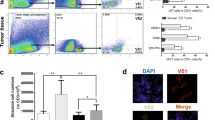
Abstract
Purpose
Invasive ductal carcinoma (IDC) and coronary artery disease (CAD), remains the greatest cause of death annually in women, driven by complex signalling pathways and shared several predisposing risk factors together. Therefore, it is important to find out the common epigenetic modifications which are responsible for possible disease progression from CAD to IDC.
Methods
CD4+T cell isolation by MACS, RT2 profiler PCR array, Gene ontology study, m6A RNA methylation, ChIP–qPCR, Q-PCR, CRISPR/Cas9-mediated knockout/overexpression, Lactate dehydrogenase release assay, RDIP–qPCR.
Results
We have identified several epigenetic regulators (e.g., VEGFA, AIMP1, etc.) which are mainly involved in inflammatory pathways in both the diseased conditions. Epitranscriptomic alterations such as m6A RNA methylation found abnormal in CD4+T helper cells in both IDC as well as CAD. CRISPR–Cas9 mediated knockout/overexpression of specific gene (BRCA1) are promising therapeutic approaches in diseased conditions by regulating m6A RNA methylation and also tumor suppressor gene P53. It also affected the R-loop formation which is vulnerable to DNA damage and BRCA1 can also induce CTL mediated cytotoxicity in breast cancer cells.
Conclusions
Therefore, by understanding the modifications of epigenetic mechanisms, their alterations and interactions will aid in the development of newer therapeutic approaches to stop the possible spread from one disease to another.











Similar content being viewed by others
Data availability
Not applicable.
References
Adams S, Gray RJ, Demaria S et al (2014) Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 32(27):2959–2966. https://doi.org/10.1200/JCO.2013.55.0491
Al-Hadid Q, Yang Y (2016) R-loop: an emerging regulator of chromatin dynamics. Acta Biochim Biophys Sin (shanghai) 48(7):623–631. https://doi.org/10.1093/abbs/gmw052
Allen MD, Jones LJ (2015) The role of inflammation in progression of breast cancer: friend or foe? (Review). Int J Oncol 47(3):797–805. https://doi.org/10.3892/ijo.2015.3075
Anan K, Morisaki T, Katano M et al (1996) Vascular endothelial growth factor and platelet-derived growth factor are potential angiogenic and metastatic factors in human breast cancer. Surgery 119(3):333–339. https://doi.org/10.1016/s0039-6060(96)80120-6
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2. https://doi.org/10.1186/1471-2105-4-2
Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L (2016) Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 13(11):674–690. https://doi.org/10.1038/nrclinonc.2016.66
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25(8):1091–1093. https://doi.org/10.1093/bioinformatics/btp101
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492 (Published correction appears in CA Cancer J Clin. 2020 Jul;70(4):313)
Brown LF, Berse B, Jackman RW et al (1995) Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in breast cancer. Hum Pathol 26(1):86–91. https://doi.org/10.1016/0046-8177(95)90119-1
Chen Y, Lin Y, Shu Y, He J, Gao W (2020) Interaction between N6-methyladenosine (m6A) modification and noncoding RNAs in cancer. Mol Cancer 19(1):94. https://doi.org/10.1186/s12943-020-01207-4 (Published 2020 May 22)
Cîmpean AM, Raica M, Suciu C, Tătucu D, Sârb S, Mureşan AM (2008) Vascular endothelial growth factor A (VEGF A) as individual prognostic factor in invasive breast carcinoma. Rom J Morphol Embryol 49(3):303–308
Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420(6917):860–867. https://doi.org/10.1038/nature01322
D’Souza M, Schou M, Skals R et al (2019) Polygenic predisposition to breast cancer and the risk of coronary artery disease. Int J Cardiol 291:145–151. https://doi.org/10.1016/j.ijcard.2019.05.051
Goel HL, Mercurio AM (2013) VEGF targets the tumour cell. Nat Rev Cancer 13(12):871–882. https://doi.org/10.1038/nrc3627
Gori JL, Hsu PD, Maeder ML, Shen S, Welstead GG, Bumcrot D (2015) Delivery and specificity of CRISPR-Cas9 genome editing technologies for human gene therapy. Hum Gene Ther 26(7):443–451. https://doi.org/10.1089/hum.2015.074
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674. https://doi.org/10.1016/j.cell.2011.02.013
Harbeck N, Gnant M (2017) Breast cancer. Lancet 389(10074):1134–1150. https://doi.org/10.1016/S0140-6736(16)31891-8
He X, Tan L, Ni J, Shen G (2021) Expression pattern of m6A regulators is significantly correlated with malignancy and antitumor immune response of breast cancer. Cancer Gene Ther 28(3–4):188–196. https://doi.org/10.1038/s41417-020-00208-1
Hong HJ, Lim HX, Song JH et al (2016) Aminoacyl-tRNA synthetase-interacting multifunctional protein 1 suppresses tumor growth in breast cancer-bearing mice by negatively regulating myeloid-derived suppressor cell functions. Cancer Immunol Immunother 65(1):61–72. https://doi.org/10.1007/s00262-015-1777-2
Horak ER, Leek R, Klenk N et al (1992) Angiogenesis, assessed by platelet/endothelial cell adhesion molecule antibodies, as indicator of node metastases and survival in breast cancer. Lancet 340(8828):1120–1124. https://doi.org/10.1016/0140-6736(92)93150-l
Jørgensen N, Hviid TVF, Nielsen LB et al (2021) Tumour-infiltrating CD4-, CD8- and FOXP3-positive immune cells as predictive markers of mortality in BRCA1- and BRCA2-associated breast cancer. Br J Cancer 125(10):1388–1398. https://doi.org/10.1038/s41416-021-01514-7
Kayton ML, Libutti SK (2001) Endothelial monocyte activating polypeptide II (EMAP II) enhances the effect of TNF on tumor-associated vasculature. Curr Opin Investig Drugs 2(1):136–138
Kim YW, Kwon C, Liu JL, Kim SH, Kim S (2012) Cancer association study of aminoacyl-tRNA synthetase signaling network in glioblastoma. PLoS ONE 7(8):e40960. https://doi.org/10.1371/journal.pone.0040960
Ko YG, Park H, Kim T, Lee JW, Park SG, Seol W, Kim JE, Lee WH, Kim SH, Park JE, Kim S (2001) A cofactor of tRNA synthetase, p43, is secreted to up-regulate proinflammatory genes. J Biol Chem 276(25):23028–23033. https://doi.org/10.1074/jbc.M101544200
Koelwyn GJ, Newman AAC, Afonso MS et al (2020) Myocardial infarction accelerates breast cancer via innate immune reprogramming. Nat Med 26(9):1452–1458. https://doi.org/10.1038/s41591-020-0964-7
Li Y, Xiao J, Bai J et al (2019) Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Mol Cancer 18(1):137. https://doi.org/10.1186/s12943-019-1066-3 (Published 2019 Sep 14)
Liu L, Liu X, Dong Z et al (2019) N6-methyladenosine-related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer 10(22):5447–5459. https://doi.org/10.7150/jca.35053 (Published 2019 Aug 29)
Loibl S, Gianni L (2017) HER2-positive breast cancer. Lancet 389(10087):2415–2429. https://doi.org/10.1016/S0140-6736(16)32417-5
Madapura HS, Salamon D, Wiman KG et al (2012) p53 contributes to T cell homeostasis through the induction of pro-apoptotic SAP. Cell Cycle 11(24):4563–4569. https://doi.org/10.4161/cc.22810
Megha T, Ferrari F, Benvenuto A et al (2002) p53 mutation in breast cancer. Correlation with cell kinetics and cell of origin. J Clin Pathol 55(6):461–466. https://doi.org/10.1136/jcp.55.6.461
