Abstract
Purpose
Toll-like receptor 4 (TLR4) is increasingly recognized for its ability to govern the etiology and prognostic outcomes of colorectal cancer (CRC) due to its profound immunomodulatory capacity. Despite widespread interest in TLR4 and CRC, no clear analysis of current literature and data exists. Therefore, translational advances have failed to move beyond conceptual ideas and suggestions.
Methods
We aimed to determine the relationship between TLR4 and CRC through a systematic review and analysis of published literature and datasets. Data were extracted from nine studies that reported survival, CRC staging and tumor progression data in relation to TLR4 expression. Primary and metastatic tumor samples with associated clinical data were identified through the Cancer Genome Atlas (TCGA) database.
Results
Systematic review identified heterogeneous relationships between TLR4 and CRC traits, with no clear theme evident across studies. A total of 448 datasets were identified through the TCGA database. Analysis of TCGA datasets revealed TLR4 mRNA expression is decreased in advanced CRC stages (P < 0.05 for normal vs Stage II, Stage III and Stage IV). Stage-dependent impact of TLR4 expression on survival outcomes were also found, with high TLR4 expression associated with poorer prognosis (stage I vs III (HR = 4.2, P = 0.008) and stage I vs IV (HR = 11.3, P < 0.001)).
Conclusion
While TLR4 mRNA expression aligned with CRC staging, it appeared to heterogeneously regulate survival outcomes depending on the stage of disease. This underscores the complex relationship between TLR4 and CRC, with unique impacts dependent on disease stage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) remains one of the most prevalent cancer diagnoses worldwide, with incidence rates in the United States of America of 37.8 per 100,000 (National Cancer and Institute: Surveillance 2021). This places CRC as the fourth most common cancer in western populations (Australian Institute and of Health and Welfare 2020; National Cancer and Institute: Surveillance 2021) which when coupled with its high mortality rates, cements this disease as a major healthcare burden. While significant advances have been made in identifying high level risk factors for CRC, heterogeneity in tumor progression and treatment response continues to challenge the understanding of its etiology (Buikhuisen et al. 2020). Few factors remain significant when traditional, largely unmodifiable risk factors (e.g. age, sex) are adjusted for, pointing to complex mechanisms governing tumor microenvironment which dictate growth trajectory and vulnerability to anti-cancer therapy (Buikhuisen et al. 2020).
The tumor microenvironment is a complex system of molecular and cellular components, produced by both host and tumor (Wang et al. 2018). The microenvironment’s contribution to prognosis and clinical outcome has proven controversial, although evidence supports both beneficial and inhibitory roles. For example, the microenvironment facilitates immune invasion and destruction of tumor tissue (Fang et al. 2014). In contrast, it also contributes to tumor development, cancer cell survival and treatment resistance (Zhao et al. 2019). Irrespective of this complexity, it is clear that infiltration of peripheral immune cells into the tumor microenvironment is related to CRC progression and prognosis. A 2019 study using the cancer genome atlas (TCGA) and gene expression omnibus (GEO) databases reported that M0 macrophages, M1 macrophages and CD4+ memory T cells were more abundant in CRC tissue compared to healthy tissues (P < 0.02) (Ge et al. 2019). Furthermore, higher infiltration of M1 macrophage populations in CRC tissue correlated with lower participant survival (P = 0.04) (Ge et al. 2019). This underscores the involvement of the host immune system in CRC.
In light of the strong immune-mediated mechanisms that appear to be linked with CRC etiology and treatment response, there has been substantial interest in the potential role of the innate immune surveillance protein, toll-like receptor 4 (TLR4). TLR4 is a pattern recognition receptor, which upon activation, initiates a strong inflammatory response (Takeda and Akira 2004). TLR4 requires the accessory proteins myeloid differentiation factor 2 (MD-2) and cluster of differentiation 14 (CD14) to efficiently bind to ligands including, LPS, heat shock proteins (Hsp70 and Hsp90) and high-mobility group protein I (HMGBI) (Cheng et al. 2015). TLR4 signaling is vital to intestinal homeostatic maintenance, as previously reviewed (Bruning et al. 2021). TLR4 is notably upregulated in the intestine under inflammatory states including in people with ulcerative colitis, and this is further linked to ulcerative colitis-associated CRC risk and development (Fukata et al. 2007). Furthermore, genetic variants of TLR4 (rs10116253, rs192791 1, rs7873784) have been linked to CRC (Huang et al. 2018a).
