the degree of differentiation (× 2 = 6.153, P = 0.0131) as well as TNM classification (× 2 = 6.216, P = 0.0447). However, no significant differences were observed in relation to age, gender, histological type, tumor size, and Lauren classification (refer to Table 2). As depicted in Fig. 1D, individuals exhibiting high UBE2L3 expression demonstrate a lower overall survival rate compared to those with low UBE2L3 expression. The CT data of the patients was collected over a 5-year follow-up period. In Fig. 2A, it was observed that Patient A did not develop metastases during the follow-up period, while Patient B was diagnosed with liver and lung metastases 24 months into the follow-up. A total of 23 patients were identified with metastases at various sites and at different times following surgery during the follow-up period. The specific patient data is depicted in Fig. 2B. Based on the IHC findings, the patients were categorized into two groups: those with metastasis and those without metastasis, and further divided into UBE2L3 high-expression group and UBE2L3 low-expression group. The findings indicated a significantly higher expression level of UBE2L3 in gastric cancer tissues compared to adjacent tissues in patients with and without metastasis, as illustrated in Fig. 2C and D. In summary, the heightened expression of UBE2L3 indicates an unfavorable prognosis in patients with GC.
A The absence of GC metastasis in patient A during the follow-up period, whereas patient B developed liver and lung metastasis at 24 months (red arrow indicates the primary lesion, yellow arrow indicates the surgical site, blue arrow indicates liver metastasis, and green arrow indicates lung metastasis). B This section presents the clinical characteristics of 23 patients who experienced metastasis at various time points following their surgical procedures. C UBE2L3 expression levels of UBE2L3 in patients with metastasis. D UBE2L3 expression levels of UBE2L3 in patients without metastasis were examined
UBE2L3 expression in GC cells and construction of UBE2L3 knockdown and overexpress cells models
The expression of UBE2L3 in GC was assessed by conducting Western blot analysis and quantitative real-time polymerase chain reaction (qRT-PCR) on five human GC cell lines and GES-1. The results indicate that UBE2L3 exhibited high expression in MKN45 and MKN28, and low expression in AGS (Fig. 3A, C). Subsequently, three UBE2L3 shRNAs were employed to suppress the expression of UBE2L3 in MKN28 and MKN45, while UBE2L3 was overexpressed in AGS to ascertain its function in GC. The transfection efficiency was assessed by observing the presence of green fluorescent proteins in MKN28, MKN45, and AGS cells. The experimental results indicated that the transfection efficiency exceeded 80% in all three cell lines (Fig. 3D, E). Furthermore, the transfection efficiency was assessed through Western blot analysis, revealing that two UBE2L3 knockdown cell lines and one UBE2L3 overexpressing cell line successfully attained the anticipated UBE2L3 protein levels (Fig. 3B). The aforementioned data suggests that the establishment of UBE2L3 knockdown and overexpression cell models has been successful, rendering them suitable for subsequent experiments.
The expression of UBE2L3 in various cell lines and the establishment of a lentivirus transfection model. A The expression of UBE2L3 protein in six cell lines. B MKN45, MKN28, and AGS cells were transfected with lentivirus, and the expression of UBE2L3 was assessed using Western blot analysis. C UBE2L3 mRNA expression of UBE2L3 was assessed in six different cell lines. D Transfection efficiency of lentivirus: the transfection efficiency of lentivirus in three cell lines was found to be above 80%. E 72 h after transfection, the fluorescence of the green fluorescent protein on the lentiviral vector was observed
UBE2L3 regulates GC cells proliferation, colony formation, invasion, and apoptosis
Based on the discovery that UBE2L3 levels are linked to the prognosis of GC, we employed the following experimental approach to validate the functional role of UBE2L3 in the malignant behavior of GC in vitro. The study revealed that UBE2L3 promotes the proliferation rate of GC cells, as demonstrated by the CCK-8 assay results presented in Fig. 4A. Furthermore, the cells’ proliferation ability was verified through the colony formation assay. A reduced number of colonies were observed in the UBE2L3 knockdown group, while the overexpression group exhibited the opposite outcome (Fig. 4B). Subsequently, the Transwell assay was used to evaluate the invasive capacity of GC cells. Likewise, the suppression of UBE2L3 hindered cellular passage through the matrigel, whereas the up-regulation of UBE2L3 facilitated increased cellular invasion through the matrigel (Fig. 4C). Finally, the results of the apoptosis assay revealed a significant increase in the apoptosis rate of the sh-UBE2L3 group, and a significant decrease in the UBE2L3 overexpression group (Fig. 4D). In conclusion, the aforementioned findings unequivocally demonstrate the significant involvement of UBE2L3 in GC.
