Abstract
Recommendations for the management of community-acquired pneumonia (CAP) advocate that, in the absence of the clinical and laboratory findings typical of bacterial CAP, antibiotics are not required. However, the true value of the clinical and laboratory predictors of pediatric CAP still needs to be assessed. This prospective cohort study in three emergency departments enrolled 142 children with radiological pneumonia. Pneumonia with lung consolidation was the primary endpoint; complicated pneumonia (bacteremia, empyema, or pleural effusion) was the secondary endpoint. We showed that three clinical signs (unilateral hypoventilation, grunting, and absence of wheezing), elevated procalcitonin (PCT), C-reactive protein (CRP), negative nasopharyngeal viral PCR, or positive blood pneumococcal PCR (P-PCR) were significantly associated with both pneumonia with consolidation and complicated pneumonia. Children with negative clinical signs and low CRP values had a low probability of having pneumonia with consolidation (13%) or complicated pneumonia (6%). Associating the three clinical signs, CRP >80 mg/L and a positive P-PCR ruled in the diagnosis of complicated pneumonia with a positive predictive value of 75%.
Conclusion: A model incorporating clinical signs and laboratory markers can effectively assess the risk of having pneumonia. Children with negative clinical signs and low CRP are at a low risk of having pneumonia. For children with positive clinical signs and high CRP, a positive blood pneumococcal PCR can more accurately confirm the diagnosis of pneumonia.
What is Known: • Distinguishing between bacterial and viral pneumonia in children is challenging. • Reducing the inappropriate use of antibiotics is a priority. |
What is New: • Children with negative clinical signs and low C-reactive protein (CRP) values have a low probability of having pneumonia. • Children with high CRP values can be tested using a pneumococcal PCR to rule in the diagnosis of pneumonia with a high positive predictive value. |
Similar content being viewed by others
Introduction
Community-acquired pneumonia (CAP) remains the leading cause of childhood mortality worldwide [2, 11, 12]. Streptococcus pneumoniae (SP) is the main pathogen of bacterial CAP [4] and of pneumonia with complications such as pleural effusion, empyema, or bacteremia [13]. The annual incidence of CAP in Europe and North America is 30–40 per 1000 children <5 years old [29, 33, 39]. The microbiological etiologies of childhood CAP vary greatly between studies: 19–65% are viruses, 19–37% are typical and atypical bacteria, and 23–33% are mixed viral–bacterial infections [4, 35, 45].
Unfortunately, a microbiological diagnosis is often impossible due to the inaccessibility of the infected sites, i.e., lung tissue or bronchoalveolar lavage, and this is further complicated by the fact that there is still no gold standard test for the diagnosis of pediatric CAP [6, 30]. Blood or pleural cultures are very specific but are only positive for CAP in about 4–10% of cases [7, 10, 43]. Blood pneumococcal PCR assays, targeting genes such as autolysin A (LytA) or pneumolysin (PLY), seem to be more sensitive than blood cultures for diagnosing pneumococcal CAP [19, 40] and are used as surrogate markers of pneumococcal pneumonia [1, 25, 47]. However, recent studies have shown that PLY PCR may lack specificity. This is because it can be positive in children with pneumococcal or S. viridans colonization, especially in nasopharyngeal PCR [19, 20, 36]. Therefore, the utilization of blood, rather than nasopharyngeal PCR targeting both PLY and LytA genes, seems to be more specific. Bronchoalveolar lavage or lung aspirates are too dangerous for routine testing. Obtaining sputum from young children is difficult and it is also often contaminated by commensals from the oropharynx. Nasopharyngeal bacterial cultures are often positive but can be unreliable due to healthy bacterial carriage. In contrast to adults [14], the rapid urine SP C-polysaccharide assay has shown high rates of false positives in children [15, 17], although recent studies have shown its possible added value [6, 24]. Antibody responses to four pneumococcal surface protein antibodies and one pneumolysin antibody have displayed encouraging results [41], but they cannot be interpreted until the second week after sampling.
In conclusion, laboratory tests are unreliable at predicting causative bacterial involvement. The British Thoracic Society’s recent guidelines for the management of CAP therefore propose that blood testing and chest radiography should not be considered routine investigations [27]. Furthermore, the recent PIDS and IDSA guidelines on pediatric CAP do not recommend the use of antibiotics in the absence of clinical or laboratory findings suggestive of bacterial infection. However, an exact definition of the findings that allow differentiation between bacterial and viral infection are not straightforward. Since there is no gold standard method for differentiating between bacterial and viral pneumonia, the majority of children are indeed treated with antibiotics, often with broad-spectrum antibiotics [31, 46], despite the high incidence of viral pneumonia, notably in children under 2 years old [3]. Developing clinical or laboratory markers that could predict the bacterial etiology of CAP in a more reliable fashion would benefit patients and physicians [6, 23]. The recent guidelines also recommend more rigorous management and follow-up of CAP patients with such complications as bacteremia or empyema.
The present study aimed to investigate approaches combining clinical and biological markers that would more accurately predict viral, bacterial, and complicated CAP.
