Abstract
Purpose
Methyltransferase-like 3 (METTL3), a key member of the m6A methyltransferase complex, is upregulated in multiple human malignancies and plays a role in regulating tumor migration. This study aimed to reveal the underlying mechanism by which METTL3 in regulates the metastasis of colorectal cancer (CRC).
Methods
We compared METTL3 expression levels in CRC tumor tissues and adjacent nontumor tissues by immunohistochemistry (IHC). The functional roles of METTL3 in CRC were assessed by real-time cell migration assays, wound-healing assays and Transwell assays. miRNA sequencing (miRNA-seq), RNA-binding protein immunoprecipitation (RIP) assays and N6-methyladenosine immunoprecipitation (MeRIP) assays were performed to confirm the molecular mechanism underlying the involvement of METTL3 in CRC cell metastasis.
Results
We found that METTL3 was overexpressed in CRC tissues. METTL3 knockdown significantly inhibited CRC cell migration and invasion, while METTL3 overexpression had the opposite effects. Furthermore, we demonstrated that METTL3 regulates miR-196b expression via an N6-methyladenosine (m6A)-pri-miR-196b-dependent mechanism and thereby promotes CRC metastasis.
Conclusion
This study shows the important role of METTL3 in CRC metastasis and provides novel insight into m6A modification in CRC metastasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors among both men and women (Sung et al. 2021; Siegel et al. 2021). Due to improvements in detection and treatment, the 5-year relative survival rate of CRC has dramatically increased (Siegel et al. 2021). Patients diagnosed early with only localized disease have a 5-year survival rate of 90%. Nevertheless, this rate drops to 72% and 14% for patients diagnosed with regional and distant-stage disease, respectively (Siegel et al. 2021). Therefore, identifying the factors involved in CRC tumorigenesis and progression is imperative to find novel potential targets for improving the clinical outcomes of patients with metastatic CRC.
N6-methyladenosine (m6A) is the most abundant chemical modification in eukaryotic messenger RNAs (mRNAs) (Meyer et al. 2012) and preferentially occurs in the consensus motif “RRACH” (R=G or A; H=A, C, or U) (Dominissini et al. 2012; Meyer et al. 2012; Bodi et al. 2010). Recent studies have shown that m6A modification may have a profound impact on multiple aspects of RNA metabolism, including the nuclear export, splicing, stability and translation of target mRNAs (Chen et al. 2020b; Kasowitz et al. 2018; Du et al. 2016; Wang et al. 2014, 2015; Roundtree et al. 2017). m6A modification has been shown to affect cell meiosis, the circadian clock and stem cell self-renewal and differentiation (Weng et al. 2018; Xu et al. 2017; Zhong et al. 2018). RNA m6A modification is dynamically regulated and involves methylation by “writers”, recognition by “readers”, and demethylation by “erasers” (Liu et al. 2014; Jia et al. 2011; Sibbritt et al. 2013). Accumulating evidence indicates that the dysregulation of m6A regulatory enzymes is associated with cancer progression (Chen et al. 2018; Visvanathan et al. 2018; Zhu et al. 2020; Hua et al. 2018). Methyltransferase-like 3 (METTL3) is an m6A methylase that functions in methylation processes (Sibbritt et al. 2013), and the dysregulation of METTL3 expression has been reported in multiple human malignancies (Cheng et al. 2019; Chen et al. 2021b; Wang et al. 2020; Hua et al. 2018; Zhou et al. 2021a). An increasing number of studies have shown that METTL3 promotes cancer cell metastasis (Chen et al. 2021b, 2018; Cheng et al. 2019; Hua et al. 2018; Wang et al. 2020; Zhou et al. 2021a). In CRC, METTL3 has been reported to regulate metastasis by enhancing the mRNA stability of SOX2, HK2, GLUT1 and YPEL5 through an m6A-IGF2BP2/3-dependent mechanism (Li et al. 2019; Zhou et al. 2021a; Shen et al. 2020). However, the underlying mechanism of METTL3 in the metastasis of CRC remains largely unclear.
miRNAs are short, noncoding RNAs that play critical roles in diverse biological processes, including proliferation, apoptosis and metastasis (Wang et al. 2017; Kim et al. 2014; Meng et al. 2013; Ling et al. 2016; Andriani et al. 2018; El Bezawy et al. 2017). Mature miRNAs are generated via a two-step processing pathway to yield 19–24 nucleotide small RNAs that regulate gene expression at the posttranscriptional level (Lee et al. 2002). The first step is precise cleavage of the stem loops embedded in the primary transcripts (pri-miRNAs) by the microprocessor complex, composed of the RNA-binding protein DGCR8 and the type III RNase DROSHA, to release pre-miRNAs (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). Then, the pre-miRNAs are exported to the cytoplasm and diced by Dicer to generate ~ 22 nt miRNA duplexes (Hutvagner et al. 2001; Bohnsack et al. 2004). pri-miRNA processing is a critical step in miRNA biogenesis. This initial event requires recognition of the junction between the stem and the flanking single-stranded RNA of the pri-miRNA hairpin by DGCR8 (Han et al. 2006). m6A methyltransferases (METTL3, METTL14), an m6A-binding protein (HNRNPA2B1) and a demethylase (FTO) have been reported to affect miRNA expression levels (Berulava et al. 2015; Alarcon et al. 2015a, b; Han et al. 2019; Wang et al. 2019; Bi et al. 2021; Peng et al. 2019; Ma et al. 2017; Chen et al. 2020a; Zhou et al. 2021b). In recent years, an increasing number of researchers have demonstrated that METTL3 promotes cancer cell proliferation, apoptosis and metastasis by accelerating miRNA maturation in an m6A-dependent manner (Han et al. 2019; Wang et al. 2019; Bi et al. 2021; Peng et al. 2019).
