Abstract
Purpose
This study aimed to explore the correlations among heavy metals concentration, histologic subtypes and molecular characteristics in patients with non-small cell lung cancer (NSCLC).
Methods
In this study, an NGS panel of 82 tumor-associated genes was used to identify genomic alternations in 180 newly diagnosed patients with NSCLC. The concentrations of 18 heavy metals in the serum samples were detected by inductively coupled plasma emission spectrometry (ICP-MS).
Results
A total of 243 somatic mutations of 25 mutant genes were identified in 115 of 148 patients with LUAD and 45 somatic mutations of 15 mutant genes were found in 24 of 32 patients with LUSC. The genomic alternations, somatic interactions, traditional serum biomarkers, and heavy metals were markedly different between patients with LUAD and LUSC. Moreover, patients with LUSC were significantly positively correlated with Ba, but not LUAD. Lastly, patients with EGFR mutations presented significant negative correlations with Cd and Sr, whereas patients with TP53 mutations showed a significant positive correlation with Pb.
Conclusion
The genomic alternations, somatic interactions, traditional serum biomarkers, and heavy metals were different between patients with LUAC and LUSC, and heavy metals (e.g., Ba, Pb, and Cd) may contribute to the tumorigenesis of NSCLC with different histological and molecular subtypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the development of personalized therapy, the sub-classification of NSCLC is ever more necessary. Previous studies have well indicated that lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) derived from distinct epithelial cells, exhibited distinct clinic-pathological features and expressed distinct biomarkers (Chen and Dhahbi 2021; Relli et al. 2019; Wang et al. 2022). Moreover, different genomic mutation landscapes between these two subtypes were also identified, which were higher mutation rates of EGFR, ALK, RET, ROS1, and KRAS in LUAD, and more prevalent mutations of PIK3CA, AKT1, and CDKN2A in LUSC (Campbell et al. 2016; Cancer Genome Atlas Research 2012, 2014). Especially, East Asian women without a smoking history tend to be developed LUAD and display a higher mutation rate of EGFR and a lower incidence of KRAS mutation (Ha et al. 2015; Li et al. 2011). What is more, prognosis also differs between these two subtypes (Wang et al. 2020). LUAD is more likely to develop in brain metastases after surgery and combined modality therapy compared with LUSC (Mamon et al. 2005; McAleese et al. 2019). Although the differences in clinic-pathological features, traditional serum biomarkers, and genomic profiles between LUAD and LUSC have been well studied, little is known about the difference in heavy metals.
Many studies have explored the association between environmental exposure to heavy metals (including As, Cd, Cr, Cu, Hg, Ni, Pb, and Zn) and lung cancer (Caffo et al. 2014; Hartwig et al. 2002; Huff et al. 2016; Kim et al. 2015). Metal dyshomeostasis could result in the activation of oncogenic signaling pathways, inhibition of the DNA repairing system, production of oxidative stress, and modification of epigenetic inheritance (Caffoet al. 2014; Hartwiget al. 2002; Huffet al. 2016; Kim et al. 2015). The concentration of Cu and the ratio of Cu/Zn derived from serum in patients with lung cancer were higher than that in healthy controls (Diez et al. 1989). The discharge amount of Zn was positively correlated with the incidence of lung cancer in Texas, USA (Coyle et al. 2006). Moreover, Huang et al. reported that the concentration of Cu in the soil is significantly positively correlated with LUAD for both sexes and with LUSC only for males (Huang et al. 2013). In addition to the effects of heavy metals on tumorigenesis, certain studies also focused on the relevance between heavy metals and prognosis. Low Cd levels in the blood (< 1.47 µg/L) were significantly correlated with improved overall survival in patients with lung cancer with stage IA (Pietrzak et al. 2021). In comparison with Fe-negative patients with LUAD, Fe-positive patients displayed improved overall survival accompanying higher numbers of M1-like pro-inflammatory tumor-associated macrophages (Thielmann et al. 2019). However, this unique influence of Fe on patients with LUSC was not observed (Thielmannet al. 2019). Collectively, both tumorigenic and prognostic effects of heavy metals showed specificities on different histological subtypes of NSCLC, whose elucidations might be useful for anti-cancer therapy.
In our study, targeted next-generation sequencing (NGS) was carried out to detect tissue DNA and ctDNA from NSCLC patients by an NGS panel of 82 tumor-associated genes. The differences in clinic-pathological features, traditional serum biomarkers, genomic alterations, somatic interactions, and the deposition of heavy metals were systematically compared between patients with LUAD and LUSC. Furthermore, we also investigated the correlations among histological subtypes, genomic alterations, traditional serum biomarkers, and heavy metals.
Materials and methods
Patients and samples collection
One hundred and eighty newly diagnosed patients with NSCLC from the Department of Oncology in the Affiliated Hospital of Chengde Medical University were recruited between April 2020 and July 2021. The pathological diagnosis was confirmed by three independent pulmonary pathologists based on the 4th edition of the World Health Organization Classification of Lung Tumors (Travis et al. 2015). Ninety-one Formalin-Fixed and Paraffin-Embedded (FFPE) tumor specimens and 89 plasma samples were collected for NGS analysis. This study was implemented according to the Code of Ethics of the World Medical Association (Declaration of Helsinki) (Gandevia and Tovell 1964).
