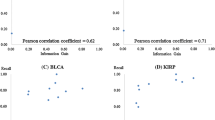
Abstract
Purpose
Omics data are crucial for medical diagnosis as it contains intrinsic biomedical information. Multi-omics integrated analysis has become a new direction for scientists to explore life mechanisms. Nevertheless, the quality of complex omics data often varies greatly due to different samples or even different omics types, it is challenging to dynamically capture the uncertainty for different kinds of omics data.
Methods
This paper proposes a uncertainty-aware dynamic integration framework for multi-omics classification. The framework consists of three modules: deep embedding, confidence prediction, and downstream tasks. The deep embedding module extract key information from multi-omics data to obtain a low-dimensional feature representation which is used to train downstream tasks. Combined with the deep embedding module, the confidence prediction module is used to dynamically capture the uncertainty of the data. We introduce “confidNet” to assign a confidence value for each type of omics data, which is used for dynamic integration between multi-omics.
Results
Compared with other integration methods, the proposed method can contain more crucial biomedical information in the obtained low-dimensional representation. Our framework realizes reliable integration among multiple omics, and it can still achieve high accuracy on small sample data sets. We have verified the effectiveness of the model in a large number of experiments.
Conclusion
Our framework can be widely applied to high-dimensional omics data and has great potential to facilitate medical decision-making and biological analysis.








Similar content being viewed by others
References
Abdi H, Williams LJ (2010) Principal component analysis. Wires Comput Stat 2(4):433–459
Amodio M, Dijk DV, Srinivasan K, Chen WS, Krishnaswamy S (2019) Exploring single-cell data with deep multitasking neural networks. Nat Methods 16(11):1139–1145
Berger B, Peng J, Singh M (2013) Computational solutions for omics data. Nat Rev Genet 14(5):333–346
Cantini Laura, Zakeri Pooya, Hernandez Celine, Naldi Aurelien, Thieffry Denis, Remy Elisabeth, Baudot Anaïs (2021) Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer. Nat Commun 12(1):1–12
Capper D, Jones DT, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE et al (2018) Dna methylation-based classification of central nervous system tumours. Nature 555(7697):469–474
Chaudhary K, Poirion OB, Lu L, Garmire LX (2017) Deep learning based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res 24(6):2017
Chaudhary K, Poirion OB, Lu L, Garmire LX (2017) Deep learning based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res 24(6):2017
Chen T, Fox E, Guestrin C (2014) Stochastic gradient hamiltonian monte carlo. In ICML, pp. 1683–1691. PMLR
Chung NC, Mirza B, Choi H, Wang J, Wang D, Ping P, Wang W (2019) Unsupervised classification of multi-omics data during cardiac remodeling using deep learning. Methods 166:66–73
Corbière C, Thome N, Bar-Hen TB, Cord M, Pérez P (2019) Addressing failure prediction by learning model confidence. NIPS 32
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the em algorithm. J R Stat Soc B 39(1):1–22
Fei-Fei L, Fergus R, Perona P (2004) Learning generative visual models from few training examples: An incremental bayesian approach tested on 101 object categories. In CVPR, pp. 178–178. IEEE
Gal, Y. and Z. Ghahramani 2016. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In ICML, pp. 1050–1059. PMLR
Gawlikowski J, Tassi CRN, Ali M, Lee J, Humt M, Feng J, Kruspe A, Triebel R, Jung J, Roscher R, Muhammad S, Wen Y, Richard B, Xiang ZX (2021) A survey of uncertainty in deep neural networks. arXiv preprint arXiv:2107.03342
Graves A (2011) Practical variational inference for neural networks. NIPS 24
Guidotti R, Monreale A, Ruggieri S, Turini F, Giannotti F, Pedreschi D (2018) A survey of methods for explaining black box models. ACM (CSUR) 51(5):1–42
Gustafsson FK, Danelljan M, Schon TB (2020) Evaluating scalable bayesian deep learning methods for robust computer vision. In CVPR, pp. 318–319
Hartigan JA, Wong MA (1979) Algorithm as 136: A k-means clustering algorithm. J R Stat Soc C-Appl 28(1):100–108
Hasin Y, Seldin M, Lusis A (2017) Multi-omics approaches to disease. Genome Biol 18(1):1–15
Hernández-Lobato JM, Adams R (2015) Probabilistic backpropagation for scalable learning of bayesian neural networks. In ICML, pp. 1861–1869. PMLR