Methods
Ethics statement
Ethical approval was obtained from the three participating hospitals’ relevant research ethics committees: Geneva’s Cantonal Research Ethics Committee; the Human Research Ethics Committee of the Canton Vaud, in Lausanne; and the Canton Valais Medical Ethics Committee, in Sion. Written informed consent was obtained from all the participants’ parents and from teenage participants themselves, before enrollment.
Design
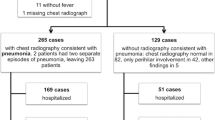
A multi-center prospective diagnostic study was conducted in the Pediatric Emergency Departments of three Swiss tertiary hospitals (Geneva, Lausanne, and Sion). We consecutively included 142 children who presented with pneumonia at one of these departments from January 2008 to July 2009. Inclusion criteria were ≥ 2 months old and ≤16 years old, fever (>38 °C), cough, increased respiratory rate or respiratory distress, and infiltrates on chest radiographs. Exclusion criteria were immunodeficiency, chronic lung or heart diseases, and hospital-acquired pneumonia.
Blinded chest radiographs were examined by pediatric radiologists. Infiltrates visible on chest radiographs were classified as infiltrates with consolidation when there was a dense opacity, with or without air bronchograms, occupying a portion of a lobe, and as infiltrates without consolidation when densities were linear and patchy in a lacy pattern, as per the Bulletin of the World Health Organization [5]. CAP with consolidation was considered a proxy for bacterial pneumonia [38] and was our primary endpoint for the first prediction model. Complicated pneumococcal CAP, defined as CAP with bacteremia, pleural effusion, or empyema, was the endpoint for the second prediction model.
Each participant was investigated with regard to the following data: age, sex, vaccination status, day care attendance, number of siblings, tobacco exposure, clinical examination (respiratory rate, respiratory distress, chest indrawing, nasal flaring, wheezing, unilateral hypoventilation, crackles, oxygen saturation), chest radiograph, WBC count, CRP and PCT levels, blood cultures, blood pneumococcal PCR (P-PCR), NP viral PCR, and mycoplasma/chlamydia PCR. The seven-valent pneumococcal vaccine (PCV-7) was in use at that time.
Laboratory methods
Blood and nasopharyngeal samples were immediately stored at −80 °C. Blood P-PCR assays targeting pneumolysin (WB-Ply) and autolysin A (WB-LytA) were performed in triplicates, as follows. DNA extraction and qPCR: a mixture containing 100 μL of whole blood, 100 μL of TE buffer (10 mM Tris–HCl pH 8, 1 mM EDTA), and 100 μL glass beads (acid washed, 212–300 μm, SIGMA) were vortexed for 1 min. Samples were centrifuged at 6000 rpm for 3 min at room temperature. DNA was extracted from the supernatant using a DNeasy kit (QIAGEN) and eluted in 70 μL water. Quantitative PCR was run on a StepOne™ Real-Time PCR System (Applied Biosystems) using an ABsolute qPCR Mix (Abgene). Reaction mixtures contained 8 μL DNA extract and 50 nM primers and probe in a total volume of 20 μL. Cycling conditions included 2 min at 50 °C, 15 min at 95 °C, followed by 50 cycles at 95 °C for 15 s and 60 °C for 80 s. Specific primers and probes for WB-LytA and WB-Ply were: LytA_F5′ACGCAATCTAGCAGATGAAGC, LytA_R5′TGTTTGGTTGGTTATTCGTGC, LytA_Probe_5′-FAM-TTTGCCGAAAACGCTTGATACAGGG [32], Ply_F5′-TGCAGAGCGTCCTTTGGTCTAT, Ply_R5′-CTCTTACTCGTGGTTTCCAACT, Ply_Probe_5′-FAM-TGGCGCCCATAAGCAACACTCGAA [8].
NP viral PCR assays were performed as per a previously published protocol [42]. These assays identified a standard panel of 24 viruses: influenza (A, B), parainfluenza (1, 2, 3), picornavirus, rhinovirus (A, B), enterovirus (A, B, C, D), human meta-pneumovirus (A, B), respiratory syncytial virus (A, B), adenovirus (A, B, C, E), and coronavirus (229E, HKU1, NL63, OC43). CRP values were determined in blood using immunometric methods (Nycocard CRP) according to the manufacturer’s instructions. Procalcitonin (PCT) values were determined using the Kryptor® (Brahms GmbH, Germany) assay according to the manufacturer’s instructions.
Statistics
Three types of predictors were analyzed: (1) clinical signs; (2) inflammatory markers, CRP, and PCT; and (3) specific blood pneumococcal, nasopharyngeal viral, and mycoplasma PCRs.
