Abstract
Purpose
Immunotherapies have largely failed as treatment options for pancreatic ductal adenocarcinoma (PDAC). In this field, clinical translational studies into personalized treatment are of fundamental importance. In our study, we model tumor-cell immune-cell interactions in a co-culture of primary human PDAC organoids and matched peripheral blood mononuclear cells (PBMCs).
Methods
Using flow cytometry, we evaluated changes in T cell subtypes upon co-culture of patient-derived PDAC organoids and matched PBMCs.
Results
After co-culturing PDAC organoids with PBMCs, we observed changes in CD4+, CD8+ and Treg cell populations. We observed favorable clinical outcome in patients whose PBMCs reacted to the co-culture with organoids.
Conclusion
This experimental model allows to investigate interactions between patient derived PDAC organoids and their PBMCs. This co-culture system could serve as a preclinical platform to guide personalized therapeutic strategies in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) still carries a dismal prognosis. Because of immune evasion and a generally immunosuppressive tumor microenvironment, PDAC is considered an immunological desert and immunotherapies have largely failed in this disease. Immune evasion is a limiting factor for PDAC progression and metastasis and is instrumental for the overall sobering prognosis (Mundry et al. 2020).
Tumor immunotherapy plays an important role in modern cancer treatment. In this regard efforts in modulating the immune system as a therapeutic strategy are broad (Balachandran et al. 2019; Schizas et al. 2020). These treatment approaches have shown promising results in some cancers, benefits in PDAC are still limited due to an extensive immunosuppression within the tumor microenvironment (Ho et al. 2020; Wiehagen et al. 2017). The immunosuppressive environment in PDAC can lead to a manipulation of tumor cells to evade immune surveillance and is one of the most important factor reasoning the failure of immunotherapy (Banerjee et al. 2018).
Considering the growing incidence of PDAC, translational studies driving our understanding of pancreatic cancer and precision oncology are urgently needed (Sung et al. 2021). A major breakthrough in the modeling of pancreatic cancer was the advent of the organoid technology (Reichert et al. 2013; Renz et al. 2018a, b).
Up to now, organoid models have been favourably established for several cancer entities (Broutier et al. 2017; Sachs et al. 2018; Seino et al. 2018; Vlachogiannis et al. 2018; Weeber et al. 2015). These results underline the importance of translational studies. Co-culture models using Peripheral Blood Mononuclear Cell (PBMCs) are applied to form a great possibility to model the disease and its tumor microenvironment (Cattaneo et al. 2020).
Here, we aimed to characterize changes in certain T cell subtypes from PBMCs in a co-culture model with primary human PDAC organoids.
Materials and methods
Materials and Methods were in part published in the thesis of Tianmiao Ma (Ma 2022).
Material
See Key Resources.
Methods
PBMCs isolation from blood samples
Pancreatic tumor specimens and PBMCs were obtained from patients undergoing surgical resection at the Surgical Department of the Ludwig-Maximilians-University (LMU) Hospital from 2020 to 2021 following approval by the Ethics Committee of LMU (#02512). Ten healthy people and 4 PDAC patients were included for blood donation and 4 matching PDAC tissue samples were obtained. The blood collection and processing of all the patients were taken by professionals blinded to the information of the patients in strict accordance with local safety regulations before surgery. Institutional review board approval was obtained.
PBMC isolation
20 mL of fresh blood were collected in a 50 mL Tube (Falcon, Corning, Mexico) for PBMC isolation diluted in PBS in a proportion of 1:1. 20 mL Biocholl (Biochrom, Berlin, Germany) was used accordingly as a control. The Biocoll was carefully overlaid by blood/PBS mixture without destroying its surface. The different components of peripheral blood were separated into different layers after centrifugation at 20 min/1200rcf/room temperature (RT). Then the intermediate phase (PBMCs) was carefully transferred to a new falcon tube and washed with PBS in proportion 1:4. PBMCs were resuspended in the washing buffer and centrifuged at 10 min/300rcf/RT. The supernatant discarded was followed by centrifugation at 10 min/200rcf/RT, and the PBMC pellets were thoroughly washed again with 20 ml of PBS. The falcon tubes contained with PBMCs were then centrifuged at 10 min/200rcf/RT, and the supernatant was rejected after that. Finally, the loose pellets of PBMCs through adequate vortexing were dissolved in RPMI 1640 medium (RPMI Gibco, Thermo Fisher Scientific, Massachusetts, USA), supplemented with 10% fetalbovine serum (FBS) and 1% penicillin–streptomycin (P/S).
Organoid culture
Organoid culture was performed according to previous described protocol (Dijkstra et al. 2018). Briefly, fresh tissue derived from surgical resection was placed in 50 ml tube (Falcon, Corning, Mexico) with 10 ml of PBS (PAN BioTECH, Germany) on ice. The tissue was transferred to a 10 cm Petri dish, (Thermo Fisher Scientific, US) minced into small fragments using a scalpel (Thermo Fisher Scientific, US), and added to a GentleMACS tube with 10 ml of prepared digestion buffer. This tube was then fixed and incubated for 2 h at RT with gentle shaking (the program was pre-set in the software). Subsequently, this sample was filtered through the cell strainer (100 μm) to a new falcon tube, added with the required volume of ice-cold PBS + 0.1% BSA to 15 ml, and centrifuged at 5 min/1000 rpm/4 °C. After discarding the supernatant, the pellets were washed with 3 ml of ACK buffer (Gibco Life Technologies, Germany) and incubated at RT for at least 3 min until the red blood cells were invisible, then washed with 5 ml of ice-cold PBS + 0.1%BSA to stop lysis and centrifuged at 5 min/1000 rpm/4 °C before the supernatant was carefully discarded. 3 ml of TrypLE (Gibco 12,563–011, Thermo Fisher Scientific) was added to dissociate the cell clusters into single ones and incubated for 5 min in a 37 °C water bath. Later, the sample was washed with 5 ml of ice-cold PBS + 0.1% BSA and centrifuged at 5 min/1000 rpm/4 °C again to obtain qualified cells for culture. The cell pellets collected after the supernatant discarded were re-suspended in matrigel (Corning, New York, USA) (50 μl/well) and incubated on ice for 5 min. The Matrigel-cell-suspension was slowly transferred into 24-well plates (Thermo Fisher Scientific, US) to form a 3D dome-like structure ensuring air bubble free plating. The cells were incubated for 10-15 min for gel solidification. Each matrigel dome was covered by 500 μl of complete medium prewarmed to 37 °C for long-time culture in the incubator. The medium was refreshed every three days during organoid culture.