DNA extraction and quality control
The methods of DNA extraction and quality control were quoted from our published paper, which was described as follows: “Genomic (g) DNA from the FFPE tumor specimens was extracted by GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany). The quantity and purity of gDNA were evaluated by Qubit® 3.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE, USA). DNA Integrity was assessed by the Agilent 2100 Bioanalyzer instrument (Agilent Technologies) via the High Sensitivity DNA Reagent (Agilent Technologies, Santa Clara, CA, USA).”(Nian et al. 2022).
Library preparation, hybridization capture, and illumina sequencing
Firstly, 300 nanogram gDNA per sample broke by an E220 focused ultrasonicator Covaris (Covaris, LLC.) to get 150–200 bp DNA fragments. Secondly, 10–100 nanogram DNA fragments were used for library construction by the KAPA library preparation kit (Kapa Biosystems Inc.; Roche Diagnostics). Thirdly, the NGS libraries were trapped by the xGen Lockdown Probe pool (Integrated DNA Technologies, Inc.), and the captured DNA fragments were amplified by 1X KAPA HiFi Hot Start Ready Mix (Kapa Biosystems Inc.; Roche Diagnostics). Lastly, the Illumina NextSeq CN500 platform with a medium flux chip (NextSeq CN500 Mid Output v2 kit; Illumina Inc.) was adopted to establish the NGS libraries (Nian et al. 2022).
Bioinformatics analysis
The methods of bioinformatics analysis were quoted from our published paper, which was described as follows: “The low-quality reads were filtered to gain clean data. All filtered reads were aligned to the human genome (University of California Santa Cruz ID: hg19) via the Burrows-Wheeler Aligner v. 0.7.12. Then, the Picard and Genome Analysis Toolkit (GATK v.3.2) method was adopted for duplicate removal, local realignment, and base quality score recalibration, which was also used to generate the quality statistics. Lastly, the VarDict was used for the authentication of single nucleotide variation (SNV) and Insertion/Deletion (InDel).
The ANNOVAR software tool was adopted for annotating somatic mutations. The candidates of somatic mutations were recognized by the following filter conditions: (i) remove mutations with coverage depth (VDP) less than 10×; (ii) remove variant sites with mutant allele frequency (MAF) > 0.001 in the 1000 Genomes databases (1,000 Genomes Project Consortium; https://www.internationalgenome.org/); (iii) retain variant sites with MAF ≥ 0.001 and < 0.1 in the 1000 Genomes databases with COSMIC evidence (http://cancer.sanger.ac.uk/cosmic); (iv) retain variations in the exonic or splicing region (10 bp upstream and downstream of splicing sites); (v) remove synonymous mutations; (vi) remove unknown variant classification; and (vii) remove the functional benign variant sites predicted by MutationTaster, PolyPhen-2 or SIFT. The biological consequences were explored by the Kyoto Encyclopedia Of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analysis via the cluster Profiler package (http://bioconductor.org/packages/release/bioc/html/ clusterProfiler.Html) (Yu et al. 2012). The correlation between the genomic alternations and their clinical consequences was defined by OncoKB Precision Oncology Database (http://oncokb.org/).”(Nian et al. 2022).
Traditional serum biomarker detection
At least 10 mL of whole blood for each patient was centrifuged at 3000 rpm/10 min at 4 °C to get the upper serum. The buffer solution (PH = 7.5) was used to prepare the standard solutions of carbohydrate antigen 125 (CA125), carcinoembryonic antigen (CEA), cytokeratin-19 fragments (CYFRA21-1), neuron-specific enolase (NSE), progesterone-releasing peptide (ProGRP), and squamous cell carcinoma antigen (SCC) adopting the serial dilution method. The levels of these traditional serum biomarkers were detected by chemiluminescence immunoassay (Gong and Zhang 2020; Qu et al. 2013). The testing equipment was an automated chemiluminescence immunoassay analyzer (COBAS 8000 E 801; Roche Diagnostics GmbH), and the detection kits of CA125, CEA, CYFRA21-1, NSE, ProGRP, and SCC were all originally imported from the Roche Diagnostics GmbH.
ICP-MS detection
The concentrations of 18 heavy metals in serum samples were detected by inductively coupled plasma mass spectrometry (ICP-MS) (Agilent 7800), including Arsenic (As), Barium (Ba), Cadmium (Cd), Cobalt (Co), Chromium (Cr), Cuprum (Cu), Gallium (Ga), Mercury (Hg), Manganese (Mn), Nickel (Ni), Plumbum (Pb), Stibium (Sb), Selenium (Se), Stannum (Sn), Strontium (Sr), Thallium (Tl), Vanadium (V), and Zinc (Zn). ICP-MS is used for multi-elemental capabilities analysis and the detection procedures are detailed in the manufacturer’s instructions (Zhao et al. 2018). In simple terms, at least 2 ml of whole blood for each patient was centrifuged at 3000 rpm/10 min to get the upper serum. The centrifugal serum was stored at − 20 °C.
Statistical analysis
The maftools package was adopted to depict genomic landscapes, lollipop plots, and spectrums of co-occurring and mutually exclusive genomic alterations via R software (R 4.0.3, R Core Team; https://www.R-Project.org). Fisher’s exact test was used to evaluate the statistical differences in categorical variables between the patients with LUAD and the patients with LUSC by R software. Continuous variables are shown as median with interquartile range (IQR) and they were compared by Mann–Whitney U test between the two groups in GraphPad Prism (v.7.0; GraphPad Software, La Jolla, CA). Correlation analysis among histologic subtypes, genomic alternations, 6 traditional serum biomarkers, and 18 heavy metals was conducted by Spearman’s Rank Correlation Analysis. A P value of less than 0.05 was considered statistically significant.
