Abstract
Purpose
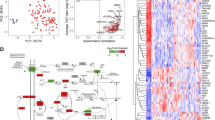
We used proteomic sequencing and experimental verification to identify the potential ferroptosis-related proteins in ameloblastoma.
Methods
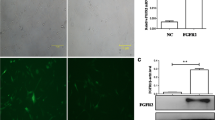
Samples of ameloblastoma (n = 14) and normal gingival tissues (n = 5) were collected for proteomic sequencing to identify differentially expressed proteins (DEPs) in ameloblastoma. Ferroptosis-related genes were downloaded from FerrDb V2, which were then compared with DEPs to obtain ferroptosis-related DEPs (FR-DEPs). A functional enrichment analysis was performed, and a protein–protein interaction network was built. The hub proteins were screened using the Cytoscape software, and potential drugs targeting them were retrieved from the DrugBank database. A hub protein was selected for immunohistochemical validation, and its expression was assessed in ameloblastomas, odontogenic keratocysts, dentigerous cysts, and normal gingival tissues. The primary ameloblastoma cells were cultured to explore the effect of the protein on the migratory properties of the tumour cells.
Results
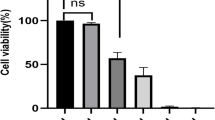
A total of 58 FR-DEPs were screened, and six hub proteins were identified: mTOR, NFE2L2, PRKCA, STAT3, EGFR, and CDH1. Immunohistochemical analysis showed that mTOR expression was upregulated in ameloblastomas compared with that in odontogenic keratocysts, dentigerous cysts, and normal gingival tissues. p-mTOR was highly expressed in ameloblastomas, with a positivity rate of 83.3%. In addition, rapamycin, an inhibitor of mTOR, can inhibit the migratory capacity of primary cultured ameloblastoma cells.
Conclusion
Our results revealed the ferroptosis-related proteins in ameloblastomas and their underlying biological processes. Additionally, mTOR was overexpressed and was found to be associated with the aggressiveness of ameloblastomas, which may be a potential target for future treatments.






Similar content being viewed by others
Data availability
All the data used in the present study are available from the corresponding author upon reasonable request.
References
Álvarez-Garcia V, Tawil Y, Wise HM, Leslie NR (2019) Mechanisms of PTEN loss in cancer: It’s all about diversity. Semin Cancer Biol 59:66–79. https://doi.org/10.1016/j.semcancer.2019.02.001
Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH (2017) Proteomics: technologies and their applications. J Chromatogr Sci 55(2):182–196. https://doi.org/10.1093/chromsci/bmw167
Ayuk, S. M., & Abrahamse, H. (2019). mTOR signaling pathway in cancer targets photodynamic therapy In Vitro. Cells, 8(5). https://doi.org/10.3390/cells8050431
Baba Y, Higa JK, Shimada BK, Horiuchi KM, Suhara T, Kobayashi M, Woo JD, Aoyagi H, Marh KS, Kitaoka H, Matsui T (2018) Protective effects of the mechanistic target of rapamycin against excess iron and ferroptosis in cardiomyocytes. Am J Physiol Heart Circ Physiol 314(3):H659-h668. https://doi.org/10.1152/ajpheart.00452.2017
Caron A, Richard D, Laplante M (2015) The roles of mTOR complexes in lipid metabolism. Annu Rev Nutr 35:321–348. https://doi.org/10.1146/annurev-nutr-071714-034355
Chaisuparat R, Yodsanga S, Montaner S, Jham BC (2013) Activation of the Akt/mTOR pathway in dentigerous cysts, odontogenic keratocysts, and ameloblastomas. Oral Surg Oral Med Oral Pathol Oral Radiol 116(3):336–342. https://doi.org/10.1016/j.oooo.2013.06.013
Chen M, Jiang Y, Sun Y (2021) KDM4A-mediated histone demethylation of SLC7A11 inhibits cell ferroptosis in osteosarcoma. Biochem Biophys Res Commun 550:77–83. https://doi.org/10.1016/j.bbrc.2021.02.137
Cui Y, Li H, Xiao T, Zhang X, Hou Y, Li H, Li X (2023) A proteomics study to explore differential proteins associated with the pathogenesis of ameloblastoma. J Oral Pathol Med. https://doi.org/10.1111/jop.13433
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B 3rd, Stockwell BR (2012) Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149(5):1060–1072. https://doi.org/10.1016/j.cell.2012.03.042