Organoid passaging
Organoids were split every 5–10 days, in a ratio of 1:2. The matrigel-organoid mixture was collected in a new falcon tube until fully washed with cold cell recovery solution and cold PBS (10 ml in total), respectively. The cell pellets were acquired after centrifugation at 5 min/1000 rpm/4 °C and the supernatant was discarded. The desired amount of matrigel (50 μl/well) was added to the dissociated cells and mixed completely on ice. 50 μl of the new matrigel-organoid mixture was pipetted into a 24-well plate, each of them was replenished with 500 μl of pre-warmed complete medium. The plate was then returned to the incubator for further culturing.
Culture PBMCs with conditioned medium derived from human pancreatic cancer cell lines
Human pancreatic cancer cell lines (Panc1 and Miapaca2) were originally received from American Type Culture Collection (ATCC). Cells were checked quarterly for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza) and authenticated. These cells were separately cultivated in a T-75 flask (Thermo Fisher Scientific, US) with 15 ml of previous described medium, detached with 5 ml 0.025% Trypsin/EDTA solution, and passaged every 3–4 days depending on cell growth. The conditioned medium samples from the supernatant of Panc1 and Miapaca2 after culturing for 72 h were gathered in a new falcon tube and filtered through a sterile 0.20 μm filter to remove cell debris, then frozen at -20 °C, respectively. The PBMCs were cultured with 50% of previous described medium and 50% of Panc1/Miapaca2-derived CM, as an experimental (EXP) group. 48 h later, PBMCs were collected separately and stained for FACS analysis.
Co-cultured PBMCs with human pancreatic cancer cell lines
Co-culture was performed with modifications as previously described (Dijkstra et al. 2018). Briefly, Panc1/Miapaca2 cells were seeded in a 6-well plate one day before being co-cultured with PBMCs. The next day, one or two vials of frozen PBMCs were thawed in a 37 °C water bath, transferred to a falcon tube, and washed with 9 ml of PBS. After centrifuging at 5 min/500 rcf/RT and discarding the supernatant, the PBMCs were resuspended in the previous described culture medium. The same amount of prepared PBMCs were added to each well at a ratio of 25:1 with Panc1/Miapaca2 cells. Waiting for 48 h, the PBMCs in the control (CON) group and the mixture of PBMCs and Panc1/Miapaca2 in the bottom 3 wells EXP group were collected respectively and then stained for flow cytometry.
Co-cultured PBMCs with organoids from PDAC patients
The co-culture of PBMCs and organoids was performed with modifications as previously described (Dijkstra et al. 2018). The PBMCs were resuspended in the organoid complete medium, added to the certain wells with organoids-matrigel mixtures, and cultured in the incubator for 48 h, which served as an EXP group. In the CON group, the same numbers of PBMCs were cultured alone in the same medium and matrigel for 48 h. The staining of PBMCs for flow cytometry was performed after cell collecting in the control group and co-culture group, respectively. Autologous co-cultures were established with PDAC patient-derived organoid lines and their matched PBMCs.
Flow Cytometry
The PBMCs were re-suspended in the desired medium at 1 × 106 cells/ml and transferred to the FACS tubes (200 μl/tube). The certain tubes in both CON and EXP group were added with 1 μl of each antibody, vortexed, and incubated in a dark chamber for 15-30 min/RT. The samples were washed with 2 ml of FACS buffer (PBS + 2%NaN3 + 5%BSA) completely and centrifuged at 5 min/500 rcf/RT. After discarding the supernatant and adding another 500 μl of FACS buffer in each tube, the samples were measured by BD LSRFortessaTM Cell Analyzer and according to the gating strategy which you can see in Supplementary Material. The data were recorded and downloaded from BD FACS Diva 8.0.1 software and imported to Flowjo10 for further analysis. The gating strategies of Memory T cells and Tregs on the FACS plots were shown in Suppl. Material Figure S2 A, B.
Statistical Analysis
The Student’s t-test to evaluate the differences between control and experimental groups or one/two-way analysis of variance (ANOVA) for multiple group comparisons were applied in all statistical analyses by GraphPad Prism 9 (Graphstad, US) based on the population of immune cells. P < 0.05 was considered to be statistically significant.
Results
Frequency of effector, effector memory and central memory T cells was higher in PBMCs of PDAC patients compared to healthy donors
First, we aimed to determine the baseline for T cell subsets in PBMCs obtained from healthy donors (HD) and PDAC patients (PDAC-D) as a prerequisite for the following experiments. The analysis revealed that the frequency of effector (Teff), memory (Tem) and central memory T cells (Tcm) of CD4+ cells was higher in PBMCs from PDAC-D than from HD (p < 0.001, Supplementary Fig. 1A). The frequencies of Tnaiv cells for both CD4+ and CD8+ were lower in PDAC-D when compared to HD (p < 0.001, Supplementary Fig. 1B). In a similar manner, frequency of CD8+ Teff, Tem and Tcm cells in PBMC (PDAC-D) group was found to be significantly higher than those in PBMC (HD) group (p < 0.05, Supplementary Fig. 1B). Frequency of regulatory T cells (Tregs) in PBMCs of PDAC-D was also higher than in healthy PBMCs (data not shown, p < 0.05).