Results
Patient characteristics
In this study, the number of newly diagnosed NSCLC patients with stages I, II, III, and IV was 0, 1, 21, and 158, respectively. In terms of histologic type, 148 patients were diagnosed with LUAD, and 32 patients were LUSC. One hundred and seventy-six patients were detected by both heavy metals and traditional serum biomarkers. Significant differences in gender, EGFR mutation, smoking history, anatomical staging, brain metastatic status, CEA level, and SCC level were found between patients with LUAD and LUSC, whereas no significant differences were observed in other demographic and clinical characteristics, including age, height, weight, BMI, TP53 mutation, the history of drinking, CA125 level, CYFRA21-1 level, NSE level, and ProGRP level (Table 1).
Differences in genomic alterations between patients with LUAD and LUSC
To delineate the landscapes of genomic alterations, the somatic mutations from the tumor tissue DNA of 91 patients and the plasma-derived ctDNA of 89 patients were analyzed by an NGS panel of 82 tumor-associated genes (Table S1). We mainly focused on protein-altering variants according to the annotation of somatic SNVs and InDels. A total of 195 somatic variants of 25 mutated genes were detected in 86 out of 91 (94.51%) tumor tissues (Supplementary Fig. 1A and Table S2), and 93 somatic variants of 17 mutated genes were identified in 54 out of 89 (60.67%) peripheral blood samples (Supplementary Fig. 1B and Table S2). Our results were consistent with previous studies that the rate of mutation detection from tumor tissue DNA was much higher than that from plasma-derived ctDNA (Cai et al. 2021). Moreover, the percentages of tissue DNA-specific identified genes, ctDNA-specific identified genes, and co-identified mutated genes were 45.16% (14/31), 19.35% (6/31), and 35.48% (11/31) in NSCLC, respectively (Supplementary Fig. 1C).
It is well established that NSCLC patients carrying mutations of driver genes benefit much from the targeted therapies, and different genomic profiles have been uncovered between LUAD and LUSC (Campbellet al. 2016; Cancer Genome Atlas Research 2012, 2014). In this study, 243 somatic mutations of 25 mutant genes were identified in 115 of 148 (77.70%) patients with LUAD (Fig. 1A, C, and Table S3), and 45 somatic mutations of 15 mutant genes were found in 24 of 32 (75.00%) patients with LUSC (Fig. 1B, D, and Table S3). Among these mutant genes, ABCB1, ERBB4, GNA11, GSTP1, ROS1, and XRCC1 were exclusively identified in patients with LUSC, whereas 16 mutant genes were uniquely present in patients with LUAD. Nine mutant genes (CDKN2A, EGFR, ERBB2, FGFR2, KIT, PIK3CA, RB1, RET, and TP53) were concurrently detected in both groups. To further compare the feasibility of genomic profiling of patients with different histologic subtypes using plasma-derived ctDNA, the consistencies of genomic profiling between tissue DNA and ctDNA in the LUAD group and the LUSC group were all analyzed by the above NGS panel. Consistent with previous studies (Cai et al. 2021), ctDNA analysis has a higher consistency of genomic profiling in patients with LUAD (40.00%, 10/25) than that in patients with LUSC (26.67%, 4/15) (Fig. 1E and F).
Somatic mutation landscape of NSCLC derived from LUAD tumor tissue DNA (n = 79) (A), LUSC tumor tissue DNA (n = 12) (B), LUAD ctDNA (n = 69) (C), and LUSC ctDNA (n = 20) (D). Patients were arranged along the x-axis. Mutant genes are ranked by mutant frequency and the right panel shows the number of samples with nonsynonymous mutations. Tumor mutation burden (TMB, mutations per Mb) is shown in the upper panel. Concordance of mutated genes between tumor tissue DNA and ctDNA in patients with LUAD (E) and LUSC (F)
Furthermore, we also found that the frequencies and sites of these concurrent mutant genes between patients with LUAD and with LUSC were markedly different (Supplementary Fig. 2 and Table S4). For example, although about half of the patients carried TP53 mutations in both LUAD (48.65%, 72/148) and LUSC (59.38%, 19/32) groups, marked differences in the mutant types and sites were observed (Supplementary Fig. 2A). The in-frame insertion and frame-shift deletion of TP53 were exclusively identified in the LUAD group, whereas Q60fs, K93N, G113R, and other nine mutant sites were specifically observed in the LUSC group.
Differences in somatic interactions between patients with LUAD and LUSC
Patients with LUAD showed a higher mutation rate of EGFR compared to those with LUSC. Interestingly, EGFR mutations and Kirsten rat sarcoma (KRAS) mutations are generally mutually exclusive, and patients with both mutations confer resistance to EGFR-TKIs (Pao et al. 2005). In our study, the somatic interactions observed in patients with LUAD were markedly different from those with LUSC. EGFR and KRAS were mutually exclusive in the LUAD group (P = 0.0016) (Fig. 2A and Table S5), whereas no mutually exclusive interactions were found in the LUSC group (Fig. 2B and Table S5). Meanwhile, marked differences in the co-occurring set of genes were also identified between these two groups. FBXW7 and CDKN2A (P = 0.0093), KRAS and STK11 (P = 0.0158), and CDA and BRAF (P = 0.0346) were significant co-occurring pairs of genes in patients with LUAD (Fig. 2A and Table S5), while KIT and ERBB2 (P = 0.0417), and GSTP1 and FGFR2 (P = 0.0417) were observed in patients with LUSC (Fig. 2B and Table S5).