Dodson M, Castro-Portuguez R, Zhang DD (2019) NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol, 23: 101107. https://doi.org/10.1016/j.redox.2019.101107
Ebeling M, Scheurer M, Sakkas A, Pietzka S, Schramm A, Wilde F (2023) BRAF inhibitors in BRAF V600E-mutated ameloblastoma: systematic review of rare cases in the literature. Med Oncol 40(6):163. https://doi.org/10.1007/s12032-023-01993-z
Goh YC, Siriwardena B, Tilakaratne WM (2021) Association of clinicopathological factors and treatment modalities in the recurrence of ameloblastoma: Analysis of 624 cases. J Oral Pathol Med 50(9):927–936. https://doi.org/10.1111/jop.13228
Hosoi H, Dilling MB, Shikata T, Liu LN, Shu L, Ashmun RA, Germain GS, Abraham RT, Houghton PJ (1999) Rapamycin causes poorly reversible inhibition of mTOR and induces p53-independent apoptosis in human rhabdomyosarcoma cells. Cancer Res 59(4):886–894
Ikenoue T, Hong S, Inoki K (2009) Monitoring mammalian target of rapamycin (mTOR) activity. Methods Enzymol 452:165–180. https://doi.org/10.1016/s0076-6879(08)03611-2
Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, Lotan TL (2018) Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol 15(4):222–234. https://doi.org/10.1038/nrurol.2018.9
Janku F, Yap TA, Meric-Bernstam F (2018) Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 15(5):273–291. https://doi.org/10.1038/nrclinonc.2018.28
Jiang X, Stockwell BR, Conrad M (2021) Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 22(4):266–282. https://doi.org/10.1038/s41580-020-00324-8
Kim YC, Guan KL (2015) mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 125(1):25–32. https://doi.org/10.1172/jci73939
Kumamoto H, Ooya K (2007) Immunohistochemical detection of phosphorylated Akt, PI3K, and PTEN in ameloblastic tumors. Oral Dis 13(5):461–467. https://doi.org/10.1111/j.1601-0825.2006.01321.x
Lei G, Zhuang L, Gan B (2021) mTORC1 and ferroptosis: Regulatory mechanisms and therapeutic potential. Bioessays, 43(8): e2100093. https://doi.org/10.1002/bies.202100093
Li N, Zhong M, Song M (2012) Expression of phosphorylated mTOR and its regulatory protein is related to biological behaviors of ameloblastoma. Int J Clin Exp Pathol 5(7):660–667
Li N, Sui J, Liu H, Zhong M, Zhang M, Wang Y, Hao F (2015) Expression of phosphorylated Akt/mTOR and clinical significance in human ameloblastoma. Int J Clin Exp Med 8(4):5236–5244
Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, Sun B, Wang G (2020) Ferroptosis: past, present and future. Cell Death Dis 11(2):88. https://doi.org/10.1038/s41419-020-2298-2
Liu GY, Sabatini DM (2020) mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 21(4):183–203. https://doi.org/10.1038/s41580-019-0199-y
Mafi S, Mansoori B, Taeb S, Sadeghi H, Abbasi R, Cho WC, Rostamzadeh D (2021) mTOR-Mediated Regulation of Immune Responses in Cancer and Tumor Microenvironment. Front Immunol, 12: 774103. https://doi.org/10.3389/fimmu.2021.774103
Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, Li B (2019) Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 12(1):34. https://doi.org/10.1186/s13045-019-0720-y
Murugan AK (2019) mTOR: role in cancer, metastasis and drug resistance. Semin Cancer Biol 59:92–111. https://doi.org/10.1016/j.semcancer.2019.07.003
Narayan B, Urs AB, Augustine J, Singh H (2020) Role of phosphatase and tensin homolog in pathogenesis of ameloblastoma: an immunohistochemical study. J Cancer Res Ther 16(3):513–516. https://doi.org/10.4103/jcrt.jcrt_528_18
Navé BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR (1999) Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J, 344 (Pt 2):427–431
Nodit L, Barnes L, Childers E, Finkelstein S, Swalsky P, Hunt J (2004) Allelic loss of tumor suppressor genes in ameloblastic tumors. Mod Pathol 17(9):1062–1067. https://doi.org/10.1038/modpathol.3800147
Ou YN, Yang YX, Deng YT, Zhang C, Hu H, Wu BS, Liu Y, Wang YJ, Zhu Y, Suckling J, Tan L, Yu JT (2021) Identification of novel drug targets for Alzheimer’s disease by integrating genetics and proteomes from brain and blood. Mol Psychiatry 26(10):6065–6073. https://doi.org/10.1038/s41380-021-01251-6
