Abstract
Aim
To establish and validate a prognostic nomogram of cholangiocarcinoma (CCA) using independent clinicopathological and genetic mutation factors.
Methods
213 patients with CCA (training cohort n = 151, validation cohort n = 62) diagnosed from 2012 to 2018 were included from multi-centers. Deep sequencing targeting 450 cancer genes was performed. Independent prognostic factors were selected by univariate and multivariate Cox analyses. The clinicopathological factors combined with (A)/without (B) the gene risk were used to establish nomograms for predicting overall survival (OS). The discriminative ability and calibration of the nomograms were assessed using C-index values, integrated discrimination improvement (IDI), decision curve analysis (DCA), and calibration plots.
Results
The clinical baseline information and gene mutations in the training and validation cohorts were similar. SMAD4, BRCA2, KRAS, NF1, and TERT were found to be related with CCA prognosis. Patients were divided into low-, median-, and high-risk groups according to the gene mutation, the OS of which was 42.7 ± 2.7 ms (95% CI 37.5–48.0), 27.5 ± 2.1 ms (95% CI 23.3–31.7), and 19.8 ± 4.0 ms (95% CI 11.8–27.8) (p < 0.001), respectively. The systemic chemotherapy improved the OS in high and median risk groups, but not in the low-risk group. The C-indexes of the nomogram A and B were 0.779 (95% CI 0.693–0.865) and 0.725 (95% CI 0.619–0.831), p < 0.01, respectively. The IDI was 0.079. The DCA showed a good performance and the prognostic accuracy was validated in the external cohort.
Conclusion
Gene risk has the potential to guide treatment decision for patients at different risks. The nomogram combined with gene risk showed a better accuracy in predicting OS of CCA than not.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cholangiocarcinoma is an epithelial cell malignancy located within the biliary tree united by late diagnosis and poor outcomes (Razumilava and Gores 2014). The incidence of CCA has increased globally over the past few decades. According to their anatomical location, CCAs are classified into intrahepatic (iCCA), perihilar (pCCA), and distal (dCCA) (Rizvi et al. 2018). The tumor-node-metastasis (TNM) staging system is critical for prognostic prediction and risk stratification (Gaag et al. 2012). Although resection and curative liver transplantation are options for selected patients with pCCA, 5-year survival rates are very low, ranged 25–40% (Mazzaferro et al. 2020). The established standard of care includes first-line (gemcitabine and cisplatin), second-line (FOLFOX), and adjuvant (capecitabine) systemic chemotherapy (Kelley et al. 2020). The clinicopathological information and treatment choice were all influencing the prognosis of CCA. Hence, the current anatomical-based staging system is not sufficient to predict the prognosis of CCA.
Prognosis differences might be related to biological heterogeneity, and molecular investigation might reveal biomarkers that can be used to predict prognosis and guide treatment for patients in different risk groups. With the improvement of detection technology, many gene mutations were found to be related to the prognosis of CCA, such as FGFR2 (Makawita et al. 2020), HAMP (Wang and Du 2021), GLUT-1 (Labib et al. 2019), KRAS (Dong et al. 2022), and so on. Many gene mutation-based target tyrosine kinase inhibitors were attempted to improve the prognosis of CCA (Andersen et al. 2012). But in fact, to guide individual treatment for patients, new biomarkers that reflect tumor heterogeneity are still needed. Meanwhile, a new prognosis evaluation system including gene mutation information should be established for CCA.
In this study, 450 cancer-related gene expression was sequenced and analysis aimed to identify and validate a gene expression signature that predicts OS in patients with CCA. Moreover, we combined genomic and clinical variables to generate a nomogram model with better predictive power than clinical risk factors for OS. An external cohort showed similar results. The discriminative ability and calibration of the nomogram with or without genomic information were evaluated using C-index values, integrated discrimination improvement (IDI), calibration plots, and decision curve analysis (DCA). Overall, we highlight the genomic information in CCA prognosis prediction and treatment selection.