In this study, we showed that METTL3 is upregulated in CRC and promotes CRC cell migration and invasion. Moreover, we first demonstrated that METTL3 can methylate pri-miR-196b, and then upregulate the expression of miR-196b, thereby increasing CRC cell migration ability.
Materials and methods
Clinical specimens
The CRC tissue microarray (TMA) slides used for immunohistochemistry (IHC) of METTL3 protein expression were purchased from Shanghai Outdo Biotech (Shanghai, China). The colon cancer TMA (HColA180Su09) contains 69 tumor tissues and 55 adjacent tissues. The rectal cancer TMA (HRec-Ade180Sur-04) contains 68 tumor tissues and 57 adjacent tissues. The CRC TMA slides used for in situ hybridization (ISH) analysis of miR-196b expression were obtained from the tumor bank of the Department of Pathology of the First Affiliated Hospital, Sun Yat-sen University (Guangzhou, China). The human sample collection procedure was approved by the Ethics Committee of the Sixth Affiliated Hospital, Sun Yat-sen University (Guangzhou, China), and written informed consent was obtained from all of the patients. All experimental protocols were carried out in accordance with the approved guidelines and were approved by the Ethics Committee of the Sixth Affiliated Hospital, Sun Yat-sen University (Guangzhou, China).
Cell culture
The human CRC cell lines HCT 116, SW480 and Caco2 were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI 1640 medium or DMEM supplemented with 10% fetal bovine serum (vol/vol) and 1% penicillin‒streptomycin (Gibco, Grand Island, NY, USA) at 37 °C in 5% CO2.
Lentiviral packaging and cell transduction
For lentiviral packaging and cell transduction, METTL3 knockdown or overexpression lentiviruses were obtained from Shanghai GeneChem Co., Ltd, China. For overexpression, cDNA was amplified by quantitative real-time PCR (qRT-PCR) and subcloned into the GV341 vector according to the manufacturer’s instructions. For stable silencing, shRNA lentiviruses (shMETTL3 with the target sequence GCCTTAACATTGCCCACTGAT) were constructed using GV248 vectors. For miR-196b, miR-196b precursor sequences were cloned into the GV309 vector. CRC cells at 20–30% confluence were plated in 24-well dishes and infected with METTL3 overexpression lentivirus (oeMETTL3), METTL3 knockdown lentivirus (shMETTL3), miR-196b overexpression lentivirus or the corresponding negative controls, respectively. Pools of stably transduced cells were generated by selection using puromycin (1 mg/ml) for 2 weeks. siMETTL3, miR-196b mimic and miR-196b inhibitor were ordered from RiboBio Co., Ltd, China (METTL3_1 with the target sequence CAAGTATGTTCACTATGAA; METTL3_2 with the target sequence GACTGCTCTTTCCTTAATA). Transfection was achieved using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocols. After transfection, the expression of METTL3 and miR-196b was validated by qRT-PCR or Western blotting (WB).
RNA extraction and qRT-PCR
Total RNA was extracted from cells using RNAiso Plus (Invitrogen, USA) according to the manufacturer’s protocol. RNA was reverse-transcribed to cDNA using the PrimeScriptTMRT Reagent Kit and random primers (Takara, Dalian, China). qRT-PCR to assess pri-miR-196b expression was performed using SYBR Premix Ex Taq™ II (Takara), and expression data were normalized to GAPDH mRNA expression according to the manufacturer's instructions. The expression of mature miR-196b was analyzed using the All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA). The small endogenous nucleolar RNU6B was used as a control for miRNA normalization. All experiments were performed in triplicate. Real-time PCR was performed using the Applied Biosystems 7900 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Gene expression △Ct values from each sample were calculated by normalizing to an internal control (RNU6B/GAPDH), and relative expression was calculated using the formula 2−△△Ct values. The primers used in this study were as follows: pri-miR-196b_F: CACCAGAACTGGTCGGTGATT and pri-miR-196b_R: TAATGAAGGCAGTGTCGTGCT; GAPDH_F: AGCCTCAAGATCATCAGC and GAPDH_R: GAGTCCTTCCACGATACC.
WB
Cells were collected from cultured dishes and lysed in RIPA lysis buffer (Cell Signaling Technology, Danvers, MA, USA) supplemented with protease inhibitors. Briefly, equal amounts of protein were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk for 1 h at room temperature and then incubated with primary antibodies overnight at 4 °C. The next day, after incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature, the signals were detected using a chemiluminescence ECL detection kit (Santa Cruz Biotechnology). Anti-METTL3 (1:1000 dilution, Abcam, Cambridge, UK) and anti-GAPDH (1:1000 dilution, Abcam, Cambridge, UK) antibodies were used.
IHC
IHC staining was performed as described previously (Huang et al. 2018; Chen et al. 2021b). Briefly, TMA slides were incubated in a dry oven at 60 °C for 2 h. After deparaffinization and rehydration, antigen retrieval was performed by boiling the sections in a 0.1 mol/L citrate acid solution (pH = 6.0). Endogenous peroxidase activity was blocked using 0.3% H2O2 for 10 min at room temperature. The TMA slides were blocked with 5% normal goat serum (BOSTER, China) for 30 min and subsequently incubated with an anti-METTL3 antibody (1:1000, Abcam, Cambridge, UK) overnight at 4 °C. A staining index (with values ranging from 0 to 12) was calculated as follows the staining intensity (0, negative staining; 1, weak staining; 2, moderate staining; and 3, strong staining) multiplied by the proportion of positively stained tumor cells (0, < 5%; 1, 5–25%; 2, 25–50%; 3, 50–75%; and 4, ≥ 75%). The median value of the total staining score was 6; thus, a score of 0–6 indicated low expression, and a score of 8–12 indicated high expression.