Peterson RT, Beal PA, Comb MJ, Schreiber SL (2000) FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem 275(10):7416–7423. https://doi.org/10.1074/jbc.275.10.7416
Popova NV, Jücker M (2021) The Role of mTOR Signaling as a Therapeutic Target in Cancer. Int J Mol Sci 22(4): https://doi.org/10.3390/ijms22041743
Qiao X, Shi J, Liu J, Liu J, Guo Y, Zhong M (2021) Recurrence rates of intraosseous ameloblastoma cases with conservative or aggressive treatment: a systematic review and meta-analysis. Front Oncol, 11: 647200. https://doi.org/10.3389/fonc.2021.647200
Qiu Y, Cao Y, Cao W, Jia Y, Lu N (2020) The application of ferroptosis in diseases. Pharmacol Res, 159: 104919. https://doi.org/10.1016/j.phrs.2020.104919
Scheper MA, Chaisuparat R, Nikitakis NG, Sauk JJ (2008) Expression and alterations of the PTEN / AKT / mTOR pathway in ameloblastomas. Oral Dis 14(6):561–568. https://doi.org/10.1111/j.1601-0825.2007.01421.x
Sham E, Leong J, Maher R, Schenberg M, Leung M, Mansour AK (2009) Mandibular ameloblastoma: clinical experience and literature review. ANZ J Surg 79(10):739–744. https://doi.org/10.1111/j.1445-2197.2009.05061.x
Vered M, Wright JM (2022) Update from the 5th edition of the world health organization classification of head and neck tumors: odontogenic and maxillofacial bone tumours. Head Neck Pathol 16(1): 63–75. https://doi.org/10.1007/s12105-021-01404-7
Wang C, Aikemu B, Shao Y, Zhang S, Yang G, Hong H, Huang L, Jia H, Yang X, Zheng M, Sun J, Li J (2022) Genomic signature of MTOR could be an immunogenicity marker in human colorectal cancer. BMC Cancer 22(1):818. https://doi.org/10.1186/s12885-022-09901-w
Wang X, Liu K, Gong H, Li D, Chu W, Zhao D, Wang X, Xu D (2021) Death by histone deacetylase inhibitor quisinostat in tongue squamous cell carcinoma via apoptosis, pyroptosis, and ferroptosis. Toxicol Appl Pharmacol, 410: 115363. https://doi.org/10.1016/j.taap.2020.115363
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res, 46(D1): D1074-d1082. https://doi.org/10.1093/nar/gkx1037
Xiong G, Ouyang S, Xie N, Xie J, Wang W, Yi C, Zhang M, Xu X, Chen D, Wang C (2022) FOSL1 promotes tumor growth and invasion in ameloblastoma. Front Oncol 12: 900108. https://doi.org/10.3389/fonc.2022.900108
Yang X, Lu Y, Zhou H, Jiang HT, Chu L (2023) Integrated proteome sequencing, bulk RNA sequencing and single-cell RNA sequencing to identify potential biomarkers in different grades of intervertebral disc degeneration. Front Cell Dev Biol 11:1136777. https://doi.org/10.3389/fcell.2023.1136777
Yu L, Luo Y, Ding X, Tang M, Gao H, Zhang R, Chen M, Liu Y, Chen Q, Ouyang Y, Wang X, Zhen H (2023) WIPI2 enhances the vulnerability of colorectal cancer cells to erastin via bioinformatics analysis and experimental verification. Front Oncol 13:1146617. https://doi.org/10.3389/fonc.2023.1146617
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing.
Funding
This study was supported by a grant-in-aid for the Project of Science and Technology of Hebei Province (no. 21377719D).
Author information
Authors and Affiliations
Contributions
Conception and design were contributed by HL and XL; Manuscript was drafted by HL and XM; Provision of study materials or patients was contributed by SM, XZ and XL; All experiments were completed by HL, XM and RY; Data analysis and interpretation were contributed by RY, SM and XZ; XL revised the manuscript and were the supporters of the study. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no confict of interest.
Ethics approval
The studies involving human participants were reviewed and approved by the ethics committees of the Hospital of Stomatology of Hebei Medical University (Shijiazhuang, China) (No. [2020]005).
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.




Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, H., Ma, X., Yang, R. et al. Identification of ferroptosis-related proteins in ameloblastoma based on proteomics analysis. J Cancer Res Clin Oncol 149, 16717–16727 (2023). https://doi.org/10.1007/s00432-023-05412-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05412-8