Methods
Clinical specimens and study design
Totally 213 CCA patients were admitted in this study from the Second Hospital of Shandong University and the Shandong Provincial Hospital, which were divided into training cohort (n = 151) and validation cohort (n = 62) according to the admitted sequence. No patient had received any anti-tumor therapy before the pathological confirmation except directly resection. On accounting of quite different prognosis of different position of CCAs, and mixed hepatocellular cholangiocarcinomas have emerged as a distinct subtype of primary liver cancer (Razumilava and Gores 2014), we excluded the iCCAs in our study to eliminate the influence of iCCAs arising in cirrhosis. The inclusion criteria included pathologically confirmed cholangiocarcinoma; ECOG ≤ 2; clinicopathological, gene mutation and the following-up information were completed. The exclusion criteria included complicated with other tumors; not confirmed by pathology; did not receive any antitumor therapy; loss to follow-up.
All patients underwent staging contrast-enhanced CT scans or MRI, if necessary 18F-fluorodeoxyglucose positron emission tomography (18FDG-PET) also was recommended. Two radiologists independently reassessed all imaging scans, and the third radiologist was involved to resolve any disagreements. We restaged all patients according to the 8th edition of the American Joint Committee on Cancer Staging Manual. Surgery was preferred if available. The systemic treatment including adjuvant chemical therapy after resection; chemical therapy alone if the tumor was un-resectable, and the first line was gemcitabine and cisplatin, followed by FOLFOX if failed; target tyrosine kinase inhibitors were attempted if gene mutation recommended; immune checkpoint inhibitors were not used commonly mostly due to non-medical insurance and low willingness-to-pay. This retrospective analysis of anonymous data was approved by the institutional ethics review boards of the Shandong Provincial Hospital (ethics approval number: LCYJ: NO. 2019-081), and informed consent was waived by the ethics review boards.
Procedures
We extracted at least 50 ng of DNA from each 40mm3 formalin-fixed, paraffin-embedded tumor sample using the QIAamp DNA FFPE Tissue Kit according to the manufacturer's protocol. The hybridization capture panel captured all coding exons of 450 tumor-related genes and selected introns of 39 commonly rearranged genes. Illumina NextSeq-500 was used to capture and sequence genes from FFPE samples and matched paracancerous samples. Sequencing results were further analyzed for single nucleotide variations (SNV), long- and short-range insertions and deletions (Indels), copy number variations (CNVs) and gene rearrangement/fusion structural variants. OrigiMed, an accredited and CLIA-certified College of American Pathologists laboratory, performed the genomic profiling using the YuanSu 450 panel (Cao et al. 2019).
Gene mutation and critical clinicopathological information
The main endpoint was the overall survival (OS). We calculated OS to death from any reason. In the training cohort, expression profiling study was performed and which the mutation frequency was higher than 5% was selected to be the potential prognostic candidates. If gene candidates were correlated with the OS were analyzed, using univariate and multivariate Cox analysis, in which both were positive confirmed to be a prognostic gene mutation in this study. A novel gene mutation score was established by the number of the prognostic gene mutations, and all the patients were divided into three risk of groups: the low-risk (gene mutation score = 0), median-risk (gene mutation score = 1) and high-risk (gene mutation score ≥ 2) group. The overall survival curve of patients in different groups was compared.
Albumin–bilirubin (ALBI) grade was a novel independent prognostic factor of CCA (Wang et al. 2018). The ALBI score is 0.66 × log10TBIL (μmol/l)− 0.085 × ALB (g/l). To increase the degree of discrimination, we modified the ALBI grade named mALBI. The mALBI grade was defined by the resulting score (grade 1: ≤ − 2.60; grade 2: − 1.39 to − 2.27; grade 3:− 2.27 to − 2.60; grade 4: > − 1.39) (Tokunaga et al. 2021).
Ca19-9 was a classical tumor biomarker of CCA (Wannhoff and Gotthardt 2019). Both the Ca19-9 level before and after resection were reported to be related to the prognosis of CCA (Jiang et al. 2021). So, we established a new tumor biomarker named Ca19-9 score, which was cumulated according the Ca19-9 before and after the resection if higher than 1000 u/ml.
Statistical analysis
Comparisons between the training and test cohort were performed using the χ2 test when appropriate. OS was calculated using the Kaplan–Meier method and hazard ratios (HRs) were calculated using a univariate Cox regression analysis. The gene mutation score as a predictor for systemic treatment efficacy was analyzed in the training cohort and validated in the external test cohort. Univariate and multivariate Cox regression analyses were performed to select the independent prognostic factors including the gene mutation score and all clinicopathological information. The p value threshold was 0.05 (p > 0.05) for removing non-significant variables from the analysis. Correspondingly, significant variables (p ≤ 0.05) remained in the final Cox model. Covariates included the gene mutation score (0, 1, ≥ 2), mALBI (1–4), Ca19-9 score (0–2), stage, systemic treatment (no, unregular, and regular systemic treatment) and surgery (radical operation or not).