ISH
ISH was performed using miRNA-196b probe from Exiqon (Exiqon A/S, Denmark). The probe was detected using anti-digoxigenin-AP (Roche, Denmark), and the hybridized probes were detected by applying the BCIP/NBT Alkaline Phosphatase Color Development Kit. No-probe controls were included for both hybridization procedures. Images were taken with a Leica DMI 4000B inverted microscope (Leica Microsystems, Wetzlar, Germany). ISH staining of the images was analyzed using Image Pro-Plus (version 5.0, Media Cybernetics, Silver Spring, MD, USA) (Xavier et al. 2005; Huang et al. 2018).
Real-time cell migration assays
Real-time cell migration assays were performed on the xCELLigence system from ACEA Biosciences. Briefly, the lower chamber of the CIM plate was filled with serum-containing medium, and the upper chamber was filled with serum-free medium. The cells were resuspended in serum-free medium, counted and seeded into the upper chamber by applying 5 × 104 cells in 100 µL of medium. After addition of the cells, the CIM plate was incubated for 30 min at room temperature and then blocked in an RTCA DP instrument. The CI values were measured automatically every 15 min for 3 days.
Wound healing assays
Wound healing assays were performed with Ibidi Culture-Insert 2 Well. Briefly, 2 × 105 cells were seeded into each well of the Culture-Insert 2 Well. The Culture-Insert 2 Well was removed after 24 h, and the cell layer was washed with PBS to remove cell debris and nonattached cells. The dish was filled with medium, and the wound-healing process was monitored under a microscope. Each assay was repeated 3 times.
Migration assays
Cell migration assays were carried out with transwell chambers (BD Bioscience, San Jose, CA, USA). HCT 116 cells (5 × 104) or SW480 cells (8 × 104) were seeded in the upper chamber in serum-free medium, and the lower chamber was filled with medium containing 10% FBS. After incubation at 37 °C in 5% CO2 for a suitable time (HCT 116 cells: 27 h, SW480 cells: 26 h), cells in the upper chambers were fixed with 4% paraformaldehyde for 15 min, stained with 0.1% crystal violet and photographed using a microscope. All studies were repeated at least three times in triplicate.
Invasion assays
Invasion assays were performed as described previously (Huang et al. 2018). Briefly, Transwell chambers precoated with Matrigel (BD Bioscience, San Jose, CA, USA) were used to perform the invasion assay. A total of 1 × 105 cells were cultured in serum-free medium in the upper chambers of each well, while medium containing 10% FBS was added to the lower chamber of the well. After 30 h, the cells were fixed with 4% polyoxymethylene and stained with crystal violet. Cells on the upper side of the membrane that had not migrated were gently wiped off, and the stained cells on the lower side were observed under a microscope. The number of migrated cells in five fields per chamber was counted, and the average values were calculated.
RIP assays
After formaldehyde-crosslinking (0.3% for 10 min), we isolated the nuclear fraction from 4 × 107 cells using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Rockford, IL, USA). The nuclear fraction was lysed in RIP Lysis Buffer (Millipore). RIP was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) following the manufacturer’s protocol. For endogenous immunoprecipitation we used 5 μg of an anti-METTL3 rabbit antibody (anti-METTL3-1#: catalog No. A301-567A; anti-METTL3-2#: catalog No. A301-568A. Bethyl, Montgomery, UK) or normal rabbit IgG as a control bound to protein A magnetic beads (Millipore). After immunoprecipitation, each immunoprecipitate was resuspend in 150 μL of proteinase K buffer to digest the proteins, and then the supernatant was transferred to a new tube. A total of 250 μL of RIP wash buffer and 400 μL of phenol:chloroform:isoamyl alcohol were added to each tube. The tubes were vortexed for 15 s and centrifuged at 14,000 rpm for 10 min. Then, the aqueous solution was precipitated and resuspended in 10 μL of RNase-free water. The RNA was analyzed by qRT-PCR.
MeRIP assays
The nuclear fraction from 4 × 107 cells was isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). RNA was extracted from the nuclear fraction using TRIzol (Invitrogen) and then fragmented into sizes between 60 and 200 nucleotides in length using Ambion® RNA Fragmentation Reagents (Thermo Scientific). The fragmented RNA was precipitated by adding one-tenth volumes of 3 M sodium acetate (pH 5.2) and glycogen (100 μg/mL final) and 2.5 volumes of 100% ethanol. Rabbit anti-m6A antibody (Synaptic Systems, Germany) and rabbit normal IgG control bound to protein A magnetic beads (Millipore) were used for immunoprecipitation. The immunoprecipitated RNA was extracted using phenol:chloroform:isoamyl alcohol. Then, the aqueous solution was precipitated and resuspended in 10 μL of RNase-free water. The RNA was analyzed by qRT-PCR.
Statistical analysis
Statistical analysis was performed using SPSS18.0 software (SPSS, IBM, Chicago, IL, USA). The data are expressed as the mean ± SD, and statistical significance was determined with Student’s t tests. Statistical comparisons between groups were analyzed using Student’s paired t test. P values less than 0.05 were considered as statistically significant. The relationships between miR-196b expression and clinicopathological features were analyzed using the chi-square test.
Results
METTL3 is upregulated in human CRC
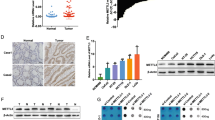
To better understand METTL3 protein expression and localization in CRC, we detected METTL3 expression in CRC tissues and adjacent tissues using IHC. We found that METTL3 expression was mainly localized in the nucleus (Fig. 1a). Furthermore, METTL3 was significantly upregulated in the CRC tissues (Fig. 1b, c). Next, we examined METTL3 expression in The Cancer Genome Atlas (TCGA) datasets (http://cancergenome.nih.gov). Consistent with our findings, METTL3 was significantly upregulated in CRC tissues compared with adjacent normal tissues (Fig. 1d, e).