As expected, we found that the constellation of immune cell subsets in PDAC patients differ from those in healthy patients.
Direct cell–cell contact is necessary for immune cells to effect immune response
Next, we investigated effects of conditioned medium (Topalian et al.) containing soluble factors (i.e. cytokines, chemokines, etc.) to induce changes in T cell subsets during cultivation without direct cell–cell contact.
Therefore, we cultured PBMCs from HD in CM from PDAC cells (Panc1/MiaPaca2). We did not observe significant changes in different subsets of CD4+ (CM-Panc1; Fig. 1A–E column 1–2, CM-MiaPaca2; Supplementary Fig. 1C) and CD8+ Tnaiv, Teff, Tem, Tcm and Treg compared to PBMCs from HD cultured alone (Fig. 1F–I, column 1–2). Taken together, cultivation of PBMCs from healthy donors in conditioned medium of PDAC cells did not induce changes in the T cell subsets of PBMCs.
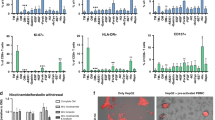
Direct immune-tumor cell contact is necessary to modulate immune response. Co-Culturing PBMCs (HD) with CM (Panc1/MiaPaca2), and Panc1 cells. A The frequency of CD4+ Tnaiv cells showed no statistically significant differences in each co-culture group. B The frequency of CD4+ Teff cells showed no statistically significant differences in each co-culture group. C The frequency of CD4+ Tem cells showed no statistically significant differences in each co-culture group. D The frequency of CD4+ Tcm cells was higher in the EXP (+ Panc1) group than that in the CON (HD) group (**P < 0.01, *P < 0.05, and *P < 0.05). E The Treg frequency in the EXP (+ Panc1) group was higher than that in the CON (HD) group (**P < 0.001). F The frequency of CD8+ Tnaiv cells showed no statistically significant differences in each co-culture group. G The frequency of CD8+ Teff cells showed no statistically significant differences in each co-culture group. H CD8+ Tem population was significantly higher in the EXP (+ Panc1) group than that in the CON (HD) group (**P < 0.01, *P < 0.05, and *P < 0.05). I The population of CD8+ Tcm cells showed no statistically significant differences in each co-culture group. J FACS plot of Memory CD4+ T cell subsets in the CON (HD) and EXP (+ CM Panc1/MiaPaca2) groups. K FACS plot of Memory CD4+ T cell subsets in the CON (HD) and EXP (+ Panc1/MiaPaca2) groups. L FACS plot of Tregs in the CON (HD) and EXP (+ CM Panc1/MiaPaca2) groups. M FACS plot of Tregs in the CON (HD) and EXP (+ Panc1/MiaPaca2) groups. N FACS plot of different Memory CD8+ T cell subsets in the CON (HD) and EXP (+ CM Panc1/MiaPaca2) groups. O FACS plot of different Memory CD8+ T cell subsets in the CON (HD) and EXP (+ Panc1/MiaPaca2) groups
Next, we co-cultivated PBMCs (HD) with Panc1/MiaPaca2 cells. In these experiments we observed changes in CD4+ Tcm and CD8+ cells. Co-culturing PBMCs with Panc1 cells (EXP-Panc1-HD) led to an increase in CD4+ Tcm cells when compared to controls (CON-HD) (p = 0.04, Fig. 1 D, column 3).
Frequency of Treg in EXP-Panc1-HD also increased after co-cultivation with cells from both cell lines (p = 0.04, Fig. 1 E, column 3). CD8+ Tem frequency in the Panc1-HD group was significantly higher than in the CON-HD group (p < 0.05, Fig. 1 H, column 3). Panc1 cells were more potent to induce effects on immune cell differentiation than MiaPaca2 cells.
Direct cell–cell contact led to changes in T cell subtype frequencies and as expected, co-cultivation of PBMCs with PDAC cells led to a higher amount of memory and regulatory T cells.
Co-culture of primary human PDAC organoids and PBMCs from HD effected heterogenous phenotype change in T cell subsets
For the following experiments we co-cultured primary human PDAC organoids directly with PBMCs from healthy donors. In these experiments, we did not observe differences in the frequency of CD4+ Teff and Tnaiv between those two groups (p > 0.05, Fig. 2A, B).
Co-culture of primary human PDAC organoids and PBMCs from HD effect heterogenous phenotype change in T cell subsets. A The frequency of CD4+ Tnaiv cells showed no statistically significant differences in co-culture group. B CD4+ Teff cells were larger in the EXP (+ Panc1) group than that in the CON group but not significantly. C The frequency of CD4+ Tem cells was lower in the EXP (+ Org) group than that in the CON (HD) group (**P < 0.01, *P < 0.05, and *P < 0.05). D The frequency of CD4+ Tcm cells was higher in the EXP (+ Org) group than that in the CON (HD) group (**P < 0.01, *P < 0.05, and *P < 0.05). E The Treg frequency in the EXP (+ Org) group was higher than that in the CON (HD) group (**P < 0.001). F CD8+ Tnaiv frequency was lower in EXP (+ Org) group than in CON (HD) group (**P < 0.01, *P < 0.05, and *P < 0.05). G The frequency of CD8+ Teff cells showed no statistically significant differences in co-culture group. H CD8+ Tem frequency was significantly higher in the EXP (+ Org) group than that in the CON (HD) group (**P < 0.01, *P < 0.05, and *P < 0.05). I The frequency of CD8+ Tcm cells showed no statistically significant differences in co-culture group. J FACS plot of different Memory CD4+ T cell subsets. K FACS plot of Tregs. L FACS plot of different Memory CD8+ T cell subsets
Co-cultivation of PBMCs from HD with PDAC organoids (PDAC-Org-HD) led to a decrease in the amount of Tem cells, while the amount of Tcm cells increased (p = 0.04, Fig. 1D). Frequency of CD4+ Tem in PBMCs from HD alone and in PDAC-Org-HD was also significantly lower (p = 0.03, Fig. 2C). In addition, the amount of CD4+ Treg increased in PDAC-Org-HD compared to PBMC-HD (p < 0.001, Fig. 1E). Concerning CD8+ Tcells, co-cultivation of PBMCs with PDAC organoids led to a decrease in the amount of Tnaiv cells as well as an increase in Tem cells in PDAC-Org-HD (Fig. 2F, H). CD8+ Teff and Tcm cells were not affected in PDAC-Org-HD (Fig. 2G, I).