The rms package in R was used to formulate nomograms. For the generation of nomograms, we used the coefficients from a multivariable Cox regression model. The discriminative ability and calibration of the nomograms with or without the gene mutation score were assessed using C-index values, integrated discrimination improvement (IDI), calibration plots, and decision curve analysis (DCA). R (version 3.2.1) and SPSS (22.0) were used for the statistical analyses. Statistical tests were conducted two-sidedly, and p values less than 0.05 were considered significant.
Results
Baseline clinical data of the subjects
In this retrospective study, 151 CCA patients were included in the training cohort and 62 patients in the validation cohort. The baseline clinical data, including but not limited age, sex, tumor stage, radical surgery or not, mALBI, Ca19-9 score, systemic treatment or not, were compared between the two groups, and no significantly difference was found. Detailed information was shown in Table 1.
The gene mutation was similar in the training and validation cohort
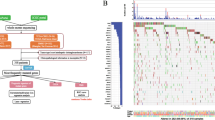
The TOP10 gene mutations in training cohort were TP53(64%), KRAS (36%), SMAD4(19%), CDKN2A (18%), ARID1A (15%), ARID2(14%), ERBB2(10%), LRP1B (12%), KMT2C (11%), and MUC16(10%); and TP53(68%), KRAS (44%), SMAD4(26%), CDKN2A (23%), ERBB2(19%), KMT2C (15%), ARID2(13%), MUC16(13%), TERT(13%), and ARID1A(11%) in the validation cohort. The common mutation included substitution/Indel, gene amplification, gene homozygous deletion, fusion/rearrangement, and truncation. Detailed information is shown in Fig. 1.
The gene mutation was correlated with the prognosis of CCA
The genes whose mutation frequency higher than 5% in the training cohort were selected to be prognostic candidates of CCA. SMAD4, MUC16, BRCA2, KRAS, ERBB2, NF1, TERT, and MDM2 were found to be associated with OS of CCA in the univariable Cox analysis, in those MUC16, ERBB2, and MDM2 did not show significant association with OS in the multivariable Cox analysis. Therefore, SMAD4, BRCA2, KRAS, NF1, and TERT formed the gene mutation score, which divided CCA patients into three groups. The detailed information is shown in Fig. 2. The OS of the low-risk group was 42.7 ± 2.7 ms (95% CI 37.5–48.0), the median-risk group was 27.5 ± 2.1 ms (95% CI 23.3–31.7), and the high-risk group was 19.8 ± 4.0 ms (95% CI 11.8–27.8) (p < 0.001), respectively. The survival curve is shown in Fig. 3a.
The gene risk can be used as a prediction of concurrent systemic chemotherapy
Considering many gene mutation was associated with OS of CCA in our study, and different treatment response revealed in patients, we analyzed the relationship with the gene mutation score and systemic chemotherapy, and interestingly found that the gene mutation score can be used as a prediction of concurrent systemic chemotherapy. The systemic chemotherapy condition of patients was divided into three kinds: no systemic treatment, unregular systemic treatment, and regular systemic treatment. The regular systemic treatment (RST) means the patient received full course chemotherapy and adjusted timely according to multi-disciplinary team suggestion. No systemic treatment (NST) means the patient received no chemotherapy for whatever reason. The unregular systemic treatment (UST) was the median condition.
The OS of NST, UST, and RST patients in the low-risk group were 45.4 ± 6.4 ms (95% CI 32.9–52.9), 41.5 ± 5.4 ms (95% CI 30.9–52.0), and 42.7 ± 3.5 ms (95% CI 35.9–49.5), (p > 0.05), respectively. In the median-risk group, the OS in those three groups were 13.5 ± 2.1 ms (95% CI 9.4–17.7), 23.0 ± 2.5 ms (95% CI 18.0–27.9), and 33.3 ± 3.1 ms (95% CI 27.2–39.4), (p < 0.05), respectively. In the high-risk group, the OS were 6.3 ± 1.5 ms (95% CI 3.5–9.2), 11.9 ± 2.4 ms (95% CI 7.2–16.6), and 26.4 ± 5.7 ms (95% CI 14.9–37.9), (p < 0.05), respectively. Systemic treatment improved the OS of CCA in the median- and high-risk groups (Fig. 3b, c), but not in the low-risk group (Fig. 3d).