TLR4 is expressed on a range of different cell types within the tumor microenvironment, including dendritic, stromal, macrophage and epithelial cells (Li J et al. 2017). The importance of site-specificity of TLR4 expression in healthy and diseased states, including CRC, is well documented (Bruning et al. 2021). Pre-clinical CRC models indicate that TLR4 has both pro- and anti- tumor roles, with expression sites being a possible differentiating factor between whether TLR4 aids in cancer destruction or survival (Li et al. 2017). To add further complexity, TLR4 has also been identified to modulate toxicity following cancer therapy, including diarrhea and pain (Wardill et al. 2016). As such, it is currently unclear whether TLR4 is beneficial, or, potentially harmful in the CRC microenvironment, and whether it is a rationale target for intervention. We therefore aimed to systematically review current published evidence and datasets to crystalize the relationship between TLR4 and CRC staging, treatment toxicity and survival.
Methods
Search strategy, study selection and data retrieval
PubMed, Cochrane Library and Embase were searched between January and February 2022 for peer-reviewed journal publications using keywords listed in Supporting Information Table 1 and were screened for inclusion based on specific criteria; original research, clinical trials and studies conducted between 2010 and 2021; archival human tissue; CRC; participant survival; tumor recurrence; prognosis; toxicity; and TLR4 expression. Exclusion criteria included: animal models; cell lines; and cancer types other than CRC. Eligible publications were reviewed with the following data being extracted manually by two independent authors (EEC, JKC) using a computer-based template: sample size; CRC stage; chemotherapy treatments; participant demographics; type of TLR4 analysis; TLR4 specific outcomes (including expression rates and site-specificity); survival data (overall survival (OS), progression-free survival (PFS) or disease-free survival (DFS)); and tumor progression data. Summary outcomes are presented in Table 1.
TCGA clinical CRC cases database extraction and statistical analysis
RNA sequencing data and associated clinical metadata with a total of 512 samples in read counts (HTSeq-Counts) of CRC were obtained from the TCGA data portal (https://portal.gdc.cancer.gov/, accessed in December 2020). Data related to TLR4 mRNA expression, CRC staging and OS were extracted. TLR4 mRNA expression was dichotomized into high and low expression using the tertile cut point. The OS curve was constructed using Kaplan–Meier and log-rank test analysis, comparing high and low TLR4 expression groups for all cases and within each CRC stage. Statistical analyses were performed using GraphPad Prism 8.3.1 (GraphPad Software Inc., CA, USA) and R. studio 1.2.5033 (Inc., Boston, MA).
Multivariate analysis was also performed to determine whether mRNA expression was associated with OS in each tumor stage where variables included tumor stage (I: IV), sex and age. To avoid using potentially biased cut-points splitting low and high TLR mRNA expressing participant groups, a two sample t-test using continuous TLR4 mRNA expression values (with no cut-point required) compared mRNA expression between alive and deceased participants. Finally, TLR4 mRNA expression between normal tumor adjacent tissue and tumor samples from different stages were analyzed with a one-way ANOVA (normal vs stage I, stage II, stage III and stage IV).
Results
180 publications were initially identified, with 9 meeting inclusion criteria for final analysis (Fig. 1). 6 publications were clinical trials with a combined participant total of 1081. The remaining 3 publications used archival tissue from previous clinical research. Only 2 publications analyzed advanced stage CRC (non-resectable tumor stage II–IV), whereas 7 publications included mixed analysis of varying CRC stage. Participant survival data was extracted from 8 publications, inclusive of DFS, PFS and OS dependent on individual study outcomes. Only 1 publication included data regarding toxicity in relation to TLR4 expression. Finally, CRC recurrence was analyzed in 3 publications. TLR4 expression in the publications was assessed using immunohistochemistry (5/9, all of which used different primary antibodies), polymerase chain reaction (PCR) (3/9) and flow cytometry (1/9). Only 4 publications included site-specific analysis of TLR4 expression in CRC (Table 1) (Cammarota et al. 2010; Eiro et al. 2013; Formica et al. 2013; Sussman et al. 2014). Of the 9 publications, 4 analyzed formalin fixed and paraffin embedded tissue blocks, 4 analyzed peripheral blood samples and 1 (Sussman et. al. 2014) analyzed tumor tissue microarray slides provided by the NCI Cancer Diagnosis Program (CDP).
