Abstract
Introduction
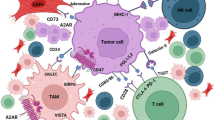
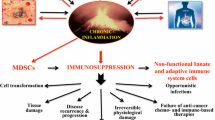
Receptor/ligand pair immune checkpoints are inhibitors that regulate immunity as vital “Don’t Find-Me” signals to the adaptive immune system, additionally, the essential goals of anti-cancer therapy. Moreover, the immune checkpoints are involved in treatment resistance in cancer therapy. The immune checkpoints as a signal from “self” and their expression on healthy cells prevent phagocytosis. Cells (e.g., senescent and/or apoptotic cells) with low immune checkpoints, such as low CD47 and/or PD-L1, are phagocytosed, which is necessary for tissue integrity and homeostasis maintenance. In other words, cancer cells induce increased CD47 expression in the tumor microenvironment (TME), avoiding their clearance by immune cells. PD-L1 and/or CD47 expression tumors have also been employed as biomarkers to guide cure prospects. Thus, targeting innate and adaptive immune checkpoints might improve the influence of the treatments on tumor cells. However, the CD47 regulation in the TME stands intricate, so much of this process has stayed a riddle. In this line, less attention has been paid to cytokines in TME. Cytokines are significant regulators of tumor immune surveillance, and they do this by controlling the actions of the immune cell. Recently, it has been suggested that different types of cytokines at TME might cooperate with others that contribute to the regulation of CD47 and/or PD-L1.
Materials and methods
The data were searched in available databases and a Web Search engine (PubMed, Scopus, and Google Scholar) using related keywords in the title, abstract, and keywords.
Conclusion
Given the significant role of pro/anti-inflammatory signaling in the TME, we discuss the present understanding of pro/anti-inflammatory signaling implications in “Don’t Eat-Me” regulation signals, particularly CD47, in the pathophysiology of cancers and come up with innovative opinions for the clinical transformation and personalized medicine.


Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- aCD47:
-
Anti-CD47 antibody
- ACs:
-
Apoptotic cells
- α-Pal/NRF-1:
-
Alpha palindromic-binding protein/nuclear respiratory factor 1
- AML:
-
Acute myeloid leukemia
- APCs:
-
Antigen-presenting cells
- CAR-T:
-
Chimeric antigen receptor T
- CD:
-
Cluster of differentiation
- CTCL:
-
Cutaneous T-cell lymphomas
- CXCL10:
-
C–X–C motif chemokine ligand 10
- DCs:
-
Dendritic cells
- EMT:
-
Epithelial–mesenchymal transition
- EVs:
-
Extracellular vesicle
- FDA:
-
Food and drug administration
- GM-CSF:
-
Granulocyte–macrophage colony-stimulating factor
- HMGB1:
-
High mobility group box 1 (
- HIF-1:
-
Hypoxia-inducible factors 1
- HPV:
-
Human papillomavirus
- HSV:
-
Herpes simplex virus
- IAP:
-
Integrin-associated protein
- IFN-γ:
-
Interferon-gamma
- IGS:
-
ImmunoGenic Surrender
- IL-1β:
-
Interleukin 1 beta
- ILs:
-
Interleukins
- INF:
-
Interferon
- ITIMs:
-
Immune-receptor tyrosine-based inhibitory motifs
- JAK–STAT1–IRF1:
-
Janus kinase–signal transducer and activator of transcription 1–interferon regulatory factor 1
- JNK:
-
C-jun N-terminal kinase
- LILRB1:
-
Leucocyte immunoglobulin-like receptor family B, member 1
- IL1RAP:
-
Interleukin 1 receptor accessory protein
- LLC:
-
Lewis lung carcinoma
- LPS:
-
Lipopolysaccharides
- LRP1:
-
LDL receptor-related protein
- MAPK:
-
Mitogen-activated protein kinase
- MHC:
-
Major histocompatibility complex
- NF-κB:
-
Nuclear factor-κB
- NK:
-
Natural killer
- NKG2D:
-
Natural killer group 2D
- NSCLC:
-
Non-small cell lung cancer
- PI3K:
-
Phosphoinositide 3-kinases
- PKC:
-
Protein kinase C
- Poly(I:C):
-
Polyinosinic–polycytidylic acid
- RBCs:
-
Red blood cells
- shRNA:
-
Small hairpin RNA