Differences in traditional serum biomarkers and heavy metals between patients with LUAD and LUSC
To further explore the differences in NSCLC with different histologic subtypes, we first compared the levels of six traditional serum biomarkers between patients with LUAD (n = 148) and with LUSC (n = 32). Expectedly, the levels of CEA and SCC were significantly different between the LUAD group and the LUSC group, and their median concentrations with IQR were as follows: CEA 13.21 (3.75–65.58) vs. 4.3 (2.36–16.15) ng/mL, and SCC 0.82 (0.51–1.38) vs. 1.21 (0.81–8.31) ng/mL (Supplementary Fig. 3 and Table S6).
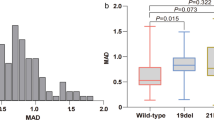
Next, we compared the concentration of the 18 heavy metals between these 2 groups, and only Ba showed significant differences (Fig. 3 and Table S6). The median concentrations with IQR for Ba in the LUAD group and the LUSC group were as follows: 36.27 (24.18–57.73) vs. 50.88 (29.62–78.54) μg/L (Fig. 3B).
Differences in traditional serum biomarkers and heavy metals between LUAD patients with and without EGFR mutations
To further complement the biological characteristics of LUAD in terms of 6 traditional serum biomarkers and 18 heavy metals, we performed comparative analyses between LUAD patients with (n = 70) and without EGFR mutations (n = 75). Interestingly, the levels of CYFRA21-1 and NSE in the EGFR positive group were all much higher than that in the EGFR negative group, whose median concentrations with IQR were as follows: CYFRA21-1 6.05 (3.505–10.08) vs. 4.12 (2.41–7.14) ng/mL, and NSE 18.34 (13.59–24.32) vs. 14.64 (12.13–19.22) ng/mL (Supplementary Fig. 4 and Table S6). Meanwhile, compared with patients without EGFR mutations, patients carrying EGFR mutations showed lower concentrations of Cd and Sr, and their median concentrations with IQR were as follows: Cd 0.29 (0.11–0.59) vs. 0.48 (0.26–1.17) μg/L, and Sr 24.78 (21.21–32.69) vs. 28.56 (22.63–39.58) μg/L (Fig. 4 and Table S6).
Correlations analysis among histologic subtypes, genomic alternations, demographic and clinical characteristics, traditional serum biomarkers, and heavy metals
To get the correlations among histologic subtypes, EGFR mutations, TP53 mutations, 5 demographic characteristics, distant metastatic status, brain metastatic status, and 18 heavy metals, the NGS analysis and the heavy metal detection were simultaneously performed on 145 patients with LUAD and 31 patients with LUSC. LUSC was significantly negatively correlated with EGFR mutations (r = − 0.3, P < 0.001), Female (r = − 0.18, P < 0.05), distant metastatic status (r = − 0.46, P < 0.001), and brain metastatic status (r = − 0.22, P < 0.01), and it was also significantly positively correlated with smoking history (r = 0.28, P < 0.001). No significant correlations were found between the LUSC and the 18 heavy metals except Ba (r = 0.16, P < 0.05) (Fig. 5). Interestingly, the EGFR mutations presented a prominent negative correlation with Cd (r = − 0.27, P < 0.001) and Sr (r = − 0.19, P < 0.01), whereas noteworthy positive correlations were also uncovered between the TP53 mutations and Pb (r = 0.18, P < 0.05) (Fig. 5). What is more, the concentration of Cd was significantly negatively correlated with females (r = − 0.4, P < 0.001), and significantly positively correlated with smoking history (r = 0.4, P < 0.001) and drinking history (r = 0.32, P < 0.001). Generally, our results were consistent with the previous finding that LUAD was more likely to occur in Chinese women carrying EGFR mutations without a history of smoking and drinking (Zhang et al. 2019). Lastly, significant correlations were also identified between most of the 18 heavy metals, such as As and Cd (r = 0.37, P < 0.001), As and Ga (r = 0.25, P < 0.001), As and Pb (r = 0.27, P < 0.001), and so on (Fig. 5).
Although CA125, CEA, CYFRA21-1, and SCC have been studied well in NSCLC, little is known about their diagnostic values combined with heavy metals in different histological and molecular subtypes. Thus, we explored the correlations among histologic types, EGFR mutations, TP53 mutations, 6 traditional serum biomarkers, and 18 heavy metals in 176 NSCLC patients. EGFR mutations were significantly positively correlated with CEA (r = 0.16, P < 0.05), and NSE (r = 0.15, P < 0.05), while TP53 mutations were significantly positively correlated with CA125 (r = 0.18, P < 0.05) and CYFRA21-1 (r = 0.25, P < 0.001). Furthermore, we also observed significant positive correlations between CA125 and Cu (r = 0.27, P < 0.001), NSE and Cu (r = 0.19, P < 0.05), SCC and Hg (r = 0.18, P < 0.05), and significant negative correlations between CA125 and Se (r = − 0.15, P < 0.05), CYFRA21-1 and Co (r = − 0.16, P < 0.05), CYFRA21-1 and Se (r = − 0.22, P < 0.01), SCC and Co (r = − 0.18, P < 0.05), and SCC and Mn (r =− 0.15, P < 0.01) (Fig. 6).