Direct contact between immune cell and tumor organoid led to higher frequency of CD8+ Tem, CD4+ Tcm and Treg cells. A lower frequency of CD4+ Tem and CD8+ Teff cells was obtained in co-culture of PDAC organoids and PBMCs from healthy donors.
Co-culture of PBMCs from PDAC patients and Panc1 cells caused heterogenous phenotype change in T cell subsets
As described above, we identified differences in subsets of T cells in PBMCs from HD co-cultivated either with cell lines or with organoids. Next, we aimed to investigate the effect of co-cultivation on PBMCs from PDAC-D with pancreatic cancer cells.
Therefore, we co-cultured PBMCs from PDAC-D with Panc1 cells. The CD4+ Tnaiv frequency in PBMCs (PDAC-D) cultured with Panc1 was not different to the CD4+ Tnaiv frequency in PBMCs (PDAC-D) cultured alone (Fig. 3A). CD4+ Tem and Teff frequencies in co-culture were both lower than those in PBMCs from PDAC donors alone (p = 0.001 and 0.016, Fig. 3B, C). The frequency of CD4+ Tcm cells in PBMCs (PDCA-D) co-cultured with Panc1 cells increased significantly (p = 0.013, Fig. 3D). No differences were seen in the frequency of Treg in PBMCs (PDAC-D) and PDAC-D-Panc1 (Fig. 3E). Frequency of CD8+ Tnaiv cells in PDAC-D-Panc1 group was higher than that in PDAC-D (p < 0.01, Fig. 3F). Moreover, there was a decrease in CD8+ Teff frequency and an increase in CD8+ Tcm frequency in the PDAC-D-Panc1 compared to PDAC-D alone (Fig. 3G, K). However, CD8+ Tem cells (PDAC-D-Panc1) were significantly lower in the co-culture group than in the control group (PDAC-D) (P = 0.002, Fig. 3H).
Co-culture of PBMCs from PDAC patients (PDAC-D) and Panc1 cells causes heterogenous phenotype change in T cell subsets. A The frequency of CD4+ Tnaiv cells showed no statistically significant differences in co-culture group. B CD4+ Teff cells were lower in the EXP (+ Panc1) group than that in the CON (P) group (**P < 0.01, *P < 0.05, and *P < 0.05). C CD4+ Tem cells were lower in the EXP (+ Panc1) group than that in the CON (P) group (**P < 0.01, *P < 0.05, and *P < 0.05). D The frequency of CD4+ Tcm cells was higher in the EXP (+ Panc1) group than in the CON (P) group (**P < 0.01, *P < 0.05, and *P < 0.05). E The frequency of CD4+ Treg cells showed no statistically significant differences in co-culture group. F The frequency of CD8+ Tnaiv cells was higher in the EXP (+ Panc1) group (**P < 0.01, *P < 0.05, and *P < 0.05). G CD8+ Teff cells were lower in the EXP (+ Panc1) group than that in the CON (P) group but not significantly. H CD8+ Tem cells were lower in the EXP (+ Panc1) group than that in the CON (P) group (**P < 0.01, *P < 0.05, and *P < 0.05). I CD8+ Tcm cells did not change significantly. J FACS plot of different Memory CD4+ T cell subsets in the CON (P) and EXP (+ Panc1) groups. K FACS plot of Tregs in the CON (PDAC D) and EXP (+ Panc1) groups without significant differences. L FACS plot of different Memory CD8+ T cell subsets in the CON (P) and EXP (+ Panc1) groups
Co-culture of Panc1 cells and PBMCs (PDAC-D) resulted in a lower frequency of CD4+ and CD8+ Tem and Teff cells than those in PBMCs from PDAC donors alone. The frequency of CD4+ Tcm cells, CD8+ Tnaiv and Tcm cells in PBMCs from PDAC donors co-cultured with Panc1 cells increased significantly.
Co-culture of matched PBMCs with primary human PDAC organoids displayed heterogenous T cell response
Next, we utilized this co-culture model employing PBMCs and primary PDAC organoids from the same patient to mimic T cell response.
In patient 1 (P1) CD4+ Tem and Teff cell population in PDAC-Org-P1 were lower than in the PBMC-P1 group. The CD4+ Tcm and Tnaiv cells in PDAC-Org-P1 were both higher than those in the PBMC-P1 (Fig. 4A). Frequency of CD4+ Tem cells decreased in the PBMC-P2 group and in PDAC-Org-P2 while frequency of CD4+ Tcm and CD4+ Tnaiv cell population slightly increased. In contrast, CD4+ Teff cells showed only minor differences in their frequency between these two groups. However, frequency of Treg cells in PDAC-Org-P2 and PDAC-Org-P3 were higher than in PBMC-P2 and PBMC-P3. We were able to detect similar changes in the CD8+ memory T cell subpopulation in PDAC-Org-P1/2 compared to autologous PBMCs (Fig. 4C).
Individual co-cultures of matched PBMCs with primary human PDAC organoids. A CD4+ Memory T cell subsets showed personal variations between the CON (P) and EXP (+ Org) groups of Patient 1, 2, 3, and 4. B Frequency of Tregs was increased in the co-cultures from Patient 1, 2, 3, and 4 compared to the control group. C CD8+ Memory T cell subsets showed personal variations between the CON (P) and EXP (+ Org) groups of Patient 1, 2, 3, and 4
In co-culture of P1 and P2, we could see an increase in CD4+ and CD8+ Tcm, Tnaiv and Treg cells as well as a decrease in CD4+ and CD8+ Tem cells. It should be noted that we did not see any differences in memory CD4+ and CD8+ T cell subtypes in co-culture of P3 and P4 compared with their controls. In contrast, there was a significant increase in frequency of Treg cells in PDAC-Org-P3 and PDAC-Org-P4 (8.15%/13.9%) compared to PBMCs alone (4.08%/5.39%) (Fig. 4B).