- CD24–Siglec-10 axis:
-
Sialic acid binding Ig-like lectin 10
- Sirf CAR-T:
-
Chimeric antigen receptor (CAR)-T cell secreting CD47 blocker signal regulatory protein α (SIRPα)-Fc fusion protein
- SIRPα:
-
Signal regulatory protein alpha
- SLAMF7:
-
Signaling lymphocytic activation molecule family member 7
- Syk:
-
Spleen tyrosine kinase
- TACE:
-
Transcatheter arterial chemoembolization
- TGF-β:
-
Transforming growth factor beta
- TLR9:
-
Toll-like receptor 9
- TME:
-
Tumor microenvironment
- TNFR1:
-
TNF receptor 1
- TNF-α:
-
Tumor necrosis-factor alpha
- TSP-1:
-
Thrombospondin-1
- VEGF:
-
Vascular endothelial growth factor
- VEGFR:
-
VEGF receptor
- WBCs:
-
White blood cells
- ZBTB28:
-
Zinc finger and BTB domain-containing protein 28
- α-Pal/NRF-1:
-
α-Pal/nuclear respiratory factor 1
References
Advani R, Flinn I, Popplewell L, Forero A, Bartlett NL, Ghosh N et al (2018) CD47 blockade by Hu5F9-G4 and Rituximab in non-Hodgkin’s Lymphoma. N Engl J Med 379(18):1711–1721. https://doi.org/10.1056/NEJMoa1807315
Ahmed KM, Li JJ (2008) NF-kappa B-mediated adaptive resistance to ionizing radiation. Free Radic Biol Med 44(1):1–13. https://doi.org/10.1016/j.freeradbiomed.2007.09.022
Alemohammad H, Najafzadeh B, Asadzadeh Z, Baghbanzadeh A, Ghorbaninezhad F, Najafzadeh A et al (2022) The importance of immune checkpoints in immune monitoring: a future paradigm shift in the treatment of cancer. Biomed Pharmacother 146:112516. https://doi.org/10.1016/j.biopha.2021.112516
Antonangeli F, Natalini A, Garassino MC, Sica A, Santoni A, Di Rosa F (2020) Regulation of PD-L1 expression by NF-κB in cancer. Front Immunol. https://doi.org/10.3389/fimmu.2020.584626
Balkwill F (2006) TNF-alpha in promotion and progression of cancer. Cancer Metastasis Rev 25(3):409–416. https://doi.org/10.1007/s10555-006-9005-3
Barkal AA, Weiskopf K, Kao KS, Gordon SR, Rosental B, Yiu YY et al (2018) Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat Immunol 19(1):76–84. https://doi.org/10.1038/s41590-017-0004-z
Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW et al (2019) CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572(7769):392–396. https://doi.org/10.1038/s41586-019-1456-0
Bartee MY, Dunlap KM, Bartee E (2017) Tumor-localized secretion of soluble PD1 enhances oncolytic virotherapy. Cancer Res 77(11):2952–2963. https://doi.org/10.1158/0008-5472.Can-16-1638
Basile MS, Mazzon E, Russo A, Mammana S, Longo A, Bonfiglio V et al (2019) Differential modulation and prognostic values of immune-escape genes in uveal melanoma. PLoS ONE 14(1):e0210276. https://doi.org/10.1371/journal.pone.0210276
Beizavi Z, Gheibihayat SM, Moghadasian H, Zare H, Yeganeh BS, Askari H et al (2021) The regulation of CD47–SIRPα signaling axis by microRNAs in combination with conventional cytotoxic drugs together with the help of nano-delivery: a choice for therapy? Mol Biol Rep 48(7):5707–5722. https://doi.org/10.1007/s11033-021-06547-y
Betancur PA, Abraham BJ, Yiu YY, Willingham SB, Khameneh F, Zarnegar M et al (2017a) A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat Commun 8:14802. https://doi.org/10.1038/ncomms14802
Betancur PA, Abraham BJ, Yiu YY, Willingham SB, Khameneh F, Zarnegar M et al (2017b) A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat Commun 8(1):14802. https://doi.org/10.1038/ncomms14802
Bian Z, Shi L, Guo YL, Lv Z, Tang C, Niu S et al (2016) CD47–SIRPα interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc Natl Acad Sci USA 113(37):E5434-5443. https://doi.org/10.1073/pnas.1521069113
Bianchi ME, Mezzapelle R (2020) The chemokine receptor CXCR4 in cell proliferation and tissue regeneration. Front Immunol 11:2109. https://doi.org/10.3389/fimmu.2020.02109