Discussion
The clinical management of patients with NSCLC is highly dependent on histologic types and molecular characteristics. Here, 148 LUAD patients and 32 LUSC patients were recruited in this study, and distinct characteristics in terms of gender, smoking history, anatomical staging, brain metastatic status, CEA level, and SCC level were found between these two groups (Relliet al. 2019). Two hundred and forty-three somatic mutations of 25 mutant genes were identified in 115 of 148 (77.70%) patients with LUAD and 45 somatic mutations of 15 mutant genes were found in 24 of 32 (75.00%) patients with LUSC. The genomic alternations, somatic interactions, traditional serum biomarkers, and heavy metals were markedly different between these two groups, further indicating that LUAD and LUSC should be classified and treated as different clinical entities. What is more, except for a significant positive correlation between LUSC and Ba, patients with EGFR mutations presented significantly negative correlations with Cd and Sr, and patients with TP53 mutations showed a significant positive correlation with Pb, suggesting that heavy metals may be regarded as supplementary characteristics for the sub-classification of NSCLC with different histological and molecular subtypes. To our knowledge, this is the first time to report these above results in the East Asian population.
NSCLC is highly heterogeneous, and different clinic-pathological features, serum biochemical markers, and genomic profiles have been identified between LUAD and LUSC (Campbellet al. 2016; Chen and Dhahbi 2021; Chen et al. 2014; Wanget al. 2020). Consistent with previous studies (Kawase et al. 2012; Wanget al. 2020; Wang et al. 2022), more males, smokers, and patients with the III stage were diagnosed with LUSC compared to patients with LUAD. Moreover, cases with LUSC have a more frequent high level of SCC than patients with LUAD (Yu et al. 2021). However, one of our findings was inconsistent with the previous reports, which was patients with LUAD were more frequently diagnosed with stage IV rather than other stages, partly due to the patients from different departments and regions (Wanget al. 2020; Wang et al. 2022).
Expectedly, the mutation rate of EGFR in the East Asian population with LUAD was markedly higher (48.65%, 72/148) compared with the mutated frequency observed in the Western population (14–21%) (Cancer Genome Atlas Research 2014; Keedy et al. 2011; Zhanget al. 2019). Despite a higher mutation rate of EGFR occurring in non-smoking women (62.5%) compared with non-smoking men (28.57%) in LUAD, the mutation rate in non-smoking women with LUAD in this study (62.5%) was lower than these women in the CHOICE study (75%) (Zhanget al. 2019). The mutant status of EGFR was independently associated with LUAD and the history of smoking, but not with the gender of females (Kosaka et al. 2004; Wei et al. 2016). This is partly because of the high percentage of non-smokers but cooks among Asian women. Consistent with the previous studies, the mutation rates of BRAF (1.35%, 2/148) and KRAS (6.08%, 9/148) in LUAD were relatively lower in comparison with the Western population (BRAF: 2–10%, KRAS: 33–38%) (Cancer Genome Atlas Research 2014; Kinno et al. 2014; Myers et al. 2015; Schmid et al. 2009). In addition, distinct somatic interactions were also observed between LUAD and LUSC in the Chinese population. FBXW7 and CDKN2A, KRAS and STK11, and CDA and BRAF were the only significant co-occurring pair of genes in patients with LUAD, while KIT and ERBB2, and GSTP1 and FGFR2 were exclusively observed in patients with LUSC. Genomic alterations in KRAS itself indicated the most common tumorigenic mutations in NSCLC, which was accompanied by a heterogeneous pattern of somatic interactions (Scheffler et al. 2019). In the LUAD patients with KRAS mutations, the most dominant co-occurring mutation was STK11 in this study, whereas co-alterations in TP53 were the biggest cluster in the Western population (Skoulidis and Heymach 2019).
Numerous studies have reported the close associations between NSCLC and environmental exposure to heavy metals, but few studies have focused on pathological cell types. Huang et al. reported that the concentration of Cu in the soil is significantly positively correlated with LUAD for both sexes and with LUSC only for males (Huanget al. 2013). Correspondingly, both the concentration of ceruloplasmin and the ratio of Cu/Zn in the serum of patients with lung cancer were much higher than those of healthy controls (Andrews 1979; Diezet al. 1989; Linder et al. 1981). However, the concentration of Cu in our study has not shown any significant differences between LUAD and LUSC, partly due to the limited number of LUSC and the regional divergence among different studies. Since the prognosis of patients with NSCLC is highly dependent on molecular characteristics, we also found a significant positive correlation between TP53 mutations and Pb, and significant negative correlations between EGFR mutations and Cd and Sr. However, there are no reports about these findings yet, and these associations need to be further studied.