Hruschka ER, Campello RJ, Freitas AA, de Carvalho ACPLF (2009) A survey of evolutionary algorithms for clustering. IEEE T Syst Man Cy C 39(2):133–155
Kim S, Kim S, Min D, Sohn K (2019) Laf-net: Locally adaptive fusion networks for stereo confidence estimation. In CVPR
Kingma DP, Welling M (2013) Auto-encoding variational bayes. arXiv preprintarXiv:1312.6114
Klodt M, Vedaldi A (2018) Supervising the new with the old: learning sfm from sfm. In ECCV, pp. 698–713
Lakshminarayanan B, Pritzel A, Blundell C (2017) Simple and scalable predictive uncertainty estimation using deep ensembles. NIPS 30
Lecun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436
Louis DN, Perry A, Reifenberger G, Von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131(6):803–820
Ma T, Zhang A (2017) Integrate multi-omic data using affinity network fusion (anf) for cancer patient clustering. In BIBM, pp. 398–403. IEEE
MacKay DJ (1992) A practical bayesian framework for backpropagation networks. Neural Comput 4(3):448–472
Nadkarni PM, Ohno-Machado L, Chapman WW (2011) Natural language processing: an introduction. J Am Med Inform Assn 18(5):544–551
Nix DA, Weigend AS (1994) Estimating the mean and variance of the target probability distribution. In ICNN’94, Volume 1, pp. 55–60. IEEE
Pawlowski N, Brock A, Lee MC, Rajchl M, Glocker B (2017) Implicit weight uncertainty in neural networks. arXiv preprintarXiv:1711.01297
Raginsky M, Rakhlin A, Telgarsky M (2017) Non-convex learning via stochastic gradient langevin dynamics: a nonasymptotic analysis. In COLT, pp. 1674–1703. PMLR
Shen R, Mo Q, Schultz N, Seshan VE, Olshen AB, Huse J, Ladanyi M (2012) Integrative subtype discovery in glioblastoma using icluster. PLoS ONE 7(4):e35236
Smith L, Gal Y (2018) Understanding measures of uncertainty for adversarial example detection. arXiv preprintarXiv:1803.08533
Vincent P, Larochelle H, Bengio Y, Manzagol PA (2008) Extracting and composing robust features with denoising autoencoders. In ICML, pp. 1096–1103
Wang B, Mezlini AM, Demir F, Fiume M, Tu Z, Brudno M, Haibe-Kains B, Goldenberg A (2014) Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 11(3):333–337
Wannenwetsch AS, Keuper M, Roth S (2017) Probflow: Joint optical flow and uncertainty estimation. In ICCV, pp. 1173–1182
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM (2013) The cancer genome atlas pan-cancer analysis project. Nat Genet 45(10):1113–1120
Wu A, Nowozin S, Meeds E, Turner RE, Hernández-Lobato JM, Gaunt AL (2018) Deterministic variational inference for robust bayesian neural networks. arXiv preprintarXiv:1810.03958
Zhang T (2001) An introduction to support vector machines and other kernel-based learning methods. AI Mag 22(2):103–103
Zhang X, Xing Y, Sun K, Guo Y (2021) Omiembed: a unified multi-task deep learning framework for multi-omics data. Cancers 13(12):3047
Zhang X, Zhang J, Sun K, Yang X, Dai C, Guo Y (2019) Integrated multi-omics analysis using variational autoencoders: application to pan-cancer classification. In BIBM, pp. 765–769. IEEE
Funding
This work was supported by the Project of State Key Laboratory of ASIC and System (No. 2021KF015); the National Natural Science Foundation of China (Grant nos. 61972456); Natural Science Foundation of Tianjin (No. 20JCYBJC00140; Key Laboratory of Universal Wireless Communications (BUPT), Ministry of Education, P.R.China (KFKT-2020101).
Author information
Authors and Affiliations
Contributions
LD and CL conceived and designed research. CL collects data and conducted experiments. RW and JC coordinated the research. CL analyzed data and wrote the manuscript. All authors read, revised, and approved the manuscript.
Corresponding author
Ethics declarations
Data availability
The TCGA data was download from https://xenabrowser.net/datapages/ The single-cell data was from the paper “Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer” (Cantini, Laura et al. 2021)
Ethics approval
This article does not contain any studies performed with human participants or animals by any of the authors.
Conflict of Interest
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Du, L., Liu, C., Wei, R. et al. Uncertainty-aware dynamic integration for multi-omics classification of tumors. J Cancer Res Clin Oncol 149, 3301–3312 (2023). https://doi.org/10.1007/s00432-022-04219-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04219-3