Our results showed an individual response in co-culture of matched PBMCs with primary human PDAC organoids compared to PBMCs cultured alone (Fig. 5A).
We observed favorable clinical outcome in patients whose PBMCs reacted to the co-culture with organoids.
We can empathize that patients with obvious changes in T cell subset in PBMCs after co-cultivation with autologous PDAC organoids (P1 and P2) were younger and showed longer free survival than those without these changes. Patients without T cell subset change in co-culture had tumor recurrence six and eleven months after surgery during follow-up (Fig. 5B).
Discussion
We evaluated differences in T cell subsets after co-culture in individual PDAC patients. Co-culture of PBMCs with established PDAC cell lines and CM guided us to optimal conditions for PBMC and PDAC organoid co-culture. CD4+ and CD8+ T cell differentiation varied in 4 patient co-cultures (EXP-PDAC-D-Org1-4 vs. PDAC-D) and Treg cell frequency was higher in all co-cultures than in their matched controls. This model reflects the heterogeneity of tumor-immune interactions in individual patients. It might serve as a preclinical model helping to guide treatment options of PDAC patients in the future.
Organoid cultures have been studied for individualized tumor response assessment in different cancers (Shi et al. 2020; van de Wetering et al. 2015). Tumor-specific T cell responses for colorectal cancer and non-small lung cancer are already using similar co-culture conditions (Dijkstra et al. 2018). However, this model with patient-derived PDAC organoids is to our knowledge not yet examined. Previous studies have examined such co-culture models partially similar but focused on lymphocyte infiltration (Tsai et al. 2018). Moreover, previous experiments did not directly focus on T cell subsets in PBMC and PDAC organoid co-culture.
Our analysis revealed that immune cell subsets in PDAC patients differ from those in healthy patients. These results are congruent with previous published data in other cancer types (Krijgsman et al. 2019; Lulu et al. 2021). We observed that conditioned medium (CM) of PDAC cells did not effect changes in T cell subsets in PBMCs detected by flow cytometry. Our observation that CM is not sufficient to influence immune cells is consistent with the fact that conditioned medium alone is not able to mimic the immune environment in tumors, especially without interactions between different cell types (Dowling & Clynes 2011).
We found increased frequencies of CD4+ Tcm and CD8+ Tem cells in PBMCs (HD) co-cultured with Panc1 cells compared to PBMCs from HD cultured alone. An activation of CD8+ T cells by tumor antigen presentation is previously described (Holokai et al. 2020). This observation can be explained by a lower activation threshold of memory T cells than naïve T cells (Liu et al. 2020; MacLeod et al. 2010). We also noticed an ascending trend in Treg cell frequency which is complementary to previous studies. Treg cells can secrete immunosuppressive cytokines such as IL-10 and TGF-ß which can increase Treg cell frequency (Oleinika et al. 2013). In case of patients PBMCs cultivated with Panc1 cells, we found individual responses in T cell subset changes. Taken together, even those established cell lines behave differently in terms of immune modulation. Besides, we also found enhanced frequency of Treg in patients’ PBMCs. Similar results were found in other solid tumors (Bates et al. 2006; Griffiths et al. 2007; Hiraoka et al. 2006).
Our results confirmed that the amount of CD4+/CD8+ Tem, Tcm, and Teff cells was higher in PBMCs from PDAC patients than in PBMCs from HD. The frequency of CD4+/CD8+ Tnaiv cells was significantly lower in patients’ PBMCs, compared to healthy PBMCs which is congruent to existing literature (Hang et al. 2019; Liu et al. 2015).
Our results show an individual response in co-culture of matched PBMCs with autologous primary human PDAC organoids compared to PBMCs cultured alone. Previous published studies can partially explain our observations. The enhancement of Treg can inhibit anti-tumor immune responses in PDAC and results in dismal disease prognosis (Bouneaud et al. 2005). Our heterogenous results in co-culture highlight the importance to better define the compositions of PDAC stroma in individual patients.
Interestingly, patients whose PBMCs respond to the co-culture with organoids were younger and showed a longer recurrence-free survival in our study compared to those without immune reaction in co-culture. Several studies have suggested that a high frequency of CD4+ and CD8+ T cells in PDAC microenvironment is associated with a better disease-free survival and/or overall survival (Balachandran et al. 2019; Lohneis et al. 2017; Wang et al. 2017). Larger sample size will be needed to underline our results more precisely.
Conclusion
In this project, we detected different T cell subset changes in co-cultures of PDAC organoids and matched PBMCs. This model carries great potential to facilitate individual treatment strategies in PDAC patients.