UBE2L3 promotes tumor growth in vivo
To validate the findings of the in vitro experiments, cell-derived xenograft (CDX) models were established in nude mice through the injection of MKN45 sh-NC and MKN45 sh-UBE2L3-3 cells. The growth of subcutaneous tumors in nude mice was monitored every 3 days, and the changes in tumor volume were measured and recorded (Fig. 5B). The results demonstrate that the tumor volume of the sh-NC group was greater than that of the sh-UBE2L3-3 group (Fig. 5A). Following the euthanasia of the nude mice, the tumor tissues were extracted, weighed, and the dimensions of each tumor were recorded. The findings indicated that the tumors in the sh-UBE2L3-3 group exhibited reduced size and weight compared to those in the sh-NC group (Fig. 5C). Finally, the findings from the hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) analysis (Fig. 5D), conducted on tumor tissues, revealed that the levels of UBE2L3 and Ki67 expression were higher in the sh-NC group compared to the sh-UBE2L3-3 group. The collective data presented above suggest that the suppression of UBE2L3 hinders the growth of gastric cancer in an in vivo setting.
UBE2L3 enhances the growth of GC in an in vivo setting. A Inhibition of UBE2L3 knockdown suppressed the growth of GC in vivo. B Graphs depicting the growth of tumors over time. C Weight of the tumor. D Immunohistochemistry (IHC) staining was performed to detect the expression of UBE2L3 and Ki67 in xenograft tumors
Conclusion
The human genome contains an estimated 40 genes that encode E2s. The genes were categorized into four classes and exhibited a conserved domain that included catalytic Cysteine residues (Stewart et al. 2016; Welsh et al. 2022). In recent years, there has been increasing research interest in the role of E2s in tumors. UBE2T, functioning as an E2 enzyme, has been identified as a crucial regulator of tumor advancement in various types of cancer, including gastric cancer (Yu et al. 2021), hepatocellular carcinoma (Sun et al. 2020), pancreatic cancer (Jiang et al. 2023), lung adenocarcinoma (Zhu et al. 2021), retinoblastoma (Xu et al. 2022), and glioblastoma (Huang et al. 2020). Furthermore, Qi et al. reported that the overexpression of UbcH5c was linked to a negative prognosis in pancreatic cancer (Qi et al. 2022). Another study has confirmed that UBE2J1 inhibits the progression of colorectal cancer by enhancing the ubiquitination and degradation of RPS3 (Wang et al. 2023). Furthermore, UBE2M and UBE2F, functioning as E2 enzymes, facilitate the transfer of NEDD8 to NEDD8 E3 ligase (Zheng et al. 2021). Several studies have demonstrated their overexpression in various types of cancer, such as hepatocellular carcinoma, breast cancer, lung adenocarcinoma, osteosarcoma, and esophageal squamous cell carcinoma (Zhou et al. 2017; Heo et al. 2020; Wang et al. 2020; Zheng et al. 2021). Moreover, an additional E2 enzyme, UBE2C, has been found to facilitate the proliferation and viability of lung carcinoma cells harboring Kras mutations. UBE2C was also found to be essential for the development of Krasg12d-induced lung tumors (Zhang et al. 2023). Regarding UBE2L3, its biological functions have been reported in hepatocellular carcinoma, cervical cancer, and lung adenocarcinoma (Tao et al. 2020; Yi et al. 2020a, b; Marrugal et al. 2023).