Mehta LS, Watson KE, Barac A et al (2018) Cardiovascular disease and breast cancer: where these entities intersect: a scientific statement from the American heart association. Circulation 137(8):e30–e66. https://doi.org/10.1161/CIR.0000000000000556 (Published correction appears in Circulation. 2019 Aug 27;140(9):e543)
Nadel J, Athanasiadou R, Lemetre C, Wijetunga NA, Broin PÓ, Sato H, Zhang Z, Jeddeloh J, Montagna C, Golden A, Seoighe C, Greally JM (2015) RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenetics Chromatin 8:46. https://doi.org/10.1186/s13072-015-00406
Neugut AI, Rosenberg DJ, Ahsan H et al (1998) Association between coronary heart disease and cancers of the breast, prostate, and colon. Cancer Epidemiol Biomarkers Prev 7(10):869–873
Park SG, Kang YS, Ahn YH, Lee SH, Kim KR, Kim KW, Koh GY, Ko YG, Kim S (2002) Dose-dependent biphasi activity of tRNA synthetase-associating factor, p43, in angiogenesis. J Biol Chem 277(47):45243–45248. https://doi.org/10.1074/jbc.M207934200
Park SG, Shin H, Shin YK, Lee Y, Choi EC, Park BJ, Kim S (2005) The novel cytokine p43 stimulates dermal fibroblast proliferation and wound repair. Am J Pathol 166(2):387–398. https://doi.org/10.1016/S0002-9440(10)62262-6
Park SG, Kang YS, Kim JY, Lee CS, Ko YG, Lee WJ, Lee KU, Yeom YI, Kim S (2006) Hormonal activity of AIMP1/p43 for glucose homeostasis. Proc Natl Acad Sci USA 103(40):14913–14918. https://doi.org/10.1073/pnas.0602045103
Rak J, Yu JL, Klement G, Kerbel RS (2000) Oncogenes and angiogenesis: signaling three-dimensional tumor growth. J Investig Dermatol Symp Proc 5(1):24–33. https://doi.org/10.1046/j.1087-0024.2000.00012.x
Rakshit S, Sunny JS, George M, Hanna LE, Sarkar K (2021) R-loop modulated epigenetic regulation in T helper cells mechanistically associates coronary artery disease and non-small cell lung cancer. Transl Oncol 14(10):101189. https://doi.org/10.1016/j.tranon.2021.101189
Sarkar K, Sadhukhan S, Han SS, Vyas YM (2014) Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood 124(23):3409–3419. https://doi.org/10.1182/blood-2014-07-587642
Shukla PC, Singh KK, Quan A, Al-Omran M, Teoh H, Lovren F, Cao L, Rovira II, Pan Y, Brezden-Masley C, Yanagawa B, Gupta A, Deng CX, Coles JG, Leong-Poi H, Stanford WL, Parker TG, Schneider MD, Finkel T, Verma S (2011) BRCA1 is an essential regulator of heart function and survival following myocardial infarction. Nat Commun 2:593. https://doi.org/10.1038/ncomms1601
Shuvalov O, Petukhov A, Daks A, Fedorova O, Ermakov A, Melino G, Barlev NA (2015) Current genome editing tools in gene therapy: new approaches to treat cancer. Curr Gene Ther 15(5):511–529. https://doi.org/10.2174/1566523215666150818110241
Taylor MD, Sadhukhan S, Kottangada P, Ramgopal A, Sarkar K, D’Silva S, Selvakumar A, Candotti F, Vyas YM (2010) Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci Transl Med 2(37):37ra44. https://doi.org/10.1126/scitranslmed.3000813
Toi M, Inada K, Suzuki H, Tominaga T (1995) Tumor angiogenesis in breast cancer: its importance as a prognostic indicator and the association with vascular endothelial growth factor expression. Breast Cancer Res Treat 36(2):193–204. https://doi.org/10.1007/BF00666040
Wang H, Yang ES, Jiang J, Nowsheen S, Xia F (2010) DNA damage-induced cytotoxicity is dissociated from BRCA1’s DNA repair function but is dependent on its cytosolic accumulation. Cancer Res 70(15):6258–6267. https://doi.org/10.1158/0008-5472.CAN-09-4713