Billerhart M, Schönhofer M, Schueffl H, Polzer W, Pichler J, Decker S et al (2021) CD47-targeted cancer immunogene therapy: Secreted SIRPα-Fc fusion protein eradicates tumors by macrophage and NK cell activation. Mo Therapy Oncolytics 23:192–204. https://doi.org/10.1016/j.omto.2021.09.005
Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M et al (2018) Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24(5):541–550. https://doi.org/10.1038/s41591-018-0014-x
Blazar BR, Lindberg FP, Ingulli E, Panoskaltsis-Mortari A, Oldenborg PA, Iizuka K et al (2001) CD47 (integrin-associated protein) engagement of dendritic cell and macrophage counterreceptors is required to prevent the clearance of donor lymphohematopoietic cells. J Exp Med 194(4):541–549. https://doi.org/10.1084/jem.194.4.541
Burger P, Hilarius-Stokman P, de Korte D, van den Berg TK, van Bruggen R (2012) CD47 functions as a molecular switch for erythrocyte phagocytosis. Blood 119(23):5512–5521. https://doi.org/10.1182/blood-2011-10-386805
Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN et al (2016) MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352(6282):227–231. https://doi.org/10.1126/science.aac9935
Catalan R, Orozco-Morales M, Hernandez-Pedro NY, Guijosa A, Colin-Gonzalez AL, Avila-Moreno F, Arrieta O (2020) CD47-SIRPalpha axis as a biomarker and therapeutic target in cancer: current perspectives and future challenges in nonsmall cell lung cancer. J Immunol Res 2020:9435030. https://doi.org/10.1155/2020/9435030
Cham LB, Torrez Dulgeroff LB, Tal MC, Adomati T, Li F, Bhat H et al (2020) Immunotherapeutic blockade of CD47 inhibitory signaling enhances innate and adaptive immune responses to viral infection. Cell Rep 31(2):107494. https://doi.org/10.1016/j.celrep.2020.03.058
Chang WT, Huang AM (2004) Alpha-Pal/NRF-1 regulates the promoter of the human integrin-associated protein/CD47 gene. J Biol Chem 279(15):14542–14550. https://doi.org/10.1074/jbc.M309825200
Chen Q, Fang X, Jiang C, Yao N, Fang X (2015) Thrombospondin promoted anti-tumor of adenovirus-mediated calreticulin in breast cancer: Relationship with anti-CD47. Biomed Pharmacother 73:109–115. https://doi.org/10.1016/j.biopha.2015.05.017
Chen J, Zheng D-X, Yu X-J, Sun H-W, Xu Y-T, Zhang Y-J, Xu J (2019a) Macrophages induce CD47 upregulation via IL-6 and correlate with poor survival in hepatocellular carcinoma patients. Oncoimmunology 8(11):e1652540. https://doi.org/10.1080/2162402X.2019.1652540
Chen Q, Wang C, Zhang X, Chen G, Hu Q, Li H et al (2019b) In situ sprayed bioresponsive immunotherapeutic gel for post-surgical cancer treatment. Nat Nanotechnol 14(1):89–97. https://doi.org/10.1038/s41565-018-0319-4
Chen H, Cong X, Wu C, Wu X, Wang J, Mao K et al (2020) Intratumoral delivery of CCL25 enhances immunotherapy against triple-negative breast cancer by recruiting CCR9(+) T cells. Sci Adv 6(5):eaax4690. https://doi.org/10.1126/sciadv.aax4690
Chen H, Yang Y, Deng Y, Wei F, Zhao Q, Liu Y et al (2022) Delivery of CD47 blocker SIRPα-Fc by CAR-T cells enhances antitumor efficacy. J Immunother Cancer 10(2):e003737. https://doi.org/10.1136/jitc-2021-003737
Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420(6917):860–867. https://doi.org/10.1038/nature01322
Cui Z, Xu D, Zhang F, Sun J, Song L, Ye W et al (2021) CD47 blockade enhances therapeutic efficacy of cisplatin against lung carcinoma in a murine model. Exp Cell Res 405(2):112677. https://doi.org/10.1016/j.yexcr.2021.112677
Dasari S, Tchounwou PB (2014) Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol 740:364–378. https://doi.org/10.1016/j.ejphar.2014.07.025
DeNardo DG, Ruffell B (2019) Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 19(6):369–382. https://doi.org/10.1038/s41577-019-0127-6
Deuse T, Hu X, Agbor-Enoh S, Jang MK, Alawi M, Saygi C et al (2021a) The SIRPalpha-CD47 immune checkpoint in NK cells. J Exp Med. https://doi.org/10.1084/jem.20200839