METTL3 is upregulated in CRC. a Representative image following IHC staining of CRC tumor tissues and adjacent normal tissues with anti-METTL3 antibody. b METTL3 protein expression in 69 colon cancer tissues and 55 adjacent normal tissues. Statistical significance was determined by two-tailed, unpaired Student’s t test. c METTL3 protein expression in 68 rectal cancer tissues and 57 adjacent normal tissues. Statistical significance was determined by two-tailed, unpaired Student’s t test. d–e METTL3 mRNA levels from 41 pairs of colon cancer tissues and 9 pairs of rectal cancer tissues from TCGA database. Statistical significance was determined by a two-tailed, paired Student’s t test. (***P < 0.001)
Decreased METTL3 expression inhibits CRC cell migration and invasion in vitro
To investigate the role of METTL3 in CRC metastasis, we silenced METTL3 in HCT 116 and SW480 cells using METTL3 short hairpin RNAs (shRNAs) and siMETTL3_1. Knockdown of METTL3 expression was confirmed by WB and qRT-PCR (Fig. 2a, Fig. S1 and Fig. S2a, b). Real-time cell migration (Fig. 2b, c) and wound-healing (Fig. 2d, e and Fig. S2c, d) assays showed that the knockdown of METTL3 dramatically suppressed CRC cell migration. Consistently, knockdown of METTL3 markedly reduced cell invasion ability (Fig. 2f and Fig. S2e, f).
Knockdown of METTL3 inhibited CRC cell migration and invasion in vitro. a WB analysis of METTL3 expression in CRC cells infected with siNC or siMETTL3-1. b–c Real-time migration of CRC cells transfected with siNC or siMETTL3. The delta cell index indicates electrical impedance. d–e Wound healing assays were performed to investigate the effects of siMETTL3 on the migration of CRC cells. f Transwell invasion assays were performed to estimate the effects of siMETTL3 on CRC cell invasion. The data are presented as the mean ± SD. Three independent assays were performed (d–e) (**P < 0.01, ***P < 0.001; Student’s t test)
Overexpression of METTL3 promotes cell migration and invasion in vitro
We then transfected CRC cells with METTL3 lentiviral vectors. The expression of METTL3 was confirmed by WB and qRT-PCR (Fig. 3a, b). Transwell migration assays showed that METTL3 overexpression substantially increased the migration of CRC cells (Fig. 3c). In addition, Transwell invasion assays showed that METTL3 overexpression increased CRC cell invasion (Fig. 3d). Our findings indicated that METTL3 overexpression increases migration and invasion in CRC cells.
Overexpression of METTL3 promoted CRC cell migration and invasion in vitro. a–b WB and qRT-PCR analysis of METTL3 expression of in CRC cells infected with oeNC or oeMETTL3. c Migration assays revealed that overexpression of METTL3 increased the migration of HCT 116 and SW480 cells. d Transwell invasion assays revealed that overexpression of METTL3 increased the invasion of HCT 116 and SW480 cells. The data are presented as the mean ± SD. Three independent assays were performed (*P < 0.05, **P < 0.01, ***P < 0.001; Student’s t test)
Knockdown of METTL3 reshapes the miRNA profile of CRC cells
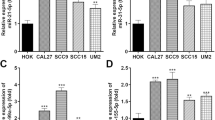
Since addition of the m6A mark acts as a key posttranscriptional modification that promotes the initiation of miRNA biogenesis, we next measured the global impact of METTL3 depletion on miRNA levels by performing miRNA-seq of RNA collected from control or METTL3 knockdown HCT116 cells. miRNA-seq revealed 179 miRNAs that were significantly differentially expressed (P < 0.05) by at least a 1.2-fold between the control and METTL3 knockdown cells. Of these miRNAs, 161 miRNAs were upregulated, and 18 miRNAs were downregulated (Fig. 4a and Table S1). We selected significantly downregulated miRNAs according to their fold-change, P value and miRNA abundance (Isoform > 300) (Fig. 4b). The expression of these miRNAs was verified by qRT-PCR, and the results showed that METTL3 knockdown significantly decreased the expression levels of miR-21-5p, miR-196b-5p, and miR-1246-5p (Fig. 4c). Previous studies have reported that m6A modification-dependent pri-miRNA processing is essential for the maturation of miR-21-5p (Liu et al. 2021; Wang et al. 2021) and miR-1246-5p (Huang et al. 2021; Peng et al. 2019), but its role in the maturation of miR-196b has not yet been investigated. Therefore, our study focused on whether METTL3 plays a role by regulating miR-196b in CRC cells.