Wang X, Lu Z, Gomez A et al (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505(7481):117–120. https://doi.org/10.1038/nature12730
Wang X, Zhao BS, Roundtree IA et al (2015) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161(6):1388–1399. https://doi.org/10.1016/j.cell.2015.05.014
Wang Y, Zhang Y, Du Y, Zhou M, Hu Y, Zhang S (2020) Emerging roles of N6-methyladenosine (m6A) modification in breast cancer. Cell Biosci 10(1):136. https://doi.org/10.1186/s13578-020-00502-3 (Published 2020 Nov 25)
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38:W214–W220. https://doi.org/10.1093/nar/gkq537
Weaver KE, Foraker RE, Alfano CM et al (2013) Cardiovascular risk factors among long-term survivors of breast, prostate, colorectal, and gynecologic cancers: a gap in survivorship care? J Cancer Surviv 7(2):253–261. https://doi.org/10.1007/s11764-013-0267-9
Weidner N, Folkman J, Pozza F et al (1992) Tumor angiogenesis: a new significant and independent prognostic indicator in early-stage breast carcinoma. J Natl Cancer Inst 84(24):1875–1887. https://doi.org/10.1093/jnci/84.24.1875
Wu J, Lu LY, Yu X (2010) The role of BRCA1 in DNA damage response. Protein Cell 1(2):117–123. https://doi.org/10.1007/s13238-010-0010-5
Xiao W, Adhikari S, Dahal U et al (2016) Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell 61(4):507–519. https://doi.org/10.1016/j.molcel.2016.01.012 (Published correction appears in Mol Cell. 2016 Mar 17;61(6):925)
Yoshiji H, Gomez DE, Shibuya M, Thorgeirsson UP (1996) Expression of vascular endothelial growth factor, its receptor, and other angiogenic factors in human breast cancer. Cancer Res 56(9):2013–2016
Acknowledgements
This research was supported by Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Govt. of India (sanction order No. “ECR/2016/000965”).
Funding
The author, Dr. Koustav Sarkar is extremely thankful for the financial help for the DST-SERB project of the Department of Science and Technology, India (Ref. No. ECR/2016/000965, Date: 20.02.2017).
Author information
Authors and Affiliations
Contributions
SR: data curation, formal analysis, methodology, and software, and writing—original draft preparation, MG: resources and supervision, LEH: resources and supervision, JSS: software and formal analysis, KVL: resources, KS: conceptualization, supervision, visualization, resources, and writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Ethical approval was obtained from Institutional Ethics Committee of SRM Medical College Hospital and Research Centre, Kattankulathur, Tamil Nadu 603204, India.
Consent to participate
Written informed consent was received from each of the control subject, breast cancer and CAD patients to collect their blood samples.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
432_2022_4130_MOESM1_ESM.tif
Supplementary file1 (TIF 2086 KB) Highest clustered gene–gene interaction network of dysregulated genes from IDC disease data set
432_2022_4130_MOESM2_ESM.tif
Supplementary file2 (TIF 1658 KB) Highest clustered gene–gene interaction network of dysregulated genes from CAD disease data set
Rights and permissions
About this article
Cite this article
Rakshit, S., Sunny, J.S., George, M. et al. T helper cell-mediated epitranscriptomic regulation via m6A RNA methylation bridges link between coronary artery disease and invasive ductal carcinoma. J Cancer Res Clin Oncol 148, 3421–3436 (2022). https://doi.org/10.1007/s00432-022-04130-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04130-x