The study revealed that the expression levels of UBE2L3 mRNA were significantly elevated in the majority of gastric cancer tissues compared to adjacent non-tumor gastric tissues, as determined through analysis of publicly available databases. Furthermore, we verified the increased expression of UBE2L3 in 125 GC tissues compared to adjacent tissue through immunohistochemistry analysis. Significantly, it was observed that elevated UBE2L3 expression was associated with the degree of differentiation, TNM classification, and prognosis in patients with GC. Furthermore, out of the 125 patients, 23 had metastasis. The expression level of UBE2L3 in GC tissues was significantly higher than that in adjacent normal tissues, both in patients with and without metastasis. Subsequently, the biological function of UBE2L3 in gastric cancer was validated through a series of laboratory methods. The findings indicated that UBE2L3 facilitated the proliferation, clone formation, and invasion of GC cell lines, while suppressing the apoptosis of GC cell lines. In vivo experiments, UBE2L3 was found to promote tumor progression. The aforementioned findings indicate that UBE2L3 functions as an oncogene in GC.
To date, research on the molecular mechanisms of UBE2L3 in diseases has primarily concentrated on the following areas. First, UBE2L3 is involved in the regulation of the NF-κB signaling pathway. It has been reported that UBE2L3 significantly enhances NF-κB activation induced by TLR7 in SLE patients through its interaction with LUBAC (Mauro et al. 2023). Taehyeung Kim and colleagues observed an up-regulation of UBE2L3 and a down-regulation of TNFAIP3 in the context of NF-κB activation, which subsequently led to the induction of inflammatory factors, suggesting a synergistic role (Kim et al. 2020). Second, the process of p53 polyubiquitination mediated by UBE2L3 has been found to decrease p53 stability during the progression of cervical cancer (Yi et al. 2020a, b). Third, UBE2L3 was involved in the process of lysophagy and the ubiquitination of lysosomes in conjunction with UBE2N (Shima et al. 2023). Finally, the suppression of UBE2L3 resulted in an elevation of non-homologous end joining and a reduction in homologous recombination during double-strand break repair, achieved through the regulation of 53BP1 protein levels (Han et al. 2014; Pozo et al. 2017). Nevertheless, further research is required to elucidate the mechanism through which UBE2L3 facilitates tumor initiation and progression in gastric cancer.
Recent clinical applications have confirmed the effectiveness of HER2 in targeted therapy for GC; however, the potential audience for this treatment remains limited. In the future, the treatment of gastric cancer is expected to involve a combination of targeted therapies and a gradual shift toward personalized treatment approaches. The identification of UBE2L3 and the prospect of further comprehensive investigation suggest the potential for significant advancements in targeted therapy for gastric cancer.
Data availability
The data generated and analyzed in this study were available.