Independent prognostic risk factors selection
All clinicopathological information and the gene risk were analyzed if associated with the OS of CCA. Items both were positively related with OS in the univariable and multivariable Cox analyses were selected to be the independent prognostic risk factors. The surgery, stage, systemic treatment, mALBI, Ca19-9 Score, and gene risk were on the official list. Detailed information is shown in Fig. 4.
The nomogram including the gene risk predicted the prognosis of CCA better than not
Nomograms were generated using multivariate analysis to predict OS in the training cohort (Fig. 5A, B). The predictors in nomogram A included surgery, stage, systemic treatment, mALBI, Ca19-9 Score, and gene risk. Meanwhile, the Nomogram B included all the clinicopathological information but excluded the gene risk to evaluate its influence in the OS prediction of CCA. The C-indexes of nomogram A and B were 0.779 (95% CI 0.693–0.865) and 0.725 (95% CI 0.619–0.831), (p < 0.01), respectively. The calibration curve of Nomogram A was closer with the ideal status than Nomogram B (Fig. 5C, D).The integrated discrimination improvement (IDI) of nomogram A to B was 0.079 (Fig. 5E). When the ratio is between 0.1 and 0.5, the net benefit ratio of Nomogram A was higher than Nomogram B (Fig. 5F). The calibration plot in the validation cohort confirmed the good prediction of the nomograms (Fig. 5G).
The comparison of Nomograms with or without gene risk and the validation information. A, B Nomogram with and without gene risk; C, D the calibration curve of Nomogram A and B; E the integrated discrimination improvement (IDI) curve of nomogram A, B; F the decision curve analysis (DCA) of Nomogram A and B; G the calibration plot in the validation cohort
Discussion
CCA is a cholangiocyte original malignancy within the biliary tree with poor prognosis, and is divided into three categories according to anatomical location as intrahepatic (iCCA), perihilar (pCCA), and distal (dCCA) with different strategies for clinical management and prognosis. Surgical treatment and liver transplantation were the first choice if available. For metastatic CCA, systemic treatment including gemcitabine and cisplatin or EGFR inhibitors was recommended. Despite great efforts have been attempted, the prognosis of CCA is not satisfactorily improved. Apart from the CCA grade, many factors were reported to be associated with CCA prognosis, such as hepatic viral infection history, the baseline liver function and Ca19-9 level, radical resection or not, and systemic treatment status. Recently, according to the cost reduction and popularizing of gene testing technical, more and more gene mutations were reported to be related with the prognosis of CCA. To better estimate the prognosis of CCA, we selected the prognosis-related clinical factors and gene mutations, and build nomograms including the gene risk score or not, compared to their accuracy and validation through the external test cohort.
Many gene mutations might occur in CCA progression. In this study, the big panel of 450 genes test showed the mutation information was similar between the training and validation cohort, and also similar to the studies reported before (Montal et al. 2020; Yoon et al. 2021). TP53, KRAS, SMAD4, and CDKN2A were the TOP4 frequency mutations both in the training and validation cohorts. SMAD4, BRCA2, KRAS, NF1, and TERT were selected to form the gene mutation score after univariable and multivariable Cox analyses. SMAD4 is reported as an independent prognostic biomarker of CCA. SMAD4 suppresses CCA proliferation, migration, invasion, and sensitivity to Pemigatinib by regulating the phosphorylation and intracellular localization of β-catenin (Liu et al. 2022). BRCA2-related CCA was uncommon. If BRCA2 is positive, Olaparib and Pembrolizumab might be helpful for advanced CCA (Zhou et al. 2022; Li et al. 2021; Costa et al. 2023). In addition to its own anti-tumor effects, Olaparib sensitizes cholangiocarcinoma cells to radiation if BRCA2 is positive (Mao et al. 2018). KRAS mutation was prevalent, and can be considered with targeted therapies (Nguyen et al. 2021). The molecular characterization of CCA could predict chemotherapy and Programmed Death 1/Programmed Death-Ligand 1 blockade responses (Yoon et al. 2021). Interestingly, we also found that the gene mutation score of CCA was associated with the response of systematic treatment (ST) in our study. In low-risk group, patients seemed not get beneficial outcome from ST, exhibiting potentiality of selecting more profitable patients to receive ST according to the gene mutation score, which might decrease the unnecessary overtreatment and improve the patients’ quality of life.