Ba is regarded as a low-toxic element, but its abnormal deposition was founded in multiple cancers (National Toxicology 1994). Yasemin et al. have reported that the concentration of Ba in the hair of patients with breast cancer is significantly lower than that of healthy controls (Benderli Cihan et al. 2011). 2.5–5 uM of Ba has independent abilities to promote anchorage-independent growth and/or invasion of keratinocytes and fibroblast (Thang et al. 2011), which was the precancerous hallmark of transformed cells (Bertotti et al. 2006; Li et al. 2008). However, no studies have reported that the concentration of Ba in patients with LUSC is significantly higher than that in patients with LUAD, which was the first time reported in the Chinese population. What is more, the interactions among different heavy metals are also important for tumorigenesis. The antineoplastic effects of As main reliance on the apoptosis of squamous cell carcinomas via producing reactive oxygen species (ROS) and activating JNK1/2 and caspase-3 (Eguchi et al. 2011; Potin et al. 2007), barium-mediated inhibition of arsenic-induced apoptosis can accelerate tumor progression in patients who exposure to both As and Ba (Yajima et al. 2012). In our study, significant correlations were observed between most of the 18 heavy metals, including As and Ba. Thus, it is interesting to explore the activation and/or inhibition of molecular mechanisms underlying these heavy metals with abnormal deposition.
In this study, the limitations are mainly reflected in four points. Firstly, NGS data of tumor tissue DNA and ctDNA were acquired from single samples without matched normal tissue samples, because the actionable genomic alterations about the clinical decision are enough to get from a single sample according to the suitable filter conditions (Adzhubei et al. 2013; Genomes Project et al. 2015; Hiltemann et al. 2015; Kircher et al. 2014; McNulty et al. 2019; Ng and Henikoff 2003; Schwarz et al. 2010; Sukhai et al. 2019; Teer et al. 2017). What we cannot ignore is that the cost of multi-type or multiregional biopsies is much more than that of a single sample. Secondly, this study only examined the concentrations of heavy metals in serum, but did not elaborate on the effects of one metal with different forms, nor elaborate on the source of exposure (e.g., soil, food, water, or air). Thirdly, the concentrations of heavy metals were detected only in serum without the matched urine, hair, and nail. Hair and nails directly store heavy metals (e.g., Co, Cr, Cu, Fe, Ni, and Zn), which makes them appropriate for monitoring the effects of heavy metals on health. Thus, it is attractive to detect the concentrations of heavy metals in serum, hair, and nail samples from one person in future studies. Lastly, further studies with more sizeable sample sizes and multi-institution are necessary to validate the generalizability of our conclusion.
Conclusions
In summary, the genomic alternations, somatic interactions, traditional serum biomarkers, and heavy metals were markedly different between patients with LUAC and LUSC, and heavy metals (e.g., Ba, Pb, and Cd) may contribute to the tumorigenesis of NSCLC with different histological and molecular subtypes.
Data availability
The raw data supporting our conclusions are included in this article. The sequencing data are available at the NCBI BioProject database PRJNA904420.
References
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7(Unit7):20. https://doi.org/10.1002/0471142905.hg0720s76
Andrews GS (1979) Studies of plasma zinc, copper, caeruloplasmin, and growth hormone: with special reference to carcinoma of the bronchus. J Clin Pathol 32:325–333. https://doi.org/10.1136/jcp.32.4.325
BenderliCihan Y, Sozen S, OzturkYildirim S (2011) Trace elements and heavy metals in hair of stage III breast cancer patients. Biol Trace Elem Res 144:360–379. https://doi.org/10.1007/s12011-011-9104-z
Bertotti A, Comoglio PM, Trusolino L (2006) Beta4 integrin activates a Shp2-Src signaling pathway that sustains HGF-induced anchorage-independent growth. J Cell Biol 175:993–1003. https://doi.org/10.1083/jcb.200605114
Caffo M, Caruso G, Fata GL et al (2014) Heavy metals and epigenetic alterations in brain tumors. Curr Genomics 15:457–463. https://doi.org/10.2174/138920291506150106151847
Cai J, Jiang H, Li S et al (2021) The landscape of actionable genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in chinese patients with non-small cell lung cancer. Front Oncol 11:751106. https://doi.org/10.3389/fonc.2021.751106
Campbell JD, Alexandrov A, Kim J et al (2016) Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 48:607–616. https://doi.org/10.1038/ng.3564
Cancer Genome Atlas Research N (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519–525. https://doi.org/10.1038/nature11404
Cancer Genome Atlas Research N (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511:543–550. https://doi.org/10.1038/nature13385
Chen JW, Dhahbi J (2021) Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods. Sci Rep 11:13323. https://doi.org/10.1038/s41598-021-92725-8
Chen Z, Fillmore CM, Hammerman PS et al (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14:535–546. https://doi.org/10.1038/nrc3775