miRNA-seq revealed the miRNA profile of CRC cells. a The volcano plot shows the differential expression of miRNAs in HCT 116 cells treated with siMETTL3 detected with miRNA-seq. b The top 3 miRNAs are listed in the table. c qRT-PCR was used to measure the expression levels of the indicated miRNAs. The data are presented as the mean ± SD. Three independent assays were performed (**P < 0.01, ***P < 0.001; Student’s t test)
METTL3-dependent m6A methylation regulates the maturation of miR-196b
To further verify that METTL3 regulates miR-196b maturation by catalyzing m6A formation in RNAs, we obtained the full sequences of pri-miR-196b and pre-miR-196b from UCSC and miRBase. Then, we analyzed whether the single-stranded RNA flanking the pri-miR-196b contains the RRACH motif for m6A modification (R=G or A; H=A, C or U) (Dominissini et al. 2012; Meyer et al. 2012; Bodi et al. 2010). SRAMP (http://www.cuilab.cn/sramp/), which is a computational predictor of mammalian m6A sites (Zhou et al. 2016), revealed m6A motifs localized precisely at the pri-miR-196b splicing site (Fig. 5a, b). qRT-PCR primers were designed to amplify the putative m6A region of pri-miR-196b (Fig. S3). Knockdown of METTL3 expression was confirmed by WB and qRT-PCR (Fig. S1). qRT-PCR showed that the knockdown of METTL3 significantly decreased the expression of miR-196b (Fig. 5c) but increased the expression of pri-miR-196b in CRC cells (Fig. 5d), while the overexpression of METTL3 increased the expression of miR-196b (Fig. 5e) but reduced the expression of pri-miR-196b in CRC cells (Fig. 5f). Furthermore, miR-196b expression positively correlated with METTL3, DGCR8 and HNRNPA2B1 expression in CRC tissues (Fig. S4). To determine whether METTL3 binds the m6A methylation region of pri-miR-196b in vitro, we conducted RIP-qRT-PCR experiments using two antibodies that map to different regions of endogenous METTL3 (anti-METTL3-1#; anti-METTL3-2#). We incubated cells in formaldehyde for in vitro crosslinking, isolated the nuclear fractions and immunoprecipitated endogenous METTL3. After METTL3 immunoprecipitation, WB (Fig. 5g, h) and qRT-PCR (Fig. 5i) revealed that METTL3 interacts with the m6A methylation region of pri-miR-196b. To determine whether m6A modification was present in the pri-miR-196b regions, we conducted MeRIP-qRT-PCR by immunoprecipitating nuclear RNA from HCT 116 and SW480 cells with an anti-m6A antibody, followed by qRT-PCR. The results showed that m6A modification was enriched at the pri-miR-196b sequence (Fig. 5j). Moreover, the level of pri-miR-196b m6A modification was significantly reduced following METTL3 silencing in SW480 cells (Fig. 5k). Taken together, these results indicated that METTL3 regulates the processing of miR-196 in an m6A-dependent manner.
METTL3 regulates the processing of miR-196b. a–b Possible m6A methylation modification sites in miR-196b. c–d qRT-PCR analysis of miR-196b and pri-miR-196b level in CRC cells treated with siNC or siMETTL3. e–f qRT-PCR analysis of miR-196b and pri-miR-196b expression in CRC cells treated with oeNC or oeMETTL3. g–h Immunoprecipitation of endogenous METTL3. i pri-miRNAs bound to endogenous METTL3 were extracted and quantified by qRT-PCR. j m6A modification level of pri-miR-196b in CRC cells assessed by MeRIP. k m6A modification level of pri-miR-196b in SW480 cells treated with shMETTL3 detected by MeRIP. The data are presented as the mean ± SD. Three independent assays were performed (c–f) (*P < 0.05, **P < 0.01, ***P < 0.001; Student’s t test)
miR-196b is required for the METTL3-induced enhancement of CRC cells migrations
To further investigate the pathological and prognostic significance of miR-196b expression in CRC patients, the miR-196b levels in TMA slides containing 283 CRC samples were quantified using ISH. Positive tissue staining was indicated by blue‒violet color (Fig. 6a). Our results showed that miR-196b was upregulated in patients with lymph node metastasis compared with those without lymph node metastasis (Fig. 6b), and miR-196b expression was correlated with TNM stage (P = 0.050), lymph node metastasis (pN) (P = 0.003) and recurrence (P = 0.028) (Table 1). To determine the role of miR-196b in CRC cell metastasis, CRC cells were stably infected with the miR-196b or scramble control lentiviral vector. Increased miR-196b expression in the cells following infection was confirmed by qRT-PCR (Fig. 6c and Fig. S5a). Real-time cell migration assays showed that overexpression of miR-196b significantly promoted CRC cells migration (Fig. 6d and Fig. S5b). Transwell invasion assays showed that ectopic miR-196b expression significantly increased the invasion of CRC cells (Fig. 6e and Fig. S5c). Taken together, these findings indicated that miR-196b increases migration and invasion of CRC cells.
miR-196b rescued the METTL3-induced migration of CRC cells. a Representative ISH images showing miR-196b. b ISH analysis of paraffin blocks of CRC specimens was followed by analysis of miR-196b levels. miR-196b expression levels in patients with CRC with lymph node metastases were significantly higher than those in patients with CRC without lymph node metastasis. c qRT-PCR analysis of miR-196b expression in CRC cells infected with pLV-miR-196b or the vector. d Real-time migration of HCT 116 cells transfected with pLV-miR-196b or the vector. The delta cell index indicates electrical impedance. e Transwell invasion assays were used to assessed the effects of miR-196b on the HCT 116 cell invasion ability. f–g The effects of miR-196b mimics or miR-196b inhibitors on the migration of SW480 cells with METTL3 knockdown or overexpression were determined by Transwell migration assays. The data are presented as the mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001; Student’s t test)
Next, we transfected miR-196b mimics into METTL3 knockdown cells, and found that the miR-196b mimics could partly increase CRC cell migration inhibited by METTL3 knockdown (Fig. 6f). Accordingly, miR-196b inhibitors partially decreased the increase in cell migration induced by METTL3 overexpression (Fig. 6g).
Discussion
Recent studies have shown the important role of m6A modification in regulating RNA metabolism and various biological processes (Chen et al. 2020b; Roundtree et al. 2017; Kasowitz et al. 2018; Du et al. 2016; Wang et al. 2014, 2015; Weng et al. 2018; Xu et al. 2017; Zhong et al. 2018). In the present study, we show that METTL3 promotes CRC migration by accelerating the maturation of pri-miR-196b in an m6A-dependent manner.