Ca19-9 was a traditional biomarker of CCA for diagnosis and prognosis (Liang et al. 2015). The preoperative CA19-9 level was associated with clinicopathological factors and overall survival. Persistent high CA19-9 level is after resection of CCA and R1 section, especially in the preoperative high-level group. To better reveal the influence of preoperative CA19-9 level and postoperative improvement of CCA prognosis, we proposed a new index named ‘CA19-9 Score’ considering both pre- and postoperative level. In our study, CA19-9 Score was significantly gradient related with OS of CCA. The high level at preoperative stage mostly related with high tumor load or lymphatic metastasis, and high level at postoperative stage mostly related with residual tumor, both were independent risk factors for OS (Lee et al. 2021).
Child–Pugh (CP) grade has been widely used to evaluate liver function and postoperative outcomes in biliary tract malignancy. (Wang et al. 2018) reported a novel alternative model of liver function, called albumin–bilirubin (ALBI) grade in 2018, showed a better prediction in extrahepatic cholangiocarcinoma. Tokunaga T. et al. modified this model by dividing the grade 2 into grade 2a and 2b, exhibited eligible prediction for second-line therapies of hepatocarcinoma (Tokunaga et al. 2021). In this study, we found that the OS of patients with middle grade liver function also quite different. For convenience to analysis, we divided the liver function by a new modified ALBI grade of grade 1, 2, 3, and 4. The lower grade usually means better nutrition and fewer hepatocellular damage, mostly stands for better prognosis. The results of Cox analysis and nomogram confirmed this.
Surgical treatment for extrahepatic CCA included pancreaticoduodenectomy (PD), bile duct segmental resection (BDR), and liver transplantation. BDR was comparable in prognosis to PD in middle bile duct cancer, with a 5-year survival rate 44% for BDR and 51% for PD (p = 0.72) (Akita et al. 2020). For patients with poor general condition, BDR was recommended as a less invasive and lower morbidity technique. Both PD and BDR of patients have better overall survival than who did not undergo curative resection and palliative chemotherapy (Saragih et al. 2022), which was verified in our study. After curative resection, microscopic residual tumor was respected as a high-risk factor of recurrence and death, and adjuvant concurrent chemo-radiation therapy could reduce this risk significantly (Lee et al. 2018). Complete surgical resection did not mean cure, the 5-yr survival can be as low as 11% (Anderson and Kim 2009). Adjuvant therapy (AT) has the potential to play a crucial role in prolonging survival and local control, especially in patients with node positive disease (Krasnick et al. 2018). In our study, systematic treatment also included epidermal growth factor receptor (EGFR) and programmed death-1(PD-1) inhibitors. Patients with systematic treatment have lower probability of recurrence and long survival. Similar to the previous study, we also found that tumor location of extrahepatic CCA does not independently predict cancer-specific survival after resection (Gaag et al. 2012).
In this study, we developed two nomograms using the clinical pathological and treatment information with or without the gene mutation score. The Nomogram with gene risk showed a better accuracy and confirmed by the validation cohort. Along with the progress and lower cost of next-generation sequencing (NGS), gene mutation test might be a routine check after CCA diagnosed, which can better guide clinicians choosing target therapeutics, decide ST or not, and predict the prognosis.
This study has limitations. Despite risk factors which have been included in this study, systemic immune inflammation index (SII) was reported as an independent risk factor of postoperative OS following curative-intent resection of eCCA with controversy (Toyoda et al. 2022; Sahara et al. 2021; Reames and Rocha 2021). C-reactive protein-to-albumin ratio (CAR) was another valuable prognostic score in patients with resected extrahepatic cholangiocarcinoma (Asakura et al. 2022). Lacking of the relevant data, those indexes were not included in this study. Another limitation is the small sample size of the validation cohort and data were from the same centers of the training cohort in different stages, not absolutely independent external validation.