Impact of TLR4 genotype and expression on CRC survival
Of the 8 publications to report on CRC survival, one reported that wild-type (WT) TLR4 genotype was beneficial to CRC participant survival rates (Tesniere et al. 2010). Metastatic CRC participants with the WT TLR4 allele had higher PFS (hazard ratio (HR): 0.73; 95% confidence interval (CI) = 0.53–1.00; P < 0.05) and OS (HR = 0.72; 95% CI = 0.52–1.01; P = 0.05) compared with participants bearing the TLR4 loss-of-function (Asp299Gly) variant post-oxaliplatin chemotherapy treatment (Tesniere et al. 2010). No differences in DFS among participants bearing the WT versus the variant TLR4 alleles were observed.
In contrast, 2 publications suggested that increased TLR4 expression is detrimental to participant survival (Cammarota et al. 2010; Wang et al. 2010). Cammarota et al. found that in mixed stage CRC tissue, participants with lower TLR4 expression in the tumor stroma compartment had improved DFS compared to participants with higher TLR4 expression (risk ratio (RR) 2.36; log-rank chi-square 4.25, P < 0.05) (Cammarota et al. 2010). Furthermore, participants with pT3 adenocarcinoma with high TLR4 expression (over 50% positive cells) relapsed sooner (14 months) compared to participants with low TLR4 expression (40 months, RR 3.15; log-rank chi-square 4.03, P < 0.05) (Cammarota et al. 2010). This is supported by Wang and colleagues, who confirmed that CRC tissue displayed expression of TLR4 in 78 of 108 samples (72%), of which 22 displayed high TLR4 expression (Wang et al. 2010). In addition, increased TLR4 expression was associated with liver metastasis (P = 0.0015) and advanced tumor stage (stage IV) (P = 0.0197). Upon univariate analysis there was no difference in 5-year DFS rate for low versus high TLR4 expression, but OS was reduced with high TLR4 expression (HR (95% CI) 2.17 (1.15–4.07), P = 0.015) (Wang et al. 2010). However, this was not retained in multivariate analysis. In contrast, when samples exhibited high expression of both TLR4 and the adapter protein MyD88, DFS and OS were poorer (HR (95% CI) 2.11 (1.05–4.23) P = 0.0352) (Wang et al. 2010).
The conflicting nature of outcomes may be reflective of the lack of site-specific TLR4 investigations throughout human CRC research. Eiro and colleagues reported TLR4 expression by fibroblasts, not tumor cells themselves, was associated with a shortened OS of CRC participants (P = 0.022). Furthermore, TLR4 expression in fibroblasts was a significant and independent factor associated with DFS (P = 0.0001), and OS (P = 0.013) (Eiro et al. 2013).
Four publications reported that TLR4 expression does not impact upon CRC survival. Formica and colleagues found that in 31 metastatic CRC participants, neutrophil TLR4 expression at baseline, or 1-month post-chemotherapy, had no association with PFS (P > 0.05) (Formica et al. 2013). This is supported by Sussman and colleagues who, in N = 279, found no associated between TLR4 expression in stromal tissue and OS after correcting for both CRC stage and grade. Furthermore, epithelial TLR4 expression was also not associated with OS (Sussman et al. 2014).