Deuse T, Hu X, Agbor-Enoh S, Jang MK, Alawi M, Saygi C et al (2021b) The SIRPα-CD47 immune checkpoint in NK cells. J Exp Med. https://doi.org/10.1084/jem.20200839
Feng M, Jiang W, Kim BYS, Zhang CC, Fu YX, Weissman IL (2019) Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer 19(10):568–586. https://doi.org/10.1038/s41568-019-0183-z
Galli S, Zlobec I, Schürch C, Perren A, Ochsenbein AF, Banz Y (2015) CD47 protein expression in acute myeloid leukemia: a tissue microarray-based analysis. Leuk Res 39(7):749–756. https://doi.org/10.1016/j.leukres.2015.04.007
Gao Q, Chen K, Gao L, Zheng Y, Yang YG (2016) Thrombospondin-1 signaling through CD47 inhibits cell cycle progression and induces senescence in endothelial cells. Cell Death Dis 7(9):e2368. https://doi.org/10.1038/cddis.2016.155
Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA et al (2017) Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep 19(6):1189–1201. https://doi.org/10.1016/j.celrep.2017.04.031
Gong J, Ji Y, Liu X, Zheng Y, Zhen Y (2022) Mithramycin suppresses tumor growth by regulating CD47 and PD-L1 expression. Biochem Pharmacol 197:114894. https://doi.org/10.1016/j.bcp.2021.114894
Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN et al (2017) PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545(7655):495–499. https://doi.org/10.1038/nature22396
Gu S, Ni T, Wang J, Liu Y, Fan Q, Wang Y et al (2018) CD47 blockade inhibits tumor progression through promoting phagocytosis of tumor cells by m2 polarized macrophages in endometrial cancer. J Immunol Res 2018:6156757. https://doi.org/10.1155/2018/6156757
Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21(3):309–322. https://doi.org/10.1016/j.ccr.2012.02.022
Harris RN (1995) Guidebook to cytokines and their receptors: Edited by N A Nicola. pp 261. Oxford University Press. 1994. £22.50 ISBN 0–19–859946–3. 23(4), 226–226. https://doi.org/10.1016/0307-4412(95)90183-3
Hayat SMG, Bianconi V, Pirro M, Jaafari MR, Hatamipour M, Sahebkar A (2020) CD47: role in the immune system and application to cancer therapy. Cell Oncol (dordr) 43(1):19–30. https://doi.org/10.1007/s13402-019-00469-5
Hsieh RC, Krishnan S, Wu RC, Boda AR, Liu A, Winkler M et al (2022) ATR-mediated CD47 and PD-L1 up-regulation restricts radiotherapy-induced immune priming and abscopal responses in colorectal cancer. Sci Immunol 7(72):9330. https://doi.org/10.1126/sciimmunol.abl9330
Hu T, Liu H, Liang Z, Wang F, Zhou C, Zheng X et al (2020) Tumor-intrinsic CD47 signal regulates glycolysis and promotes colorectal cancer cell growth and metastasis. Theranostics 10(9):4056–4072. https://doi.org/10.7150/thno.40860
Ivashkiv LB (2018) IFNgamma: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol 18(9):545–558. https://doi.org/10.1038/s41577-018-0029-z
Iwamura H, Saito Y, Sato-Hashimoto M, Ohnishi H, Murata Y, Okazawa H et al (2011) Essential roles of SIRPα in homeostatic regulation of skin dendritic cells. Immunol Lett 135(1–2):100–107. https://doi.org/10.1016/j.imlet.2010.10.004
Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R et al (2009) CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138(2):271–285. https://doi.org/10.1016/j.cell.2009.05.046
Jalil AR, Andrechak JC, Discher DE (2020) Macrophage checkpoint blockade: results from initial clinical trials, binding analyses, and CD47–SIRPα structure-function. Antib Ther 3(2):80–94. https://doi.org/10.1093/abt/tbaa006
Jalil AR, Tobin MP, Discher DE (2022) Suppressing or enhancing macrophage engulfment through the use of CD47 and related peptides. Bioconjug Chem. https://doi.org/10.1021/acs.bioconjchem.2c00019
Johnson LDS, Banerjee S, Kruglov O, Viller NN, Horwitz SM, Lesokhin A et al (2019) Targeting CD47 in Sézary syndrome with SIRPαFc. Blood Adv 3(7):1145–1153. https://doi.org/10.1182/bloodadvances.2018030577