References
Al-Batran S-E, Homann N, Pauligk C, Goetze TO, Meiler J, Kasper S, Kopp H-G, Mayer F, Haag GM, Luley K (2019) Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. The Lancet 393(10184):1948–1957
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Cats A, Jansen EP, van Grieken NC, Sikorska K, Lind P, Nordsmark M, Kranenbarg EM-K, Boot H, Trip AK, Swellengrebel HM (2018) Chemotherapy versus chemoradiotherapy after surgery and preoperative chemotherapy for resectable gastric cancer (CRITICS): an international, open-label, randomised phase 3 trial. Lancet Oncol 19(5):616–628
Clague MJ, Heride C, Urbé S (2015) The demographics of the ubiquitin system. Trends Cell Biol 25(7):417–426
Cui Z, Sun S, Li J, Li J, Sha T, He J, Zuo L (2022) UBE2L3 promotes squamous cell carcinoma progression in the oral cavity and hypopharynx via activating the NF-κB signaling by increasing IκBα degradation. Cell Biol Int 46(5):806–818
Ferlay J, Ervik M, Colombet M, et al. (Published 2020. Accessed September 30, 2021) Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer
Fernandez-Jimenez N, Bilbao JR (2019) Mendelian randomization analysis of celiac GWAS reveals a blood expression signature with diagnostic potential in absence of gluten consumption. Hum Mol Genet 28(18):3037–3042
Han X, Zhang L, Chung J, Mayca Pozo F, Tran A, Seachrist DD, Jacobberger JW, Keri RA, Gilmore H, Zhang Y (2014) UbcH7 regulates 53BP1 stability and DSB repair. Proc Natl Acad Sci 111(49):17456–17461
Heo MJ, Kang SH, Kim YS, Lee JM, Yu J, Kim HR, Lim H, Kim KM, Jung J, Jeong LS (2020) UBC12-mediated SREBP-1 neddylation worsens metastatic tumor prognosis. Int J Cancer 147(9):2550–2563
Huang P, Guo Y, Zhao Z, Ning W, Wang H, Gu C, Zhang M, Qu Y, Zhang H, Song Y (2020) UBE2T promotes glioblastoma invasion and migration via stabilizing GRP78 and regulating EMT. Aging (albany NY) 12(11):10275
Jiang X, Ma Y, Wang T, Zhou H, Wang K, Shi W, Qin L, Guan J, Li L, Long B (2023) Targeting UBE2T potentiates gemcitabine efficacy in pancreatic cancer by regulating pyrimidine metabolism and replication stress. Gastroenterology 164(7):1232–1247
Kawazoe A, Fukuoka S, Nakamura Y, Kuboki Y, Wakabayashi M, Nomura S, Mikamoto Y, Shima H, Fujishiro N, Higuchi T (2020) Lenvatinib plus pembrolizumab in patients with advanced gastric cancer in the first-line or second-line setting (EPOC1706): an open-label, single-arm, phase 2 trial. Lancet Oncol 21(8):1057–1065
Kim T, Bae S-C, Kang C (2020) Synergistic activation of NF-κB by TNFAIP3 (A20) reduction and UBE2L3 (UBCH7) augment that synergistically elevate lupus risk. Arthritis Res Ther 22:1–10
Ma X, Qi W, Yang F, Pan H (2022) UBE2L3 promotes lung adenocarcinoma invasion and metastasis through the GSK-3β/Snail signaling pathway. Am J Transl Res 14(7):4549
Marrugal Á, Ferrer I, Quintanal-Villalonga Á, Ojeda L, Pastor MD, García-Luján R, Carnero A, Paz-Ares L, Molina-Pinelo S (2023) Inhibition of HSP90 in driver oncogene-defined lung adenocarcinoma cell lines: key proteins underpinning therapeutic efficacy. Int J Mol Sci 24(18):13830
Mauro D, Manou-Stathopoulou S, Rivellese F, Sciacca E, Goldmann K, Tsang V, Lucey-Clayton I, Pagani S, Alam F, Pyne D, Rajakariar R, Gordon PA, Whiteford J, Bombardieri M, Pitzalis C, Lewis MJ (2023a) UBE2L3 regulates TLR7-induced B cell autoreactivity in systemic lupus erythematosus. J Autoimmun 136:103023