Key Resources
Reagents, chemicals, and buffer | |
|---|---|
1 × Phosphate-buffered saline (PBS) | PAN BioTECH, Germany |
A83-01 | Tocris Bioscience, England |
ACK Lysing buffer | Gibco Life Technologies, Germany |
Advanced DMEM/F-12 | Gibco Life Technologies, Germany |
B27 supplement | Gibco Life Technologies, Germany |
Biocoll | Biocell Technology, Germany |
Bovine serum albumin (BSA) | Biomol, Germany |
CASY Ton | OMNI Life Science, German |
Collagenase Type II | Thermofischer, US |
Dimethylsulfoxide (DMSO) | ROTH, Germany |
EGF recombinant human protein | Gibco Life Technologies, Germany |
Fetal Bovine Serum (FBS) | Thermo Fisher Scientific, US |
FGF-10 recombinant human protein | Peprotech, Germany |
Fixation buffer | Invivogen, US |
GlutaMAX supplement | Gibco Life Technologies, Germany |
BD Golgistop | BD Biosciences, US |
Matrigel (growth factor reduced) | Sigma Aldrich, US |
Penicillin/Streptomycin (P/S) | PAN BioTECH, Germany |
Perm buffer | Invivogen, US |
Primocin | Invivogen, US |
Protein Transport Inhibitor | BD Golgistop, US |
Recombinant Human R-Spondin 1 protein | R&D systems, US |
Recovery cell culture freezing medium | Thermo Fisher Scientific, US |
ROCK inhibitor (Y-27632) | Sigma Aldrich, US |
RPMI1640 | Gibco Life Technologies, Germany |
TrypLE express enzyme (1X) | Gibco Life Technologies, Germany |
Wnt3a recombinant human protein | R&D systems, US |
Antibodies | |
CD45 BUV650 | BD Bioscience, US |
CD3 PerCP-Cy5.5 | Biolegend, US |
CD4 BUV395 | BD Bioscience, US |
CD8 APC-H7 | BD Bioscience, US |
CD25 PE | BD Bioscience, US |
CD127 BV421 | BD Bioscience, US |
CD197 BV421 | BD Bioscience, US |
CD45RO PE-Cy7 | BD Bioscience, US |
Data availability
Not applicable.
Abbreviations
- BSA:
-
Bovine serum albumin
- CON:
-
Control
- DCs:
-
Dendritic cells
- DMEM:
-
Dulbecco's modified eagle medium
- DMSO:
-
Dimethyl sulfoxide
- EXP:
-
Experiment
- FACS:
-
Fluorescence activated cell sorter
- FBS:
-
Fetal bovine serum
- HD:
-
Healthy donors
- IFN-γ:
-
Interferon-gamma
- IL:
-
Interleukin
- Org:
-
Organoid
- P:
-
Patient
- P/S:
-
Penicillin/streptomycin
- PBMC:
-
Peripheral blood mononuclear cell
- PBS:
-
Phosphate-buffered saline
- PDAC:
-
Pancreatic ductal adenocarcinoma
- Rcf:
-
Relative centrifugal force
- rpm:
-
Revolutions per minute
- RPMI 1640:
-
Roswell Park Memorial Institute 1640
- RT:
-
Room temperature
- Tcm cells:
-
Central memory T cells
- Teff cells:
-
Effector T cells
- Tem cells:
-
Effector memory T cells
- TGF-β:
-
Transforming growth factor beta
- Tnaiv cells:
-
Naïve T cells
- Tregs:
-
Regulatory T cells
References
Balachandran VP, Beatty GL, Dougan SK (2019) Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology 156(7):2056–2072. https://doi.org/10.1053/j.gastro.2018.12.038
Banerjee K, Kumar S, Ross KA, Gautam S, Poelaert B, Nasser MW, Aithal A, Bhatia R, Wannemuehler MJ, Narasimhan B, Solheim JC, Batra SK, Jain M (2018) Emerging trends in the immunotherapy of pancreatic cancer. Cancer Lett 417:35–46. https://doi.org/10.1016/j.canlet.2017.12.012
Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, Banham AH (2006) Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol 24(34):5373–5380. https://doi.org/10.1200/jco.2006.05.9584
Bouneaud C, Garcia Z, Kourilsky P, Pannetier C (2005) Lineage relationships, homeostasis, and recall capacities of central- and effector-memory CD8 T cells in vivo. J Exp Med 201(4):579–590. https://doi.org/10.1084/jem.20040876
Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, JNM, I. J., Wigmore, S. J., Saeb-Parsy, K., Huch, M. (2017) Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 23(12):1424–1435. https://doi.org/10.1038/nm.4438
Cattaneo CM, Dijkstra KK, Fanchi LF, Kelderman S, Kaing S, van Rooij N, van den Brink S, Schumacher TN, Voest EE (2020) Tumor organoid-T-cell coculture systems. Nat Protoc 15(1):15–39. https://doi.org/10.1038/s41596-019-0232-9
Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, van Rooij N, van Leerdam ME, Depla A, Smit EF, Hartemink KJ, de Groot R, Wolkers MC, Sachs N, Snaebjornsson P, Voest EE (2018) Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 174(6):1586-1598.e1512. https://doi.org/10.1016/j.cell.2018.07.009
Dowling P, Clynes M (2011) Conditioned media from cell lines: a complementary model to clinical specimens for the discovery of disease-specific biomarkers. Proteomics 11(4):794–804. https://doi.org/10.1002/pmic.201000530
Griffiths RW, Elkord E, Gilham DE, Ramani V, Clarke N, Stern PL, Hawkins RE (2007) Frequency of regulatory T cells in renal cell carcinoma patients and investigation of correlation with survival. Cancer Immunol Immunother 56(11):1743–1753. https://doi.org/10.1007/s00262-007-0318-z
Hang J, Huang J, Zhou S, Wu L, Zhu Y, Zhu L, Zhou H, Xu K, Jiang H, Yang X (2019) The clinical implication of CD45RA(+) naïve T cells and CD45RO(+) memory T cells in advanced pancreatic cancer: a proxy for tumor biology and outcome prediction. Cancer Med 8(3):1326–1335. https://doi.org/10.1002/cam4.1988
Hiraoka N, Onozato K, Kosuge T, Hirohashi S (2006) Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin Cancer Res 12(18):5423–5434. https://doi.org/10.1158/1078-0432.Ccr-06-0369