More recently, Zhang and colleagues found that in an advanced CRC cohort (N = 94) post-standard Fluorouracil-based adjuvant chemotherapy and radical surgery, the measured level of TLR4 expression was independent of DFS; hence no impact of TLR4 on overall DFS (Zhang et al. 2019). In addition, TLR4 was not a significant factor in survival outcomes following univariate or multivariate analyses (Zhang et al. 2019). However, high amounts of Fusobacterium (Fn), an anaerobic bacterium known to activate the TLR4 pathway in CRC cells, correlated with poor DFS (P = 0.028) (Zhang et al. 2019). Finally, Gray and colleagues analyzed previously collected tissues from two large-scale clinical trials, the SCOT (ISRCTN59757862) trial and COIN (ISRCTN27286448) trial (Gray et al. 2019). Data generated from SCOT showed no association of any TLR4 single nucleotide polymorphism (SNP) with survival (Gray et al. 2019). There was also no association of the TLR4 SNP, rs867228, with DFS in cases with functional polymorphisms (Gray et al. 2019). Data from COIN showed no association of any tested TLR4 SNP with OS by either log-rank test or univariate or multivariable Cox regression (Gray et al. 2019).
CRC recurrence
Three publications reported on TLR4s contribution to CRC recurrence, with 2 publications identifying a detrimental role of TLR4 in CRC recurrence (Wang et al. 2010; Zhang et al. 2019). Wang and colleagues (2010) report upon 5 year follow-up of 108 mixed stage CRC participants, 53 participants had tumor recurrence (DFS rate: 49%), with participants exhibiting high expression of TLR4 and its accessory protein MyD88 displaying increased recurrence rates compared to those with low expression (TLR4 + MyD88 (low vs high) 5-year DFS HR (95% CI) = 2.25 (1.27–3.99) P = 0.0053) (Wang et al. 2010). Furthermore, participants with CRC and liver metastasis showed higher TLR4 and MyD88 expression versus CRC without liver metastasis (Wang et al. 2010). Among the 14 liver metastases obtained by hepatectomy, 12 were TLR4 positive and 6 showed a high expression (Wang et al. 2010). These findings are supported by Zhang and colleagues who showed high expression of TLR4 (P = 0.036) were more likely detected in participants with CRC recurrence, compared with participants without recurrence (Zhang et al. 2019).
In contrast, Eiro and colleagues observed that recurrence was dependent on the site of TLR4 expression, not its overall quantitative expression such that TLR4 expression by tumor cells was associated with a lower rate of recurrence in tumors from left colon/rectum compared to those from right colon/rectum (P = 0.028) (Eiro et al. 2013). Further, TLR4 expression by fibroblasts was associated with a high rate of recurrence (P = 0.0001) in left colon/rectum tumors (Eiro et al. 2013).
Toxicity post-chemotherapy in participants with CRC
Only 1 publication investigated the role of TLR4 in relation to post-chemotherapy toxicity outcomes, including diarrhea and nausea. Wong and colleagues investigated a cohort of 46 advanced stage CRC (stage III–IV), treated with first cycle of irinotecan-based chemotherapy (irinotecan monotherapy or in combination with fluorouracil and leucovorin—IFL regimen) (Wong et al. 2021). Participants the variant TLR4 SNPs rs4986790 and rs4986791 had more severe diarrhea (50%) compared to those without the variants (15%) (Wong et al. 2021). When looking at diarrhea of all severities, all participants (100%) with the variant TLR4 SNPs developed diarrhea, compared to only 50% of those without the variants (20 participants each, rs4986790, P = 0.012 vs. rs4986791, P = 0.012).(Wong et al. 2021) There was no association with nausea (Wong et al. 2021).
TCGA database results
TLR4 expression differs due to cancer stage
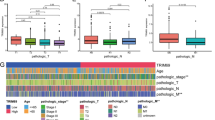
Summary of participant clinical data is presented in supporting information Table 2. Although TLR4 expression was not statistically different between normal and stage I, significantly higher TLR4 expression was observed in normal tissues vs Stage II, Stage III and Stage IV (Fig. 2A).