We found that METTL3 expression is significantly upregulated in CRC tissues and promotes CRC cell migration and invasion. These results are consistent with those reported by other studies (Li et al. 2019; Zhou et al. 2021a; Chen et al. 2021a; Pan et al. 2022). Chen et al. identified METTL3 as the most essential m6A regulatory enzyme that is overexpressed in CRC (Chen et al. 2021a). Pan et al. reported that m6A and METTL3 levels were substantially elevated in CRC tissues, and patients with CRC with high m6A or METTL3 levels exhibited shorter overall survival (Pan et al. 2022). In CRC, METTL3 has been reported to regulate metastasis by m6A-dependent posttranscriptional modification of SOX2, HK2, GLUT1, YPEL5 and CRB3 (Li et al. 2019; Zhou et al. 2021a; Shen et al. 2020; Pan et al. 2022). (Li et al. 2019; Zhou et al. 2021a) However, METTL3 has also been reported as a tumor suppressor in CRC. Deng et al. found that METTL3 is significantly associated with longer survival time and suppresses CRC cell proliferation, migration and invasion through p38/ERK pathways (Deng et al. 2019). These contradictory conclusions reached in previous studies must be related to the use of different modification sites, various m6A-binding readers and multiple downstream targets.
miRNAs are short noncoding RNAs that play critical roles in diverse biological processes, and their aberrant expression has been associated with numerous human diseases (Andriani et al. 2018; Meng et al. 2013; Wang et al. 2017; El Bezawy et al. 2017; Kim et al. 2014; Ling et al. 2016). The m6A methyltransferase, m6A-binding proteins and demethylase have been reported to affect miRNA expression levels (Berulava et al. 2015; Alarcon et al. 2015a, b; Han et al. 2019; Wang et al. 2019; Bi et al. 2021; Peng et al. 2019). METTL3 was found to methylate pri-miRNAs, which marked them for recognition and processing by the microprocessor complex protein DGCR8 (Alarcon et al. 2015b). To investigate whether METTL3 regulates CRC metastasis through miRNAs, we measured the global impact of METTL3 depletion on miRNA levels. The miRNA-seq results showed that the expression levels of miR-21, miR-196b and miR-1246 were most significantly downregulated after METLL3 knockdown. Previous studies have reported that m6A modification-dependent pri-miRNA processing is essential for the maturation of miR-21-5p (Liu et al. 2021; Wang et al. 2021) and miR-1246-5p (Huang et al. 2021; Peng et al. 2019), but its role in the maturation of miR-196b has not yet been investigated.
To investigate whether METTL3 regulates miR-196b expression through m6A modification, we first analyzed the pri-miR-196b sequence using SRAMP. The results of bioinformatic prediction showed several RRACH motifs in the pri-miR-196b sequence. We also found that the RRACH motif is not present in the pre-miR-196b regions but is located in single-stranded RNA flanking the pri-miRNA hairpin. This finding is consistent with the results of the work of Alarcon et al., who used the Finding Informative Regulatory Elements algorithm to identify overrepresentation of the GGAC motif in pri-miRNA sequences; that study demonstrated that the GGAC motif is not enriched in pre-miRNA sequences (Alarcon et al. 2015b). A previous study showed that METTL3 methylates the single-stranded RNA flanking the pri-miRNA hairpin, marking these structures for recognition and processing by DGCR8, and that METTL3 depletion reduced the binding of DGCR8 to pri-miRNAs and resulted in a reduction in mature miRNAs and the concomitant accumulation of unprocessed pri-miRNAs (Alarcon et al. 2015b). Therefore, we hypothesized that METTL3 targets pri-miR-196b for m6A modification, and promotes pri-miR-196b processing. To assess our hypothesis, we designed PCR primers to amplify the putative m6A region of pri-miR-196b. The qRT-PCR results showed that METTL3 knockdown significantly decreased the expression of miR-196b but increased the expression of pri-miR-196b in CRC cells, while the overexpression of METTL3 increased the expression of miR-196b but reduced the expression of pri-miR-196b in CRC cells. Furthermore, our results revealed that METTL3 interacts with the m6A methylation region of pri-miR-196b, and the m6A modification level of pri-miR-196b was significantly reduced following the silencing of METTL3. Taken together, our results showed that METTL3 regulates miR-196b expression through m6A modification.
An increasing number of studies have shown that miR-196b has dual functions as both a tumor promoter (Yu et al. 2016; Liao et al. 2012) and a suppressor of cancer progression (How et al. 2013; Tellez et al. 2016). In the present study, we confirmed that miR-196b is upregulated in CRC and that overexpression of miR-196b promotes CRC cell migration and invasion, which is consistent with previous reports of miR-196b as an onco-miRNA in CRC (Stiegelbauer et al. 2017). Furthermore, our in vitro gain-of-function and rescue experiments substantiated that METTL3 promotes CRC migration invasion, at least partially, in an miR-196b-dependent manner.
Conclusions
Taken together, our results show that METTL3 expression is significantly increased in CRC and that METTL3 promotes CRC cell migration and invasion. Moreover, we first demonstrate that METTL3 promotes CRC metastasis by accelerating pri-miR-196b maturation in an m6A-dependent manner; this finding may provide a potential therapeutic target for antimetastatic strategies against CRC.