Coyle YM, Minahjuddin AT, Hynan LS et al (2006) An ecological study of the association of metal air pollutants with lung cancer incidence in Texas. J Thorac Oncol 1:654–661
Diez M, Cerdan FJ, Arroyo M et al (1989) Use of the copper/zinc ratio in the diagnosis of lung cancer. Cancer 63:726–730. https://doi.org/10.1002/1097-0142(19890215)63:4%3c726::aid-cncr2820630421%3e3.0.co;2-p
Eguchi R, Fujimori Y, Takeda H et al (2011) Arsenic trioxide induces apoptosis through JNK and ERK in human mesothelioma cells. J Cell Physiol 226:762–768. https://doi.org/10.1002/jcp.22397
Gandevia B, Tovell A (1964) Declaration of Helsinki. Med J Aust 2:320–321
Genomes Project C, Auton A, Brooks LD et al (2015) A global reference for human genetic variation. Nature 526:68–74. https://doi.org/10.1038/nature15393
Gong X, Zhang H (2020) Diagnostic and prognostic values of anti-elicobacter pylori antibody combined with serum CA724, CA19-9 and CEA for young patients with early gastric cancer. J Clin Lab Anal 34(7):e23268. https://doi.org/10.1002/jcla.23268
Ha SY, Choi SJ, Cho JH et al (2015) Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget 6:5465–5474. https://doi.org/10.18632/oncotarget.2925
Hartwig A, Asmuss M, Ehleben I et al (2002) Interference by toxic metal ions with DNA repair processes and cell cycle control: molecular mechanisms. Environ Health Perspect 110(Suppl 5):797–799. https://doi.org/10.1289/ehp.02110s5797
Hiltemann S, Jenster G, Trapman J et al (2015) Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res 25:1382–1390. https://doi.org/10.1101/gr.183053.114
Huang HH, Huang JY, Lung CC et al (2013) Cell-type specificity of lung cancer associated with low-dose soil heavy metal contamination in Taiwan: an ecological study. BMC Public Health 13:330. https://doi.org/10.1186/1471-2458-13-330
Huff MO, Todd SL, Smith AL et al (2016) Arsenite and cadmium activate MAPK/ERK via membrane estrogen receptors and G-protein coupled estrogen receptor signaling in human lung adenocarcinoma cells. Toxicol Sci 152:62–71. https://doi.org/10.1093/toxsci/kfw064
Kawase A, Yoshida J, Ishii G et al (2012) Differences between squamous cell carcinoma and adenocarcinoma of the lung: are adenocarcinoma and squamous cell carcinoma prognostically equal? Jpn J Clin Oncol 42:189–195. https://doi.org/10.1093/jjco/hyr188
Keedy VL, Temin S, Somerfield MR et al (2011) American society of clinical oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol 29:2121–2127. https://doi.org/10.1200/JCO.2010.31.8923
Kim HS, Kim YJ, Seo YR (2015) An overview of carcinogenic heavy metal: molecular toxicity mechanism and prevention. J Cancer Prev 20:232–240. https://doi.org/10.15430/JCP.2015.20.4.232
Kinno T, Tsuta K, Shiraishi K et al (2014) Clinicopathological features of nonsmall cell lung carcinomas with BRAF mutations. Ann Oncol 25:138–142. https://doi.org/10.1093/annonc/mdt495
Kircher M, Witten DM, Jain P et al (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–315. https://doi.org/10.1038/ng.2892
Kosaka T, Yatabe Y, Endoh H et al (2004) Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 64:8919–8923. https://doi.org/10.1158/0008-5472.CAN-04-2818
Li Z, Zhao J, Du Y et al (2008) Down-regulation of 14–3-3zeta suppresses anchorage-independent growth of lung cancer cells through anoikis activation. Proc Natl Acad Sci U S A 105:162–167. https://doi.org/10.1073/pnas.0710905105
Li C, Fang R, Sun Y et al (2011) Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS One 6:e28204. https://doi.org/10.1371/journal.pone.0028204
Linder MC, Moor JR, Wright K (1981) Ceruloplasmin assays in diagnosis and treatment of human lung, breast, and gastrointestinal cancers. J Natl Cancer Inst 67:263–275
Mamon HJ, Yeap BY, Janne PA et al (2005) High risk of brain metastases in surgically staged IIIA non-small-cell lung cancer patients treated with surgery, chemotherapy, and radiation. J Clin Oncol 23:1530–1537. https://doi.org/10.1200/JCO.2005.04.123
McAleese J, Taylor A, Walls GM et al (2019) Differential relapse patterns for non-small cell lung cancer subtypes adenocarcinoma and squamous cell carcinoma: implications for radiation oncology. Clin Oncol (r Coll Radiol) 31:711–719. https://doi.org/10.1016/j.clon.2019.07.008
McNulty SN, Parikh BA, Duncavage EJ et al (2019) Optimization of population frequency cutoffs for filtering common germline polymorphisms from tumor-only next-generation sequencing data. J Mol Diagn 21:903–912. https://doi.org/10.1016/j.jmoldx.2019.05.005
Myers MB, McKim KL, Meng F et al (2015) Low-frequency KRAS mutations are prevalent in lung adenocarcinomas. Per Med 12:83–98. https://doi.org/10.2217/pme.14.69
National Toxicology P (1994) NTP toxicology and carcinogenesis studies of barium chloride dihydrate (CAS No. 10326–27-9) in F344/N rats and B6C3F1 mice (drinking water studies). Natl Toxicol Program Tech Rep Ser 432:1–285
Ng PC, Henikoff S (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814. https://doi.org/10.1093/nar/gkg509
Nian R, Jiang H, Zhao J et al (2022) Differences in actionable genomic alterations between brain metastases and non-brain metastases in patients with non-small cell lung cancer. Int J Oncol. https://doi.org/10.3892/ijo.2022.5390
Pao W, Wang TY, Riely GJ et al (2005) KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2:e17. https://doi.org/10.1371/journal.pmed.0020017