A Comparison of TLR4 expression between stage specific tumor and adjacent normal tissues from TCGA cohort. One-way ANOVA was performed by comparing solid tissue normal vs stage I, stage II, stage III, and stage IV participants. Statistical significance was represented as P < 0.05. B, C Assessment of TLR4 mRNA expression using the tertile cut-point. (B) Kaplan–Meier curves of overall survival (OS) in TCGA cohort. (C) Bar plot depicting the stage distribution of the cohort
TLR4 expression is associated with survival in respect to tumor stage
Number of participants per tumor stage is presented in Fig. 2C. OS of participants with CRC with respect to TLR4 expression (low vs high) was conducted. TLR4 expression was not a significant prognostic factor (HR = 1.1, P = 0.64) when all stages were combined (Fig. 2B) or compared between stages (Fig. 3). In contrast, multivariate analysis revealed high TLR4 expression prior to treatment conferred worse prognosis, with the strength of the effect increasing with tumor stage (stage I vs II (HR = 2.2, P = 0.138), stage I vs III (HR = 4.2, P = 0.008) and stage I vs IV (HR = 11.3, P < 0.001); Fig. 4). Sex and age had no impact on OS (Fig. 4). In stage I disease, those that were alive had lower TLR4 expression at diagnosis (P = 0.034). For all other stages TLR4 expression at diagnosis was higher in those still alive (P = 0.035) (Fig. 5).
Comparison of TLR4 expression in Fragments per Kilobase of transcript, per Million mapped reads (FPKM) with respect to OS. Analysis of TLR4 expression using two sample t-test based on participants’ survival in A stage I, B stage II, C stage III, and D stage IV participants. Statistical significance was represented as P < 0.05
Discussion
TLR4 is an attractive target for controlling cancer development and optimizing treatment response due to its potent regulation of systemic immune responses. Our analysis exposes the significant heterogeneity in CRC outcomes linked with TLR4 expression. We have shown that TLR4 expression decreases with increasing CRC tumor stage at prognosis, and appears to have stage-dependent associations with participant outcomes. We highlight two novel findings related to high TLR4 expression in early- and late-stage CRC being; (1) in stage I CRC results in worse participant outcomes, and (2) in stage IV CRC results in improved participant outcomes. With TLR4 expression decreasing in higher grade CRC, this potential reduction of innate immune signaling may prove to be the causative mechanism behind unfavorable treatment responses and reduced survival.
TLR4 expression relative to tumor stage is well documented in the literature (Li et al. 2019; Omrane et al. 2014). These patterns of TLR4 expression reflect its core physiological mechanism of inducing inflammation, a process known to be carcinogenic. Our data showed a significant decrease in TLR4 expression in later stage CRC (stages II–IV) compared to normal tissue. This decrease in TLR4 expression was not found in stage I tumors, suggesting that the slightly higher TLR4 expression in early CRC may align with the well-defined concept that inflammatory processes are involved in the early development of CRC (Karin and Greten 2005). However, our analysis did show that non-tumor comparative tissue had the highest TLR4 expression. As this tissue was primarily collected from adjacent tissue in the same participants, systemic inflammatory responses may have impacted on interpretation. The finding that TLR4 expression decreases with tumor growth is also consistent with the current understanding of tumor development, with tumors often adapting to evade immune detection and control. Activation of the receptor, programmed death 1 (PD-1), has been found to inhibit immune control of tumor growth, with the PD-1 ligand, PD-L1, being significantly upregulated in solid tumors like CRC (Hino et al. 2010). Therefore, this upregulation of PD-L1 is suggested to play a crucial role in the tumors ability to evade host immune system (Dong et al. 2002). This is of particular interest in the context of TLR4 research, as PD-L1 has also been shown to block the cytolytic activity of PD-1+ tumor infiltrating CD4+ and CD8+ T cells, which are reliant on dendritic cell -TLR4 interaction (Brahmer et al. 2012; Fife et al. 2009). In addition, Xiao et. al. (2016) reported that inhibition of TLR4 signaling via a blocking antibody significantly reduced the number of PD-1+ B cells in human hepatoma tissues, where PD-1+ B cell populations promoted cancer growth (Xiao et al. 2016). Furthermore, Huang (2018) found that improvement in clinical outcome is resultant of cytosolic HMGB1 triggering dendritic cell maturation through TLR4 activation, whereby consequently recruiting PD-1+ tumor-infiltrating lymphocytes to the tumor site (Huang et al. 2018b). These findings highlight the importance of TLR4 to this particular tumor kill pathway and outlines the importance for TLR4 expression for improved clinical outcomes of people living with CRC.