Ho WJ, Jaffee EM, Zheng L (2020) The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat Rev Clin Oncol 17(9):527–540. https://doi.org/10.1038/s41571-020-0363-5
Holmes S, He M, Xu T, Lee PP (2005) Memory T cells have gene expression patterns intermediate between naive and effector. Proc Natl Acad Sci U S A 102(15):5519–5523. https://doi.org/10.1073/pnas.0501437102
Holokai L, Chakrabarti J, Lundy J, Croagh D, Adhikary P, Richards SS, Woodson C, Steele N, Kuester R, Scott A, Khreiss M, Frankel T, Merchant J, Jenkins BJ, Wang J, Shroff RT, Ahmad SA, Zavros Y (2020) Murine- and Human-Derived Autologous Organoid/Immune Cell Co-Cultures as Pre-Clinical Models of Pancreatic Ductal Adenocarcinoma. Cancers (Basel), 12(12). https://doi.org/10.3390/cancers12123816
Krijgsman D, de Vries NL, Skovbo A, Andersen MN, Swets M, Bastiaannet E, Vahrmeijer AL, van de Velde CJH, Heemskerk MHM, Hokland M, Kuppen PJK (2019) Characterization of circulating T-, NK-, and NKT cell subsets in patients with colorectal cancer: the peripheral blood immune cell profile. Cancer Immunol Immunother 68(6):1011–1024. https://doi.org/10.1007/s00262-019-02343-7
Liu HL, Guan CJ, Wu YJ, Hu MG, Zhao ZM, Liu R (2015) Clinical Significance of Preoperative CD8+ Central Memory T Cells for Operable Pancreatic Adenocarcinoma. Dig Surg 32(6):433–438. https://doi.org/10.1159/000440681
Liu Q, Sun Z, Chen L (2020) Memory T cells: strategies for optimizing tumor immunotherapy. Protein Cell 11(8):549–564. https://doi.org/10.1007/s13238-020-00707-9
Lohneis P, Sinn M, Bischoff S, Jühling A, Pelzer U, Wislocka L, Bahra M, Sinn BV, Denkert C, Oettle H, Bläker H, Riess H, Jöhrens K, Striefler JK (2017) Cytotoxic tumour-infiltrating T lymphocytes influence outcome in resected pancreatic ductal adenocarcinoma. Eur J Cancer 83:290–301. https://doi.org/10.1016/j.ejca.2017.06.016
Lulu AM, Cummings KL, Jeffery ED, Myers PT, Underwood D, Lacy RM, Chianese-Bullock KA, Slingluff CL Jr, Modesitt SC, Engelhard VH (2021) Characteristics of Immune Memory and Effector Activity to Cancer-Expressed MHC Class I Phosphopeptides Differ in Healthy Donors and Ovarian Cancer Patients. Cancer Immunol Res 9(11):1327–1341. https://doi.org/10.1158/2326-6066.Cir-21-0111
Ma T (2022) Tumor-immune-interaction in a pancreatic cancer organoid co-culture model LMU University]. Munich
MacLeod MK, Kappler JW, Marrack P (2010) Memory CD4 T cells: generation, reactivation and re-assignment. Immunology 130(1):10–15. https://doi.org/10.1111/j.1365-2567.2010.03260.x
Mohebbi B, Ashtibaghaei K, Hashemi M, Hashemi M, Asadzadeh Aghdaei H, Zali MR (2019) Conditioned Medium from Cultured Colorectal Cancer Cells Affects Peripheral Blood Mononuclear Cells Inflammatory Phenotype in Vitro. Iran J Med Sci 44(4):334–341. https://doi.org/10.30476/ijms.2019.44959
Mundry CS, Eberle KC, Singh PK, Hollingsworth MA, Mehla K (2020) Local and systemic immunosuppression in pancreatic cancer: Targeting the stalwarts in tumor's arsenal. Biochim Biophys Acta Rev Cancer 1874(1):188387. https://doi.org/10.1016/j.bbcan.2020.188387
Okada SL, Simmons RM, Franke-Welch S, Nguyen TH, Korman AJ, Dillon SR, Gilbertson DG (2018) Conditioned media from the renal cell carcinoma cell line 786.O drives human blood monocytes to a monocytic myeloid-derived suppressor cell phenotype. Cell Immunol 323:49–58. https://doi.org/10.1016/j.cellimm.2017.10.014
Oleinika K, Nibbs RJ, Graham GJ, Fraser AR (2013) Suppression, subversion and escape: the role of regulatory T cells in cancer progression. Clin Exp Immunol 171(1):36–45. https://doi.org/10.1111/j.1365-2249.2012.04657.x
Peschke, K., Jakubowsky, H., Schäfer, A., Maurer, C., Lange, S., Orben, F., Bernad, R., Harder, F. N., Eiber, M., Öllinger, R., Steiger, K., Schlitter, M., Weichert, W., Mayr, U., Phillip, V., Schlag, C., Schmid, R. M., Braren, R. F., Kong, B., Reichert, M. (2022). Identification of treatment-induced vulnerabilities in pancreatic cancer patients using functional model systems. EMBO Mol Med, 14(4), e14876. https://doi.org/10.15252/emmm.202114876
Quante AS, Ming C, Rottmann M, Engel J, Boeck S, Heinemann V, Westphalen CB, Strauch K (2016) Projections of cancer incidence and cancer-related deaths in Germany by 2020 and 2030. Cancer Med 5(9):2649–2656. https://doi.org/10.1002/cam4.767
Rauth, S., Karmakar, S., Batra, S. K., & Ponnusamy, M. P. (2021). Recent advances in organoid development and applications in disease modeling. Biochim Biophys Acta Rev Cancer, 1875(2), 188527. https://doi.org/10.1016/j.bbcan.2021.188527
Reichert M, Takano S, Heeg S, Bakir B, Botta GP, Rustgi AK (2013) Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc 8(7):1354–1365. https://doi.org/10.1038/nprot.2013.079
Renz BW, Takahashi R, Tanaka T, Macchini M, Hayakawa Y, Dantes Z, Maurer HC, Chen X, Jiang Z, Westphalen CB, Ilmer M, Valenti G, Mohanta SK, Habenicht AJR, Middelhoff M, Chu T, Nagar K, Tailor Y, Casadei R, Wang TC (2018a) β2 Adrenergic-Neurotrophin Feedforward Loop Promotes Pancreatic Cancer. Cancer Cell 33(1):75-90.e77. https://doi.org/10.1016/j.ccell.2017.11.007