Pozo FM, Tang J, Bonk KW, Keri RA, Yao X, Zhang Y (2017) Regulatory cross-talk determines the cellular levels of 53BP1 protein, a critical factor in DNA repair. J Biol Chem 292(14):5992–6003
Qi S, Guan X, Zhang J, Yu D, Yu X, Li Q, Yin W, Cheng X-D, Zhang W, Qin J-J (2022) Targeting E2 ubiquitin-conjugating enzyme UbcH5c by small molecule inhibitor suppresses pancreatic cancer growth and metastasis. Mol Cancer 21(1):70
Sexton RE, Al Hallak MN, Diab M, Azmi AS (2020) Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev 39:1179–1203
Shima T, Ogura M, Matsuda R, Nakamura S, Jin N, Yoshimori T, Kuma A (2023) The TMEM192-mKeima probe specifically assays lysophagy and reveals its initial steps. J Cell Biol 222(12):e202204048
Smyth EC, Nilsson M, Grabsch HI, van Grieken NCT, Lordick F (2020) Gastric cancer. The Lancet 396(10251):635–648
Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS (2016) E2 enzymes: more than just middle men. Cell Res 26(4):423–440
Sun J, Zhu Z, Li W, Shen M, Cao C, Sun Q, Guo Z, Liu L, Wu D (2020) UBE2T-regulated H2AX monoubiquitination induces hepatocellular carcinoma radioresistance by facilitating CHK1 activation. J Exp Clin Cancer Res 39(1):1–18
Tan Z (2019) Recent advances in the surgical treatment of advanced gastric cancer: a review. Med Sci Monitor 25:3537
Tao N-N, Zhang Z-Z, Ren J-H, Zhang J, Zhou Y-J, Wong VKW, Law BYK, Cheng S-T, Zhou H-Z, Chen W-X (2020) Overexpression of ubiquitin-conjugating enzyme E2 L3 in hepatocellular carcinoma potentiates apoptosis evasion by inhibiting the GSK3β/p65 pathway. Cancer Lett 481:1–14
Wang S, Xian J, Li L, Jiang Y, Liu Y, Cai L, Hoffman RM, Jia L, Zhao H, Zhang Y (2020) NEDD8-conjugating enzyme UBC12 as a novel therapeutic target in esophageal squamous cell carcinoma. Signal Transduct Target Ther 5(1):123
Wang T, Jin C, Yang P, Chen Z, Ji J, Sun Q, Yang S, Feng Y, Tang J, Sun Y (2023) UBE2J1 inhibits colorectal cancer progression by promoting ubiquitination and degradation of RPS3. Oncogene 42(9):651–664
Wei CJ, Cui P, Li H, Lang WJ, Liu GY, Ma XF (2019) Shared genes between Alzheimer’s disease and ischemic stroke. CNS Neurosci Ther 25(8):855–864
Weinberg J, Whitcomb E, Bohm A, Chekkilla UK, Taylor A (2024) The E3 ligase SMURF1 stabilizes p27 via UbcH7 catalyzed K29-linked ubiquitin chains to promote cell migration SMURF1-UbcH7 K29 ubiquitination of p27 and cell migration. J Biol Chem
Welsh KA, Bolhuis DL, Nederstigt AE, Boyer J, Temple BR, Bonacci T, Gu L, Ordureau A, Harper JW, Steimel JP (2022) Functional conservation and divergence of the helix-turn-helix motif of E2 ubiquitin-conjugating enzymes. EMBO J 41(3):e108823
Xu N, Cui Y, Shi H, Guo G, Sun F, Jian T, Rao H (2022) UBE2T/STAT3 signaling promotes the proliferation and tumorigenesis in retinoblastoma. Invest Ophthalmol vis Sci 63(9):20–20
Yi SA, Kim GW, Yoo J, Han J-W, Kwon SH (2020a) HP1γ sensitizes cervical cancer cells to cisplatin through the suppression of UBE2L3. Int J Mol Sci 21(17):5976
Yi SA, Lee DH, Kim GW, Ryu H-W, Park JW, Lee J, Han J, Park JH, Oh H, Lee J (2020b) HPV-mediated nuclear export of HP1γ drives cervical tumorigenesis by downregulation of p53. Cell Death Differ 27(9):2537–2551