While our findings suggest a likely relationship between TLR4 expression and tumor stage, the relationship between TLR4 and long-term outcome was less clear cut in both our systematic review and genetic analyses. When looking at all tumor stages, there was no significant impact on OS in low vs high TLR4 expressing tumors. This contradicts existing data, as a metanalysis of 212 people living with CRC found that high TLR4 expression associated with a significantly reduced OS and poorer prognosis (HR (95% CI) 2.30 (1.41, 3.75), P = 0.001) (Hao et al. 2018). However, this analysis did not classify the cohort based on CRC stage which may have masked some findings and increased bias towards advanced stage disease. While our initial analyses showed no effect of TLR4 expression on OS, analysis of this relationship within specific tumor stages revealed that TLR4 may in fact have an impact but, in a stage-specific manner. Specifically, we showed that TLR4 expression in Stage IV disease was higher in tumors from people still alive compared to those that were not. While we weren’t able to show this in our longitudinal OS analyses, this may reflect the lack of power when breaking down our cohort of 488 into specific stages.
This heterogeneity in how TLR4 may act to regulate overall survival for Stage I vs Stage IV disease is likely to reflect the differences in how these disease stages are treated. Stage I disease is almost always treated with surgery, but no cytotoxic therapy, whereas stage IV disease will certainly contain cytotoxic therapy. TLR4 is considered to exert its impact on treatment outcomes via its ability to modulate immunogenic cell death (Fang et al. 2014; Kroemer et al. 2013). Immunogenic cell death acts in concert with direct cytotoxicity, and collectively results in more thorough tumor clearance, and thus long-term survival. As such, higher TLR4 expression would theoretically confer a larger immune response and thus better response in late-stage CRC. This is supported by the Isambert et al. study (2013) which found that increased activation of TLR4 via a lipid A analogue (OM-174) enhanced inflammatory anti-tumor response in metastatic CRC and improved clinical outcomes (Isambert et al. 2013). Furthermore, data from Huang and colleagues (2018) showed improved DFS in people living with late-stage rectal cancer with increased activation of TLR4 via HGMB1 binding (Huang et al. 2018b).
Despite new interpretation of stage-specific roles of TLR4, we must acknowledge some limitations of our approach. Firstly, the studies included within the literature review were varied, often with low sample sizes and differing approaches to measuring TLR4 expression. Furthermore, our genetic analysis relied on previously collected data and exhibited low power when analyzing within the specific CRC stages. It is also important to acknowledge that we relied solely on TLR4 tumor-expression data; whereas evidence from pre-clinical work suggests expression of TLR4 in host tissues (typically non-cancerous) may be critical in setting immune tone of host and thus response (Li et al. 2017). Nonetheless, our findings indicate a general trend towards higher TLR4 expression being associated with favorable OS outcomes in stage IV CRC suggesting its ability to induce immunogenic cell death is critical in CRC prognosis.
References
Australian Institute of Health and Welfare (2020) National bowel cancer screening program monitoring report 2020, Cancer series no. 128. Cat. no. CAN 133.
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, Pitot HC, Hamid O, Bhatia S, Martins R, Eaton K, Chen S, Salay TM, Alaparthy S, Grosso JF, Korman AJ, Parker SM, Agrawal S, Goldberg SM, Pardoll DM, Gupta A, Wigginton JM (2012) Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 366(26):2455–2465
Bruning EE, Coller JK, Wardill HR, Bowen JM (2021) Site-specific contribution of toll-like receptor 4 to intestinal homeostasis and inflammatory disease. J Cell Physiol 236(2):877–888
Buikhuisen JY, Torang A, Medema JP (2020) Exploring and modelling colon cancer inter-tumour heterogeneity: opportunities and challenges. Oncogenesis 9(7):66