Conclusion
The gene risk is a reliable prognostic tool for OS in patients with CCA and might be able to predict which patients benefit from concurrent chemotherapy. The nomogram combined with the clinicopathological and gene risk showed a better accuracy in predicting the prognosis of CCA. It has the potential to guide treatment decisions for patients at different risks and predicts the prognosis of CCA patients.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Akita M, Ajiki T, Ueno K et al (2020) Benefits and limitations of middle bile duct segmental resection for extrahepatic cholangiocarcinoma. Hepatobiliary Pancreat Dis Int 19(2):147–152
Andersen JB, Spee B, Blechacz BR et al (2012) Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 142(4):1021-1031.e1015
Anderson C, Kim R (2009) Adjuvant therapy for resected extrahepatic cholangiocarcinoma: a review of the literature and future directions. Cancer Treat Rev 35(4):322–327
Asakura R, Yanagimoto H, Ajiki T et al (2022) Prognostic impact of inflammation-based scores for extrahepatic cholangiocarcinoma. Dig Surg 39(2–3):65–74
Cao J, Chen L, Li H et al (2019) An accurate and comprehensive clinical sequencing assay for cancer targeted and immunotherapies. Oncologist 24(12):e1294–e1302
Costa BA, de Lara PT, Park W, Keane F, Harding JJ, Khalil DN (2023) Durable response after olaparib treatment for perihilar cholangiocarcinoma with germline BRCA2 mutation. Oncol Res Treat. https://doi.org/10.1159/000529919
Dong L, Lu D, Chen R et al (2022) Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell 40(1):70-87.e15
Jiang T, Lyu SC, Zhou L et al (2021) Carbohydrate antigen 19–9 as a novel prognostic biomarker in distal cholangiocarcinoma. World J Gastrointest Surg 13(9):1025–1038
Kelley RK, Bridgewater J, Gores GJ, Zhu AX (2020) Systemic therapies for intrahepatic cholangiocarcinoma. J Hepatol 72(2):353–363
Krasnick BA, Jin LX, Davidson JT et al (2018) Adjuvant therapy is associated with improved survival after curative resection for hilar cholangiocarcinoma: a multi-institution analysis from the US extrahepatic biliary malignancy consortium. J Surg Oncol 117(3):363–371
Labib PL, Goodchild G, Pereira SP (2019) Molecular pathogenesis of cholangiocarcinoma. BMC Cancer 19(1):185
Lee J, Kang SH, Noh OK et al (2018) Adjuvant concurrent chemoradiation therapy in patients with microscopic residual tumor after curative resection for extrahepatic cholangiocarcinoma. Clin Transl Oncol 20(8):1011–1017
Lee JW, Lee JH, Park Y et al (2021) Prognostic impact of perioperative CA19–9 levels in patients with resected perihilar cholangiocarcinoma. J Clin Med 10(7):1345
Li W, Ma Z, Fu X et al (2021) Olaparib effectively treats local recurrence of extrahepatic cholangiocarcinoma in a patient harboring a BRCA2-inactivating mutation: a case report. Ann Transl Med 9(18):1487
Liang B, Zhong L, He Q et al (2015) Diagnostic accuracy of serum CA19-9 in patients with cholangiocarcinoma: a systematic review and meta-analysis. Med Sci Monit 21:3555–3563
Liu J, Ren G, Li K et al (2022) The Smad4-MYO18A-PP1A complex regulates β-catenin phosphorylation and pemigatinib resistance by inhibiting PAK1 in cholangiocarcinoma. Cell Death Differ 29(4):818–831
Makawita S, Abou-Alfa KG, Roychowdhury S et al (2020) Infigratinib in patients with advanced cholangiocarcinoma with FGFR2 gene fusions/translocations: the PROOF 301 trial. Future Oncol (london, England) 16(30):2375–2384
Mao Y, Huang X, Shuang Z et al (2018) PARP inhibitor olaparib sensitizes cholangiocarcinoma cells to radiation. Cancer Med 7(4):1285–1296
Mazzaferro V, Gorgen A, Roayaie S, Droz Dit Busset M, Sapisochin G (2020) Liver resection and transplantation for intrahepatic cholangiocarcinoma. J Hepatol 72(2):364–377
Montal R, Sia D, Montironi C et al (2020) Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J Hepatol 73(2):315–327
Nguyen MLT, Toan NL, Bozko M, Bui KC, Bozko P (2021) Cholangiocarcinoma therapeutics: an update. Curr Cancer Drug Targets 21(6):457–475