Yonemura Y, Canbay E, Li Y, Coccolini F, Glehen O, Sugarbaker P, Morris D, Moran B, Gonzaletz-Moreno S, Deraco M (2016) A comprehensive treatment for peritoneal metastases from gastric cancer with curative intent. Eur J Surg Oncol 42(8):1123–1131
Yu Z, Jiang X, Qin L, Deng H, Wang J, Ren W, Li H, Zhao L, Liu H, Yan H (2021) A novel UBE2T inhibitor suppresses Wnt/β-catenin signaling hyperactivation and gastric cancer progression by blocking RACK1 ubiquitination. Oncogene 40(5):1027–1042
Zeng Z, Sun Q-Q, Zhang W, Wen Q-W, Wang T-H, Qin W, Xiao D-M, Zhang Z, Huang H, Mo Y-J (2021) Assessment of genetic polymorphisms within nuclear factor-κB signaling pathway genes in rheumatoid arthritis: evidence for replication and genetic interaction. Int Immunopharmacol 100:108089
Zhang S, You X, Zheng Y, Shen Y, Xiong X, Sun Y (2023) The UBE2C/CDH1/DEPTOR axis is an oncogene and tumor suppressor cascade in lung cancer cells. J Clin Investig 133(4)
Zheng Y-C, Guo Y-J, Wang B, Wang C, Mamun M, Gao Y, Liu H-M (2021) Targeting neddylation E2s: a novel therapeutic strategy in cancer. J Hematol Oncol 14:1–13
Zhou W, Xu J, Li H, Xu M, Chen ZJ, Wei W, Pan Z, Sun Y (2017) Neddylation E2 UBE2F promotes the survival of lung cancer cells by activating CRL5 to degrade NOXA via the K11 linkage. Clin Cancer Res 23(4):1104–1116
Zhou L, Ren JH, Cheng ST, Xu HM, Chen WX, Chen DP, Wong VKW, Law BYK, Liu Y, Cai XF (2019) A functional variant in ubiquitin conjugating enzyme E2 L3 contributes to hepatitis B virus infection and maintains covalently closed circular DNA stability by inducing degradation of apolipoprotein B mRNA editing enzyme catalytic subunit 3A. Hepatology 69(5):1885–1902
Zhu J, Ao H, Liu M, Cao K, Ma J (2021) UBE2T promotes autophagy via the p53/AMPK/mTOR signaling pathway in lung adenocarcinoma. J Transl Med 19:1–17
Funding
This research was supported by Fundamental Research Funds for the Central Universities of Lanzhou University (lzujbky-2022-sp08); Medical Innovation and Development Project of Lanzhou University (lzuyxcx-2022-154, lzuyxcx-2022-141); Major Science and Technology Project of Gansu Province (20ZD7FA003, 22ZD6FA050, 22JR9KA002); Project of Gansu Provincial Department of Education (2021jyjbgs-02); Project of Gansu Provincial Development and Reform Commission (2020–2022); Natural Science Foundation of Gansu Province (22JR5RA983, 21JR1RA135, 23JRRA1001); Gansu key research and development program (23YFFA0037); Scientific research training program for Cuiying students in the Second Hospital of Lanzhou University (CYXZ2023-47).
Author information
Authors and Affiliations
Contributions
Conceptualization, YZ and YL; writing—original draft preparation, XZ; writing—review and editing, YW, ML, FW, CH, CH, ZL, FT WH, and YZ; visualization, XZ. All the authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare that they have no competing interest associated with the manuscript.
Ethical approval
This study was approved by the Ethics committee of the Second Hospital of Lanzhou University (D2022-236).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Wei, Y., Wu, F. et al. UBE2L3 expression in human gastric cancer and its clinical significance. J Cancer Res Clin Oncol 150, 210 (2024). https://doi.org/10.1007/s00432-024-05669-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00432-024-05669-7