Pietrzak S, Wojcik J, Baszuk P et al (2021) Influence of the levels of arsenic, cadmium, mercury and lead on overall survival in lung cancer. Biomolecules. https://doi.org/10.3390/biom11081160
Potin S, Bertoglio J, Breard J (2007) Involvement of a Rho-ROCK-JNK pathway in arsenic trioxide-induced apoptosis in chronic myelogenous leukemia cells. FEBS Lett 581:118–124. https://doi.org/10.1016/j.febslet.2006.12.016
Qu S, Liu J, Luo J et al (2013) A rapid and highly sensitive portable chemiluminescent immunosensor of carcinoembryonic antigen based on immunomagnetic separation in human serum. Anal Chim Acta 766:94–99. https://doi.org/10.1016/j.aca.2012.12.043
Relli V, Trerotola M, Guerra E et al (2019) Abandoning the notion of non-small cell lung cancer. Trends Mol Med 25:585–594. https://doi.org/10.1016/j.molmed.2019.04.012
Scheffler M, Ihle MA, Hein R et al (2019) K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J Thorac Oncol 14:606–616. https://doi.org/10.1016/j.jtho.2018.12.013
Schmid K, Oehl N, Wrba F et al (2009) EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res 15:4554–4560. https://doi.org/10.1158/1078-0432.CCR-09-0089
Schwarz JM, Rodelsperger C, Schuelke M et al (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7:575–576. https://doi.org/10.1038/nmeth0810-575
Skoulidis F, Heymach JV (2019) Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 19:495–509. https://doi.org/10.1038/s41568-019-0179-8
Sukhai MA, Misyura M, Thomas M et al (2019) Somatic tumor variant filtration strategies to optimize tumor-only molecular profiling using targeted next-generation sequencing panels. J Mol Diagn 21:261–273. https://doi.org/10.1016/j.jmoldx.2018.09.008
Teer JK, Zhang Y, Chen L et al (2017) Evaluating somatic tumor mutation detection without matched normal samples. Hum Genomics 11:22. https://doi.org/10.1186/s40246-017-0118-2
Thang ND, Yajima I, Kumasaka MY et al (2011) Barium promotes anchorage-independent growth and invasion of human HaCaT keratinocytes via activation of c-SRC kinase. PLoS One 6:e25636. https://doi.org/10.1371/journal.pone.0025636
Thielmann CM, Costa da Silva M, Muley T et al (2019) Iron accumulation in tumor-associated macrophages marks an improved overall survival in patients with lung adenocarcinoma. Sci Rep 9:11326. https://doi.org/10.1038/s41598-019-47833-x
Travis WD, Brambilla E, Nicholson AG et al (2015) The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10:1243–1260. https://doi.org/10.1097/JTO.0000000000000630
Wang BY, Huang JY, Chen HC et al (2020) The comparison between adenocarcinoma and squamous cell carcinoma in lung cancer patients. J Cancer Res Clin Oncol 146:43–52. https://doi.org/10.1007/s00432-019-03079-8
Wang W, Liu H, Li G (2022) What’s the difference between lung adenocarcinoma and lung squamous cell carcinoma? Evidence from a retrospective analysis in a cohort of Chinese patients. Front Endocrinol (lausanne) 13:947443. https://doi.org/10.3389/fendo.2022.947443
Wei WE, Mao NQ, Ning SF et al (2016) An analysis of EGFR mutations among 1506 cases of non-small cell lung cancer patients in Guangxi, China. PLoS One 11:e0168795. https://doi.org/10.1371/journal.pone.0168795
Yajima I, Uemura N, Nizam S et al (2012) Barium inhibits arsenic-mediated apoptotic cell death in human squamous cell carcinoma cells. Arch Toxicol 86:961–973. https://doi.org/10.1007/s00204-012-0848-9
Yu G, Wang LG, Han Y et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Yu J, Du F, Yang L et al (2021) Identification of potential serum biomarkers for simultaneously classifying lung adenocarcinoma, squamous cell carcinoma and small cell carcinoma. Cancer Biomark 30:331–342. https://doi.org/10.3233/CBM-201440
Zhang XC, Wang J, Shao GG et al (2019) Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun 10:1772. https://doi.org/10.1038/s41467-019-09762-1
Zhao S, Cao S, Luo L et al (2018) A preliminary investigation of metal element profiles in the serum of patients with bloodstream infections using inductively-coupled plasma mass spectrometry (ICP-MS). Clin Chim Acta 485:323–332. https://doi.org/10.1016/j.cca.2018.07.013
Acknowledgements
We would like to thank the Hebei Natural Science Foundation and the Hebei Health Commission for their support.
Funding
This research was supported by the Hebei Natural Science Foundation (No. H2020406050) and the Hebei Health Commission (No. 20211664).
Author information
Authors and Affiliations
Contributions
DM, HT, HJ, and CZ: contributed to the study conception and design. Material preparation and data collection were performed by DM, GT, XL, YZ, GG, DW, LB, and XL. Data analysis were performed by MW, LJ, SW, HJ, and CZ. The first draft of the manuscript was written by HJ and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article was approved by the ethics committee of Affiliated Hospital of Chengde Medical University.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publish
Written informed consent was obtained from all participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mu, D., Tang, H., Teng, G. et al. Differences of genomic alterations and heavy metals in non-small cell lung cancer with different histological subtypes. J Cancer Res Clin Oncol 149, 9999–10013 (2023). https://doi.org/10.1007/s00432-023-04929-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04929-2