References
Alarcon CR, Goodarzi H, Lee H et al (2015a) HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162(6):1299–1308. https://doi.org/10.1016/j.cell.2015.08.011
Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF (2015b) N6-methyladenosine marks primary microRNAs for processing. Nature 519(7544):482–485. https://doi.org/10.1038/nature14281
Andriani F, Majorini MT, Mano M et al (2018) MiR-16 regulates the pro-tumorigenic potential of lung fibroblasts through the inhibition of HGF production in an FGFR-1- and MEK1-dependent manner. J Hematol Oncol 11(1):45. https://doi.org/10.1186/s13045-018-0594-4
Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B (2015) N6-adenosine methylation in MiRNAs. PLoS One 10(2):e0118438. https://doi.org/10.1371/journal.pone.0118438
Bi X, Lv X, Liu D et al (2021) METTL3 promotes the initiation and metastasis of ovarian cancer by inhibiting CCNG2 expression via promoting the maturation of pri-microRNA-1246. Cell Death Discov 7(1):237. https://doi.org/10.1038/s41420-021-00600-2
Bodi Z, Button JD, Grierson D, Fray RG (2010) Yeast targets for mRNA methylation. Nucleic Acids Res 38(16):5327–5335. https://doi.org/10.1093/nar/gkq266
Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10(2):185–191. https://doi.org/10.1261/rna.5167604
Chen M, Wei L, Law CT et al (2018) RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 67(6):2254–2270. https://doi.org/10.1002/hep.29683
Chen X, Xu M, Xu X et al (2020a) METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol Ther 28(2):599–612. https://doi.org/10.1016/j.ymthe.2019.11.016
Chen ZH, Chen TQ, Zeng ZC et al (2020b) Nuclear export of chimeric mRNAs depends on an lncRNA-triggered autoregulatory loop in blood malignancies. Cell Death Dis 11(7):566. https://doi.org/10.1038/s41419-020-02795-1
Chen H, Gao S, Liu W et al (2021) RNA N(6)-methyladenosine methyltransferase METTL3 facilitates colorectal cancer by activating the m(6)A-GLUT1-mTORC1 axis and is a therapeutic target. Gastroenterology 160(4):1284-1300 e1216. https://doi.org/10.1053/j.gastro.2020.11.013
Chen X, Huang L, Yang T et al (2021b) METTL3 promotes esophageal squamous cell carcinoma metastasis through enhancing GLS2 expression. Front Oncol 11:667451. https://doi.org/10.3389/fonc.2021.667451
Cheng M, Sheng L, Gao Q et al (2019) The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene 38(19):3667–3680. https://doi.org/10.1038/s41388-019-0683-z
Deng R, Cheng Y, Ye S et al (2019) m(6)A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther 12:4391–4402. https://doi.org/10.2147/OTT.S201052
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432(7014):231–235. https://doi.org/10.1038/nature03049
Dominissini D, Moshitch-Moshkovitz S, Schwartz S et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397):201–206. https://doi.org/10.1038/nature11112
Du H, Zhao Y, He J et al (2016) YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 7:12626. https://doi.org/10.1038/ncomms12626
El Bezawy R, De Cesare M, Pennati M et al (2017) Antitumor activity of miR-34a in peritoneal mesothelioma relies on c-MET and AXL inhibition: persistent activation of ERK and AKT signaling as a possible cytoprotective mechanism. J Hematol Oncol 10(1):19. https://doi.org/10.1186/s13045-016-0387-6
Gregory RI, Yan KP, Amuthan G et al (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432(7014):235–240. https://doi.org/10.1038/nature03120
Han J, Lee Y, Yeom KH et al (2004) The drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18(24):3016–3027. https://doi.org/10.1101/gad.1262504
Han J, Lee Y, Yeom KH et al (2006) Molecular basis for the recognition of primary microRNAs by the drosha-DGCR8 complex. Cell 125(5):887–901. https://doi.org/10.1016/j.cell.2006.03.043
Han J, Wang JZ, Yang X et al (2019) METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer 18(1):110. https://doi.org/10.1186/s12943-019-1036-9
How C, Hui AB, Alajez NM et al (2013) MicroRNA-196b regulates the homeobox B7-vascular endothelial growth factor axis in cervical cancer. PLoS One 8(7):e67846. https://doi.org/10.1371/journal.pone.0067846
Hua W, Zhao Y, Jin X et al (2018) METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol 151(2):356–365. https://doi.org/10.1016/j.ygyno.2018.09.015
Huang L, Wen C, Yang X et al (2018) PEAK1, acting as a tumor promoter in colorectal cancer, is regulated by the EGFR/KRas signaling axis and miR-181d. Cell Death Dis 9(3):271. https://doi.org/10.1038/s41419-018-0320-8
Huang S, Luo S, Gong C et al (2021) MTTL3 upregulates microRNA-1246 to promote occurrence and progression of NSCLC via targeting paternally expressed gene 3. Mol Ther Nucleic Acids 24:542–553. https://doi.org/10.1016/j.omtn.2021.02.020
Hutvagner G, McLachlan J, Pasquinelli AE et al (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293(5531):834–838. https://doi.org/10.1126/science.1062961
Jia G, Fu Y, Zhao X et al (2011) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7(12):885–887. https://doi.org/10.1038/nchembio.687
Kasowitz SD, Ma J, Anderson SJ et al (2018) Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 14(5):e1007412. https://doi.org/10.1371/journal.pgen.1007412
Kim J, Zhang Y, Skalski M et al (2014) microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res 74(5):1541–1553. https://doi.org/10.1158/0008-5472.CAN-13-1449
Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21(17):4663–4670. https://doi.org/10.1093/emboj/cdf476
Li T, Hu PS, Zuo Z et al (2019) METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer 18(1):112. https://doi.org/10.1186/s12943-019-1038-7
Liao YL, Hu LY, Tsai KW et al (2012) Transcriptional regulation of miR-196b by ETS2 in gastric cancer cells. Carcinogenesis 33(4):760–769. https://doi.org/10.1093/carcin/bgs023
Ling H, Pickard K, Ivan C et al (2016) The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut 65(6):977–989. https://doi.org/10.1136/gutjnl-2015-309372
Liu J, Yue Y, Han D et al (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10(2):93–95. https://doi.org/10.1038/nchembio.1432