Kamijo H, Miyagaki T, Takahashi-Shishido N, Nakajima R, Oka T, Suga H et al (2020) Thrombospondin-1 promotes tumor progression in cutaneous T-cell lymphoma via CD47. Leukemia 34(3):845–856. https://doi.org/10.1038/s41375-019-0622-6
Kaur S, Soto-Pantoja DR, Stein EV, Liu C, Elkahloun AG, Pendrak ML et al (2013) Thrombospondin-1 signaling through CD47 inhibits self-renewal by regulating c-Myc and other stem cell transcription factors. Sci Rep 3:1673. https://doi.org/10.1038/srep01673
Kaur S, Elkahloun AG, Singh SP, Arakelyan A, Roberts DD (2018) A function-blocking CD47 antibody modulates extracellular vesicle-mediated intercellular signaling between breast carcinoma cells and endothelial cells. J Cell Commun Signal 12(1):157–170
Kumari N, Dwarakanath BS, Das A, Bhatt AN (2016) Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol 37(9):11553–11572. https://doi.org/10.1007/s13277-016-5098-7
Li L, Gong Y, Tang J, Yan C, Li L, Peng W et al (2022) ZBTB28 inhibits breast cancer by activating IFNAR and dual blocking CD24 and CD47 to enhance macrophages phagocytosis. Cell Mol Life Sci 79(2):83. https://doi.org/10.1007/s00018-021-04124-x
Lian S, Xie R, Ye Y, Lu Y, Cheng Y, Xie X et al (2019a) Dual blockage of both PD-L1 and CD47 enhances immunotherapy against circulating tumor cells. Sci Rep 9(1):4532. https://doi.org/10.1038/s41598-019-40241-1
Lian S, Xie R, Ye Y, Xie X, Li S, Lu Y et al (2019b) Simultaneous blocking of CD47 and PD-L1 increases innate and adaptive cancer immune responses and cytokine release. EBioMedicine 42:281–295. https://doi.org/10.1016/j.ebiom.2019.03.018
Liu J, Wang L, Zhao F, Tseng S, Narayanan C, Shura L et al (2015) Pre-clinical development of a humanized anti-CD47 antibody with anti-cancer therapeutic potential. PLoS ONE 10(9):e0137345. https://doi.org/10.1371/journal.pone.0137345
Liu Q, Yu S, Li A, Xu H, Han X, Wu K (2017) Targeting interlukin-6 to relieve immunosuppression in tumor microenvironment. Tumour Biol 39(6):1010428317712445. https://doi.org/10.1177/1010428317712445
Liu F, Dai M, Xu Q, Zhu X, Zhou Y, Jiang S et al (2018) SRSF10-mediated IL1RAP alternative splicing regulates cervical cancer oncogenesis via mIL1RAP-NF-κB-CD47 axis. Oncogene 37(18):2394–2409. https://doi.org/10.1038/s41388-017-0119-6
Lo J, Lau EY, Ching RH, Cheng BY, Ma MK, Ng IO, Lee TK (2015) Nuclear factor kappa B-mediated CD47 up-regulation promotes sorafenib resistance and its blockade synergizes the effect of sorafenib in hepatocellular carcinoma in mice. Hepatology 62(2):534–545. https://doi.org/10.1002/hep.27859
Locksley RM, Killeen N, Lenardo MJ (2001) The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104(4):487–501. https://doi.org/10.1016/s0092-8674(01)00237-9
Lopez-Dee Z, Pidcock K, Gutierrez LS (2011) Thrombospondin-1: multiple paths to inflammation. Mediators Inflamm 2011:296069. https://doi.org/10.1155/2011/296069
Lu D, Ni Z, Liu X, Feng S, Dong X, Shi X et al (2019) Beyond T cells: understanding the role of PD-1/PD-L1 in tumor-associated macrophages. J Immunol Res 2019:1919082. https://doi.org/10.1155/2019/1919082
Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD Jr et al (2009) CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138(2):286–299. https://doi.org/10.1016/j.cell.2009.05.045
Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P (2017) Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 14(7):399–416. https://doi.org/10.1038/nrclinonc.2016.217
Maute R, Xu J, Weissman IL (2022) CD47–SIRPα-targeted therapeutics: status and prospects. Immunooncol Technol 13:100070. https://doi.org/10.1016/j.iotech.2022.100070