Cammarota R, Bertolini V, Pennesi G, Bucci EO, Gottardi O, Garlanda C, Laghi L, Barberis MC, Sessa F, Noonan DM, Albini A (2010) The tumor microenvironment of colorectal cancer: stromal TLR-4 expression as a potential prognostic marker. J Transl Med 8:112
Cheng Z, Taylor B, Ourthiague D, A, H, (2015) Distinct single-cell signaling characteristics are conferred by the MyD88 and TRIF pathways during TLR4 activation. Sci Signal 8:ra69
Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L (2002) Tumor-associated B7–H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med 8(8):793–800
Eiro NGL, Gonzalez L, Fernandez-Garcia B, Andicoechea A, Barbon E, Garcia-Muniz J, Vizoso F (2013) Toll-like receptor-4 expression by stromal fibroblasts is associated with poor prognosis in colorectal cancer. J Immunother 36(6):342–349
Fang H, Ang B, Xu X, Huang X, Wu Y, Sun Y, Wang W, Li N, Cao X, Wan T (2014) TLR4 is essential for dendritic cell activation and anti-tumor T-cell response enhancement by DAMPs released from chemically stressed cancer cells. Cell Mol Immunol 11(2):150–159
Fife BT, Pauken KE, Eagar TN, Obu T, Wu J, Tang Q, Azuma M, Krummel MF, Bluestone JA (2009) Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat Immunol 10(11):1185–1192
Formica V, Cereda V, di Bari MG, Grenga I, Tesauro M, Raffaele P, Ferroni P, Guadagni F, Roselli M (2013) Peripheral CD45RO, PD-1, and TLR4 expression in metastatic colorectal cancer patients treated with bevacizumab, fluorouracil, and irinotecan (FOLFIRI-B). Med Oncol 30(4):743
Fukata M, Chen A, Vamadevan A, Cohen J, Breglio K, Krishnareddy S, Xu R, Harpaz N, Dannenberg A, Subbaramaiah K, Cooper H, Itzkowitz S, Abreu M (2007) Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology 133:1869–1881
Ge P, Wang W, Li L, Zhang G, Gao Z, Tang Z, Dang X, Wu Y (2019) Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed Pharmacother 118:109228
Gray V, Briggs S, Palles C, Jaeger E, Iveson T, Kerr R, Saunders MP, Paul J, Harkin A, McQueen J, Summers MG, Johnstone E, Wang H, Gatcombe L, Maughan TS, Kaplan R, Escott-Price V, Al-Tassan NA, Meyer BF, Wakil SM, Houlston RS, Cheadle JP, Tomlinson I, Church DN (2019) Pattern recognition receptor polymorphisms as predictors of oxaliplatin benefit in colorectal cancer. J Natl Cancer Inst 111(8):828–836
Hao B, Chen Z, Baochen B, Miaomei Y, Yao S, Feng Y, Yu Y, Pan L, Di D, Luo G, Zhang X (2018) Role of TLR4 as a prognostic factor for survival in various cancers: a meta-analysis. Oncotarget 9:13088–13099
Hino R, Kabashima K, Kato Y, Yagi H, Nakamura M, Honjo T, Okazaki T, Tokura Y (2010) Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer 116(7):1757–1766
Huang BZ, Tsilidis KK, Smith MW, Hoffman-Bolton J, Visvanathan K, Platz EA, Joshu CE (2018a) Polymorphisms in genes related to inflammation and obesity and colorectal adenoma risk. Mol Carcinog 57(10):1278–1288
Huang CY, Chiang SF, Ke TW, Chen TW, Lan YC, You YS, Shiau AC, Chen WT, Chao KSC (2018b) Cytosolic high-mobility group box protein 1 (HMGB1) and/or PD-1+ TILs in the tumor microenvironment may be contributing prognostic biomarkers for patients with locally advanced rectal cancer who have undergone neoadjuvant chemoradiotherapy. Cancer Immunol Immunother 67(4):551–562
Isambert N, Fumoleau P, Paul C, Ferrand C, Zanetta S, Bauer J, Ragot K, Lizard G, Jeannin JF, Bardou M (2013) Phase I study of OM-174, a lipid A analogue, with assessment of immunological response, in patients with refractory solid tumors. BMC Cancer 13(172):172
Karin M, Greten FR (2005) NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 5(10):749–759
Kroemer G, Galluzzi L, Kepp O, Zitvogel L (2013) Immunogenic cell death in cancer therapy. Annu Rev Immunol 31:51–72
Li J, Yang F, Wei F, Ren X (2017) The role of toll-like receptor 4 in tumor microenvironment. Oncotarget 8:66656–66667
Li N, Xu H, Ou Y, Feng Z, Zhang Q, Zhu Q, Cai Z (2019) LPS-induced CXCR7 expression promotes gastric Cancer proliferation and migration via the TLR4/MD-2 pathway. Diagn Pathol 14(1):3
National Cancer Institute: Surveillance, E, and End Results Program (SEER) (2021) SEER Cancer Stat Facts: Colorectal Cancer, Bethesda, MD2022.