Liu E, Lv L, Zhan Y et al (2021) METTL3/N6-methyladenosine/ miR-21-5p promotes obstructive renal fibrosis by regulating inflammation through SPRY1/ERK/NF-kappaB pathway activation. J Cell Mol Med 25(16):7660–7674. https://doi.org/10.1111/jcmm.16603
Ma JZ, Yang F, Zhou CC et al (2017) METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 65(2):529–543. https://doi.org/10.1002/hep.28885
Meng X, Wu J, Pan C et al (2013) Genetic and epigenetic down-regulation of microRNA-212 promotes colorectal tumor metastasis via dysregulation of MnSOD. Gastroenterology 145(2):426-436 e421–426. https://doi.org/10.1053/j.gastro.2013.04.004
Meyer KD, Saletore Y, Zumbo P et al (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149(7):1635–1646. https://doi.org/10.1016/j.cell.2012.05.003
Pan J, Liu F, Xiao X et al (2022) METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res 41(1):19. https://doi.org/10.1186/s13046-021-02227-8
Peng W, Li J, Chen R et al (2019) Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res 38(1):393. https://doi.org/10.1186/s13046-019-1408-4
Roundtree IA, Luo GZ, Zhang Z et al (2017) YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. https://doi.org/10.7554/eLife.31311
Shen C, Xuan B, Yan T et al (2020) m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer 19(1):72. https://doi.org/10.1186/s12943-020-01190-w
Sibbritt T, Patel HR, Preiss T (2013) Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA 4(4):397–422. https://doi.org/10.1002/wrna.1166
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics 2021. CA Cancer J Clin 71(1):7–33. https://doi.org/10.3322/caac.21654
Stiegelbauer V, Vychytilova-Faltejskova P, Karbiener M et al (2017) miR-196b-5p regulates colorectal cancer cell migration and metastases through interaction with HOXB7 and GALNT5. Clin Cancer Res 23(17):5255–5266. https://doi.org/10.1158/1078-0432.CCR-17-0023
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Tellez CS, Juri DE, Do K et al (2016) miR-196b Is epigenetically silenced during the premalignant stage of lung carcinogenesis. Cancer Res 76(16):4741–4751. https://doi.org/10.1158/0008-5472.CAN-15-3367
Visvanathan A, Patil V, Arora A et al (2018) Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37(4):522–533. https://doi.org/10.1038/onc.2017.351
Wang X, Lu Z, Gomez A et al (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481):117–120. https://doi.org/10.1038/nature12730
Wang X, Zhao BS, Roundtree IA et al (2015) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161(6):1388–1399. https://doi.org/10.1016/j.cell.2015.05.014
Wang X, Zuo D, Yuan Y et al (2017) MicroRNA-183 promotes cell proliferation via regulating programmed cell death 6 in pediatric acute myeloid leukemia. J Cancer Res Clin Oncol 143(1):169–180. https://doi.org/10.1007/s00432-016-2277-2
Wang H, Deng Q, Lv Z et al (2019) N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer 18(1):181. https://doi.org/10.1186/s12943-019-1108-x
Wang Q, Chen C, Ding Q et al (2020) METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69(7):1193–1205. https://doi.org/10.1136/gutjnl-2019-319639
Wang H, Song X, Song C, Wang X, Cao H (2021) m(6)A-seq analysis of microRNAs reveals that the N6-methyladenosine modification of miR-21-5p affects its target expression. Arch Biochem Biophys 711:109023. https://doi.org/10.1016/j.abb.2021.109023
Weng H, Huang H, Wu H et al (2018) METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22(2):191-205 e199. https://doi.org/10.1016/j.stem.2017.11.016
Xavier LL, Viola GG, Ferraz AC et al (2005) A simple and fast densitometric method for the analysis of tyrosine hydroxylase immunoreactivity in the substantia nigra pars compacta and in the ventral tegmental area. Brain Res Brain Res Protoc 16(1–3):58–64. https://doi.org/10.1016/j.brainresprot.2005.10.002
Xu K, Yang Y, Feng GH et al (2017) Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res 27(9):1100–1114. https://doi.org/10.1038/cr.2017.100
Yu SL, Lee DC, Sohn HA et al (2016) Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear Factor-kappa B activity in non-small cell lung cancer cells. Mol Carcinog 55(12):1915–1926. https://doi.org/10.1002/mc.22439
Zhong X, Yu J, Frazier K et al (2018) Circadian clock regulation of hepatic lipid metabolism by modulation of m(6)A mRNA methylation. Cell Rep 25(7):1816-1828 e1814. https://doi.org/10.1016/j.celrep.2018.10.068
Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q (2016) SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res 44(10):e91. https://doi.org/10.1093/nar/gkw104
Zhou D, Tang W, Xu Y et al (2021a) METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Mol Oncol 15(8):2172–2184. https://doi.org/10.1002/1878-0261.12898
Zhou G, Yan K, Liu J et al (2021b) FTO promotes tumour proliferation in bladder cancer via the FTO/miR-576/CDK6 axis in an m6A-dependent manner. Cell Death Discov 7(1):329. https://doi.org/10.1038/s41420-021-00724-5
Zhu S, Wang JZ, Chen D et al (2020) An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun 11(1):1685. https://doi.org/10.1038/s41467-020-15403-9
Funding
This work was supported by the National Natural Science Foundation of China (81802953) and Key discipline project of Guangdong Province (2019-GDXK-0016).
Author information
Authors and Affiliations
Contributions
LH, XY and SL: designed and supervised the study. LH: wrote the first draft of the manuscript. LH, DL, YZ, XC, JC and CW: performed the experiments and analyzed the data. HL and XY: contributed the research material. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This study was approved by the Ethics Committee of the Sixth Affiliated Hospital, Sun Yat-sen University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, L., Liang, D., Zhang, Y. et al. METTL3 promotes colorectal cancer metastasis by promoting the maturation of pri-microRNA-196b. J Cancer Res Clin Oncol 149, 5095–5108 (2023). https://doi.org/10.1007/s00432-022-04429-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04429-9