Métayer LE, Vilalta A, Burke GAA, Brown GC (2017) Anti-CD47 antibodies induce phagocytosis of live, malignant B cells by macrophages via the Fc domain, resulting in cell death by phagoptosis. Oncotarget 8(37):60892–60903. https://doi.org/10.18632/oncotarget.18492
Mezzapelle R, De Marchis F, Passera C, Leo M, Brambilla F, Colombo F et al (2021) CXCR4 engagement triggers CD47 internalization and antitumor immunization in a mouse model of mesothelioma. EMBO Mol Med 13(6):e12344. https://doi.org/10.15252/emmm.202012344
Mezzapelle R, Leo M, Caprioglio F, Colley LS, Lamarca A, Sabatino L et al (2022) CXCR4/CXCL12 activities in the tumor microenvironment and implications for tumor immunotherapy. Cancers (basel). https://doi.org/10.3390/cancers14092314
Moon JW, Kong SK, Kim BS, Kim HJ, Lim H, Noh K et al (2017) IFNγ induces PD-L1 overexpression by JAK2/STAT1/IRF-1 signaling in EBV-positive gastric carcinoma. Sci Rep 7(1):17810. https://doi.org/10.1038/s41598-017-18132-0
Okazawa H, Motegi S-I, Ohyama N, Ohnishi H, Tomizawa T, Kaneko Y et al (2005) Negative regulation of phagocytosis in macrophages by the CD47-SHPS-1 system. J Immunol 174(4):2004–2011. https://doi.org/10.4049/jimmunol.174.4.2004
Oldenborg PA (2013) CD47: a cell surface glycoprotein which regulates multiple functions of hematopoietic cells in health and disease. ISRN Hematol 2013:614619. https://doi.org/10.1155/2013/614619
Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP (2000) Role of CD47 as a marker of self on red blood cells. Science 288(5473):2051–2054. https://doi.org/10.1126/science.288.5473.2051
Ozpiskin OM, Zhang L, Li JJ (2019) Immune targets in the tumor microenvironment treated by radiotherapy. Theranostics 9(5):1215–1231. https://doi.org/10.7150/thno.32648
Parker BS, Rautela J, Hertzog PJ (2016) Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer 16(3):131–144. https://doi.org/10.1038/nrc.2016.14
Passaro C, Alayo Q, De Laura I, McNulty J, Grauwet K, Ito H et al (2019) Arming an oncolytic herpes simplex virus type 1 with a single-chain fragment variable antibody against PD-1 for experimental glioblastoma therapy. Clin Cancer Res 25(1):290–299. https://doi.org/10.1158/1078-0432.Ccr-18-2311
Peña-Martínez P, Ramakrishnan R, Högberg C, Jansson C, Nord DG, Järås M (2021) IL4 promotes phagocytosis of murine leukemia cells counteracted by CD47 upregulation. Haematologica. https://doi.org/10.3324/haematol.2020.270421
Peng S, Xiao F, Chen M, Gao H (2022) Tumor-microenvironment-responsive nanomedicine for enhanced cancer immunotherapy. Adv Sci 9(1):2103836. https://doi.org/10.1002/advs.202103836
Platanias LC (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5(5):375–386. https://doi.org/10.1038/nri1604
Rathe SK, Popescu FE, Johnson JE, Watson AL, Marko TA, Moriarity BS et al (2019) Identification of candidate neoantigens produced by fusion transcripts in human osteosarcomas. Sci Rep 9(1):358. https://doi.org/10.1038/s41598-018-36840-z
Rendtlew Danielsen JM, Knudsen LM, Dahl IM, Lodahl M, Rasmussen T (2007) Dysregulation of CD47 and the ligands thrombospondin 1 and 2 in multiple myeloma. Br J Haematol 138(6):756–760. https://doi.org/10.1111/j.1365-2141.2007.06729.x
Rosell R, Lord RV, Taron M, Reguart N (2002) DNA repair and cisplatin resistance in non-small-cell lung cancer. Lung Cancer 38(3):217–227. https://doi.org/10.1016/s0169-5002(02)00224-6
Schroder K, Hertzog PJ, Ravasi T, Hume DA (2004) Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75(2):163–189. https://doi.org/10.1189/jlb.0603252
Sedger LM, McDermott MF (2014) TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants—past, present and future. Cytokine Growth Factor Rev 25(4):453–472. https://doi.org/10.1016/j.cytogfr.2014.07.016
Shain KH, Landowski TH, Dalton WS (2000) The tumor microenvironment as a determinant of cancer cell survival: a possible mechanism for de novo drug resistance. Curr Opin Oncol 12(6):557–563. https://doi.org/10.1097/00001622-200011000-00008