Razumilava N, Gores GJ (2014) Cholangiocarcinoma. Lancet (london, England) 383(9935):2168–2179
Reames BN, Rocha FG (2021) Early recurrence following resection of distal cholangiocarcinoma: a new tool for the toolbox. Ann Surg Oncol 28(8):4069–4071
Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ (2018) Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 15(2):95–111
Sahara K, Tsilimigras DI, Pawlik TM (2021) ASO author reflections: development and validation of distal cholangiocarcinoma early recurrence (DICER) score: results from the US extrahepatic biliary malignancy consortium. Ann Surg Oncol 28(8):4214–4215
Saragih P, Makmun D, Kurniawan J, Rinaldi I (2022) One year survival of extrahepatic cholangiocarcinoma patients who did not undergo curative resection and paliative chemotherapy and its associated factors. Acta Med Indones 54(1):35–41
Tokunaga T, Tanaka M, Tanaka K et al (2021) Modified albumin-bilirubin grade to predict eligibility for second-line therapies at progression on sorafenib therapy in hepatocellular carcinoma patients. Int J Clin Oncol 26(5):922–932
Toyoda J, Sahara K, Maithel SK et al (2022) Prognostic utility of systemic immune-inflammation index after resection of extrahepatic cholangiocarcinoma: results from the US extrahepatic biliary malignancy consortium. Ann Surg Oncol 29(12):7605–7614
van der Gaag NA, Kloek JJ, de Bakker JK et al (2012) Survival analysis and prognostic nomogram for patients undergoing resection of extrahepatic cholangiocarcinoma. Ann Oncol 23(10):2642–2649
Wang Z, Du Y (2021) Identification of a novel mutation gene signature HAMP for cholangiocarcinoma through comprehensive TCGA and GEO data mining. Int Immunopharmacol 99:108039
Wang Y, Pang Q, Jin H et al (2018) Albumin-bilirubin grade as a novel predictor of survival in advanced extrahepatic cholangiocarcinoma. Gastroenterol Res Pract 2018:8902146
Wannhoff A, Gotthardt DN (2019) Recent developments in the research on biomarkers of cholangiocarcinoma in primary sclerosing cholangitis. Clin Res Hepatol Gastroenterol 43(3):236–243
Yoon JG, Kim MH, Jang M et al (2021) Molecular characterization of biliary tract cancer predicts chemotherapy and programmed death 1/programmed death-ligand 1 blockade responses. Hepatology (baltimore, MD) 74(4):1914–1931
Zhou T, Mahn R, Möhring C et al (2022) Case Report: Sustained complete remission on combination therapy with olaparib and pembrolizumab in BRCA2-mutated and PD-L1-positive metastatic cholangiocarcinoma after platinum derivate. Front Oncol 12:933943
Acknowledgements
The authors would like to thank Ying Lian for her contribution to the statistics analysis.
Funding
Youth Foundation of the Second Hospital of Shandong University (2022YP68), Foundation of Science and Technology Research Institute of Shandong University (6010221007). Natural Science Foundation of Shandong Province (ZR2022MH169).
Author information
Authors and Affiliations
Contributions
WW and CW wrote the main manuscript, CW, LYX, PLL, GBL, and YSL collected the samples and information of patients. KW, XYF, and WFW test the 450 genes. WW, JW, and MZH analyzed the data. WW and SSZ prepared the images. SFX was responsible for the study design and data. Editing was performed by WW, CW, GBL, and SFX. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None.
Ethical approval and consent to participate
The requirement for informed consent was waived by the Institutional Review Board for the use of discarded samples acquired at Shandong Provincial Hospital (ethics approval number: LCYJ: NO. 2019–081).
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, W., Wu, C., Xu, L. et al. Development and validation of a gene expression-based nomogram to predict the prognosis of patients with cholangiocarcinoma. J Cancer Res Clin Oncol 149, 9577–9586 (2023). https://doi.org/10.1007/s00432-023-04858-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04858-0