Omrane I, Baroudi O, Kourda N, Bignon YJ, Uhrhammer N, Desrichard A, Medimegh I, Ayari H, Stambouli N, Mezlini A, Bouzayenne H, Marrakchi R, Benammar-Elgaaid A, Bougatef K (2014) Positive link between variant toll-like receptor 4 (Asp299Gly and Thr399Ile) and colorectal cancer patients with advanced stage and lymph node metastasis. Tumour Biol 35(1):545–551
Sussman DA, Santaolalla R, Bejarano PA, Garcia-Buitrago MT, Perez MT, Abreu M, Clarke J (2014) In silico and Ex vivo approaches identify a role for toll-like receptor 4 in colorectal cancer. J Exp Clin Cancer Res 33:45
Takeda K, Akira S (2004) TLR signaling pathways. Semin Immunol 16(1):3–9
Tesniere A, Schlemmer F, Boige V, Kepp O, Martins I, Ghiringhelli F, Aymeric L, Michaud M, Apetoh L, Barault L, Mendiboure J, Pignon JP, Jooste V, van Endert P, Ducreux M, Zitvogel L, Piard F, Kroemer G (2010) Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 29(4):482–491
Wang JJ, Lei KF, Han F (2018) Tumor microenvironment: recent advances in various cancer treatments. Eur Rev Med Pharmacol Sci 22:3855–3864
Wang EL, Qian ZR, Nakasono M, Tanahashi T, Yoshimoto K, Bando Y, Kudo E, Shimada M, Sano T (2010) High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer 102(5):908–915
Wardill HR, Gibson RJ, Van Sebille YZ, Secombe KR, Coller JK, White IA, Manavis J, Hutchinson MR, Staikopoulos V, Logan RM, Bowen JM (2016) Irinotecan-induced gastrointestinal dysfunction and pain are mediated by common TLR4-dependent mechanisms. Mol Cancer Ther 15(6):1376–1386
Wong DVT, Holanda RBF, Cajado AG, Bandeira AM, Pereira JFB, Amorim JO, Torres CS, Ferreira LMM, Lopes MHS, Oliveira RTG, Pereira AF, Sant’Ana RO, Arruda LM, Ribeiro-Junior HL, Pinheiro RF, Almeida PRC, Carvalho RF, Chaves FF, Rocha-Filho DR, Cunha FQ, Lima-Junior RCP (2021) TLR4 deficiency upregulates TLR9 expression and enhances irinotecan-related intestinal mucositis and late-onset diarrhoea. Br J Pharmacol 178(20):4193–4209
Xiao X, Lao XM, Chen MM, Liu RX, Wei Y, Ouyang FZ, Chen DP, Zhao XY, Zhao Q, Li XF, Liu CL, Zheng L, Kuang DM (2016) PD-1hi identifies a novel regulatory B-cell population in human hepatoma that promotes disease progression. Cancer Discov 6(5):546–559
Zhang S, Yang Y, Weng W, Guo B, Cai G, Ma Y, Cai S (2019) Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res 38(1):14
Zhao J, Meng Z, Xie C, Yang C, Liu Z, Wu S, Wang B, Fan P, Jin X, Wu H (2019) B7-H3 is regulated by BRD4 and promotes TLR4 expression in pancreatic ductal adenocarcinoma. Int J Biochem Cell Biol 108:84–91
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. Elise E Crame is funded by an Australian Government RTP Scholarship, (2019–2022), Hannah R Wardill the recipient of a National Health and Medical Research Council CJ Martin Biomedical Research Fellowship, (2018– 2022).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Literature review material preparation, data collection and analysis were performed by EEC and TCGA data collection and analysis were performed by SN. The first draft of the manuscript was written by EEC and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Crame, E.E., Nourmohammadi, S., Wardill, H.R. et al. Contribution of TLR4 to colorectal tumor microenvironment, etiology and prognosis. J Cancer Res Clin Oncol 149, 3009–3021 (2023). https://doi.org/10.1007/s00432-022-04199-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04199-4