Sockolosky JT, Dougan M, Ingram JR, Ho CC, Kauke MJ, Almo SC et al (2016) Durable antitumor responses to CD47 blockade require adaptive immune stimulation. Proc Natl Acad Sci USA 113(19):E2646-2654. https://doi.org/10.1073/pnas.1604268113
Sosale NG, Rouhiparkouhi T, Bradshaw AM, Dimova R, Lipowsky R, Discher DE (2015) Cell rigidity and shape override CD47’s “self”-signaling in phagocytosis by hyperactivating myosin-II. Blood 125(3):542–552. https://doi.org/10.1182/blood-2014-06-585299
Tajbakhsh A, Gheibi Hayat SM, Movahedpour A, Savardashtaki A, Loveless R, Barreto GE et al (2021) The complex roles of efferocytosis in cancer development, metastasis, and treatment. Biomed Pharmacother 140:111776. https://doi.org/10.1016/j.biopha.2021.111776
Tan Y, Chen H, Zhang J, Cai L, Jin S, Song D et al (2022) Platinum(IV) complexes as inhibitors of CD47–SIRPα axis for chemoimmunotherapy of cancer. Eur J Med Chem 229:114047. https://doi.org/10.1016/j.ejmech.2021.114047
Taverna S, Tonacci A, Ferraro M, Cammarata G, Cuttitta G, Bucchieri S et al (2022) High mobility group box 1: biological functions and relevance in oxidative stress related chronic diseases. Cells. https://doi.org/10.3390/cells11050849
Terme M, Ullrich E, Aymeric L, Meinhardt K, Desbois M, Delahaye N et al (2011) IL-18 induces PD-1-dependent immunosuppression in cancer. Cancer Res 71(16):5393–5399. https://doi.org/10.1158/0008-5472.Can-11-0993
Theruvath J, Menard M, Smith BAH, Linde MH, Coles GL, Dalton GN et al (2022) Anti-GD2 synergizes with CD47 blockade to mediate tumor eradication. Nat Med 28(2):333–344. https://doi.org/10.1038/s41591-021-01625-x
Topalian SL, Drake CG, Pardoll DM (2012) Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol 24(2):207–212. https://doi.org/10.1016/j.coi.2011.12.009
Vandchali NR, Moadab F, Taghizadeh E, Tajbakhsh A, Gheibihayat SM (2021) CD47 functionalization of nanoparticles as a poly(ethylene glycol) alternative: a novel approach to improve drug delivery. Curr Drug Targets 22(15):1750–1759. https://doi.org/10.2174/1389450122666210204203514
Viant C, Fenis A, Chicanne G, Payrastre B, Ugolini S, Vivier E (2014) SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat Commun 5:5108. https://doi.org/10.1038/ncomms6108
Wang G, Kang X, Chen KS, Jehng T, Jones L, Chen J et al (2020) An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat Commun 11(1):1395. https://doi.org/10.1038/s41467-020-15229-5
Wang Y, Zhao C, Liu Y, Wang C, Jiang H, Hu Y, Wu J (2022) Recent advances of tumor therapy based on the CD47–SIRPα axis. Mol Pharm 19(5):1273–1293. https://doi.org/10.1021/acs.molpharmaceut.2c00073
Watson NB, Schneider KM, Massa PT (2015) SHP-1-dependent macrophage differentiation exacerbates virus-induced myositis. J Immunol 194(6):2796–2809. https://doi.org/10.4049/jimmunol.1402210
Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B et al (2015) Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 16(4):375–384. https://doi.org/10.1016/s1470-2045(15)70076-8
Weiskopf K, Jahchan NS, Schnorr PJ, Cristea S, Ring AM, Maute RL et al (2016) CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest 126(7):2610–2620. https://doi.org/10.1172/jci81603
Wensveen FM, Jelencic V, Polic B (2018) NKG2D: a master regulator of immune cell responsiveness. Front Immunol 9:441. https://doi.org/10.3389/fimmu.2018.00441
Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS et al (2012) The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci USA 109(17):6662–6667. https://doi.org/10.1073/pnas.1121623109
Wu ZH, Shi Y, Tibbetts RS, Miyamoto S (2006) Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311(5764):1141–1146. https://doi.org/10.1126/science.1121513