Renz BW, Tanaka T, Sunagawa M, Takahashi R, Jiang Z, Macchini M, Dantes Z, Valenti G, White RA, Middelhoff MA, Ilmer M, Oberstein PE, Angele MK, Deng H, Hayakawa Y, Westphalen CB, Werner J, Remotti H, Reichert M, Wang TC (2018b) Cholinergic Signaling via Muscarinic Receptors Directly and Indirectly Suppresses Pancreatic Tumorigenesis and Cancer Stemness. Cancer Discov 8(11):1458–1473. https://doi.org/10.1158/2159-8290.Cd-18-0046
Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, Korving J, van Boxtel R, Duarte AA, Lelieveld D, van Hoeck A, Ernst RF, Blokzijl F, Nijman IJ, Hoogstraat M, Clevers H (2018) A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 172(1–2):373-386.e310. https://doi.org/10.1016/j.cell.2017.11.010
Schizas, D., Charalampakis, N., Kole, C., Economopoulou, P., Koustas, E., Gkotsis, E., Ziogas, D., Psyrri, A., & Karamouzis, M. V. (2020). Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev, 86, 102016. https://doi.org/10.1016/j.ctrv.2020.102016
Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, Ohta Y, Matano M, Nanki K, Kawasaki K, Takahashi S, Sugimoto S, Iwasaki E, Takagi J, Itoi T, Kitago M, Kitagawa Y, Kanai T, Sato T (2018) Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 22(3):454-467.e456. https://doi.org/10.1016/j.stem.2017.12.009
Shen, Y., Pu, K., Zheng, K., Ma, X., Qin, J., Jiang, L., & Li, J. (2019). Differentially Expressed microRNAs in MIA PaCa-2 and PANC-1 Pancreas Ductal Adenocarcinoma Cell Lines are Involved in Cancer Stem Cell Regulation. Int J Mol Sci, 20(18). https://doi.org/10.3390/ijms20184473
Shi R, Radulovich N, Ng C, Liu N, Notsuda H, Cabanero M, Martins-Filho SN, Raghavan V, Li Q, Mer AS, Rosen JC, Li M, Wang YH, Tamblyn L, Pham NA, Haibe-Kains B, Liu G, Moghal N, Tsao MS (2020) Organoid Cultures as Preclinical Models of Non-Small Cell Lung Cancer. Clin Cancer Res 26(5):1162–1174. https://doi.org/10.1158/1078-0432.Ccr-19-1376
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Sznol M (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366(26):2443–2454. https://doi.org/10.1056/NEJMoa1200690
Tsai S, McOlash L, Palen K, Johnson B, Duris C, Yang Q, Dwinell MB, Hunt B, Evans DB, Gershan J, James MA (2018) Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 18(1):335. https://doi.org/10.1186/s12885-018-4238-4
van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, van Sluis P, Li VS, Seepo S, Sekhar Pedamallu C, Clevers H (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161(4):933–945. https://doi.org/10.1016/j.cell.2015.03.053
Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh DM, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Valeri N (2018) Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359(6378):920–926. https://doi.org/10.1126/science.aao2774
Wang Z, Zhao J, Zhao H, A, S., Liu, Z., Zhang, Y., Liu, X., & Wang, F. (2017) Infiltrating CD4/CD8 high T cells shows good prognostic impact in pancreatic cancer. Int J Clin Exp Pathol 10(8):8820–8828
Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, Gadellaa-van Hooijdonk CG, van der Velden DL, Peeper DS, Cuppen EP, Vries RG, Clevers H, Voest EE (2015) Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A 112(43):13308–13311. https://doi.org/10.1073/pnas.1516689112
Wiehagen KR, Girgis NM, Yamada DH, Smith AA, Chan SR, Grewal IS, Quigley M, Verona RI (2017) Combination of CD40 Agonism and CSF-1R Blockade Reconditions Tumor-Associated Macrophages and Drives Potent Antitumor Immunity. Cancer Immunol Res 5(12):1109–1121. https://doi.org/10.1158/2326-6066.Cir-17-0258
Xu H, Lyu X, Yi M, Zhao W, Song Y, Wu K (2018) Organoid technology and applications in cancer research. J Hematol Oncol 11(1):116. https://doi.org/10.1186/s13045-018-0662-9
Funding
Open Access funding enabled and organized by Projekt DEAL. This project was financially supported by the Foederprogramm fuer Foschung und Lehre (FoeFoLe +) of the LMU Munich and the German Cancer Consortium (DKTK), German Cancer Research Centre (DKFZ) to Mathilda Knoblauch. Moreover we greatly thank the China Scholarship Council (CSC) for supporting the research and work of Tianmiao Ma. Maximilian Reichert acknowledges financial support by the German Cancer Aid (Max-Eder Program 111273 and Project 70114328) and the German Research Foundation (DFG, SFB1321 Modeling and Targeting Pancreatic Cancer, Projects S01 and TP12, Project ID329628492 and Project RE 3723/4-1). M.R. was supported by the Federal Ministry of Education and Research (BMBF), SATURN3 “Spatial and Temporal Resolution of Intratumoral Heterogeneity in 3 hard-to-treat Cancers” (Project 01KD2206P). The authors declare that no further funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
MK, TM, MI, AB and BWR planned the experiments, MK and TM performed the experiments. MK, TM, MI, AB and BWR performed the data analysis, all authors (MK, TM, IB, DK, FH, SS, KH, CBW, MR, MKA, IR, AB, JW, MR MI and BWR) edited the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of LMU (#02512).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Knoblauch, M., Ma, T., Beirith, I. et al. In-vitro model to mimic T cell subset change in human PDAC organoid co-culture. J Cancer Res Clin Oncol 149, 13051–13064 (2023). https://doi.org/10.1007/s00432-023-05100-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05100-7