Yang H, Shao R, Huang H, Wang X, Rong Z, Lin Y (2019) Engineering macrophages to phagocytose cancer cells by blocking the CD47/SIRPɑ axis. Cancer Med 8(9):4245–4253. https://doi.org/10.1002/cam4.2332
Yang Y, Yang Z, Yang Y (2021) Potential role of CD47-directed bispecific antibodies in cancer immunotherapy. Front Immunol 12:686031. https://doi.org/10.3389/fimmu.2021.686031
Ye ZH, Jiang XM, Huang MY, Xu YL, Chen YC, Yuan LW et al (2021) Regulation of CD47 expression by interferon-gamma in cancer cells. Transl Oncol 14(9):101162. https://doi.org/10.1016/j.tranon.2021.101162
Zamora AE, Crawford JC, Thomas PG (2018) Hitting the target: how T cells detect and eliminate tumors. J Immunol 200(2):392–399. https://doi.org/10.4049/jimmunol.1701413
Zhang H, Lu H, Xiang L, Bullen JW, Zhang C, Samanta D et al (2015) HIF-1 regulates CD47 expression in breast cancer cells to promote evasion of phagocytosis and maintenance of cancer stem cells. Proc Natl Acad Sci USA 112(45):E6215-6223. https://doi.org/10.1073/pnas.1520032112
Zhang X, Wang Y, Fan J, Chen W, Luan J, Mei X et al (2019) Blocking CD47 efficiently potentiated therapeutic effects of anti-angiogenic therapy in non-small cell lung cancer. J Immunother Cancer 7(1):346. https://doi.org/10.1186/s40425-019-0812-9
Zhang W, Liu Y, Yan Z, Yang H, Sun W, Yao Y et al (2020) IL-6 promotes PD-L1 expression in monocytes and macrophages by decreasing protein tyrosine phosphatase receptor type O expression in human hepatocellular carcinoma. J Immunother Cancer. https://doi.org/10.1136/jitc-2019-000285
Zhang Y, Qian J, Gu C, Yang Y (2021) Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther 6(1):78. https://doi.org/10.1038/s41392-021-00486-7
Zhang B, Li W, Fan D, Tian W, Zhou J, Ji Z, Song Y (2022) Advances in the study of CD47-based bispecific antibody in cancer immunotherapy. Immunology 167(1):15–27. https://doi.org/10.1111/imm.13498
Zhao J, Zhong S, Niu X, Jiang J, Zhang R, Li Q (2019) The MHC class I-LILRB1 signalling axis as a promising target in cancer therapy. Scand J Immunol 90(5):e12804. https://doi.org/10.1111/sji.12804
Zhong C, Wang L, Hu S, Huang C, Xia Z, Liao J et al (2021) Poly(I:C) enhances the efficacy of phagocytosis checkpoint blockade immunotherapy by inducing IL-6 production. J Leukoc Biol 110(6):1197–1208. https://doi.org/10.1002/jlb.5ma0421-013r
Zhou T, Damsky W, Weizman O-E, McGeary MK, Hartmann KP, Rosen CE et al (2020) IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature 583(7817):609–614. https://doi.org/10.1038/s41586-020-2422-6
Zhou X, Liu X, Huang L (2021) Macrophage-mediated tumor cell phagocytosis: opportunity for nanomedicine intervention. Adv Funct Mater. https://doi.org/10.1002/adfm.202006220
Acknowledgements
Thanks to guidance and advice from "The Pharmaceutical Sciences Research Center, Shiraz University of Medical Sciences".
Funding
This research received no specific grant from any funding agency in public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AZK, ZS, MA, SMGH and AT drafted the manuscript. AT presented the idea. AZK, ZS, MA, and AT performed the literature search, and SMG prepared Figs. 1 and 2. AZK, ZS, MA, and AT edited the manuscript. YG, AT, and AS supervised the whole study.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no competing interest in this article. All authors reviewed the manuscript.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Karizak, A.Z., Salmasi, Z., Gheibihayat, S.M. et al. Understanding the regulation of “Don’t Eat-Me” signals by inflammatory signaling pathways in the tumor microenvironment for more effective therapy. J Cancer Res Clin Oncol 149, 511–529 (2023). https://doi.org/10.1007/s00432-022-04452-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04452-w