Univariate and multivariate logistic regressions were calculated using STATA® 11.0 software (StataCorp, Texas, USA). These tested the associations between CAP with or without consolidation and CAP with or without complications and clinical signs, CRP and PCR, two types of blood P-PCR targeted for WB-LytA and WB-Ply, and nasopharyngeal viral PCR. Medians were expressed using interquartile ranges (IQRs). Measures of effect were expressed using odds ratios (ORs) with 95% confidence intervals (CIs). Non-parametric Wilcoxon–Mann–Whitney rank-sum tests were used for continuous variables, assuming non-normal distributions. For categorical data, Pearson’s Chi-squared test and univariate logistic regression were used to establish the presence or absence of general associations. Stepwise multivariate logistic regression analyzed potential confounders. The p value significance level was set at 0.05.
The performance of each type of predictor was expressed using sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV), positive and negative likelihood ratios, and odd ratios. We developed two models involving three diagnostic steps: (1) clinical signs, (2) inflammatory markers (CRP), and (3) specific blood pneumococcal PCR. Likelihood ratios and the Fagan nomogram [22] were used to calculate post-test probabilities step by step. Post-test probabilities were then calculated after each diagnostic step (clinical, CRP, PCR) and expressed in a pre- and post-test flow diagram.
Results
Of 142 children with pneumonia included in this study, from 3 months to 15 years old (median 3.15 years), 50 (35%) had CAP with consolidation and 92 (65%) had CAP without consolidation. Twenty-six (18%) children presented with complicated pneumonia, including 8 bacteremias, 10 empyemas, and 15 pleural effusions. In this cohort, SP was the only bacterium responsible for bacteremia (positive blood culture), pleural effusion, or empyema (positive pleural culture or PCR). Indeed, no staphylococcus, group A streptococcus, or haemophilus were detected.
A total of 44 patients were blood P-PCR positive (41 for WB-Ply, 16 for WB-LytA, and 13 for both), 8 were NP mycoplasma-PCR positive, and 51 were NP viral PCR positive. Some overlapping PCRs were found: 17 were pneumococcal and viral positive; 3 were pneumococcal and mycoplasma positive.
Table 1 shows the baseline characteristics of CAP with and without consolidation and of pneumonia with or without complications. Children presenting with CAP with consolidation or a complication were significantly older. No significant differences were noted in the distribution of other baseline variables.
The clinical model was built using the only three clinical signs that were significantly associated with CAP with consolidation: no wheezing (no need for bronchodilators), unilateral hypoventilation, and grunting. Having at least two of the three clinical signs was very significantly associated with pneumonia with consolidation (OR = 2.64 [95% CI 1.18–5.92], p < 0.018); thus, the model seemed appropriate for use as the first step in post-test estimation.
PCT and CRP values showed excellent p values (≤ 0.001) for distinguishing CAP with consolidation. A negative test for NP viral PCR was associated with CAP with consolidation (p = 0.012). Blood P-PCR only showed very strong p values (≤ 0.002) for discriminating between complicated and uncomplicated pneumonia, but not between CAP with or without consolidation. Even after controlling for potential confounders (age, day-care, tobacco, and vaccination), multivariate logistic regression showed that these significant associations between clinical and biological variables and CAP with consolidation remained significant.
A comparison of CRP and PCT values using specific microbiological PCR results (Table 2) showed significant differences. Children with positive pneumococcal WB-LytA presented very high median CRP and PCT values, significantly different to WB-LytA negative ones. Positive NP viral PCR was associated with lower median CRP values than negative NP viral cases. Positive mycoplasma NP PCR was associated with low PCT values, significantly different from negative mycoplasma NP PCR. In this study, PCT levels predicted Mycoplasma pneumoniae better than CRP did, and CRP levels distinguished viruses better than PCT did.
Performance testing of different markers (Table 3) showed that the clinical model worked very well for CAP with consolidation and complicated pneumonia (both specificities: 84%); however, it was not sensitive enough. The most sensitive biomarkers of pneumonia with consolidation were obtained by adding CRP and PCT at low cut-offs: the sensitivity of the clinical model with CRP was 92% and with PCT was 90%. Maximum specificity was found by combining high inflammatory markers (CRP or PCT) with positive pneumococcal PCR (WB-LytA). Adding a negative NP, viral PCR result increased its specificity even more.
Regarding biomarkers of complicated pneumonia (with bacteremia, pleural effusion, or empyema), maximal sensitivity (92.3%) was achieved using CRP or PCT (with the same low cut-offs). However, optimal specificity was achieved by associating high levels of these inflammatory markers with blood pneumococcal PCR (WB-LytA) (specificity 96.6%) or, for an even better result, also associating a negative NP viral PCR (specificity = 98.3%).
A model was constructed to associate clinical findings (two of three clinical signs) as a first step, adding CRP as a second step, and blood pneumococcal PCR as a third step. CRP was used instead of PCT due to its stronger association with the diagnosis of pneumonia (higher OR). Figure 1 shows how the post-test probability of complicated pneumonia increased from 18.3% (known prevalence = pre-test probability for this population: 26/142) to 40.6% with a positive clinical model result, to 52.0% with a CRP above 80 mg/L, and to a much stronger 85.7% if WB-LytA was positive. In contrast, it decreased to 11.8% if clinical signs were absent and to below 3% if CRP was <80 mg/L. The prediction of pneumonia with consolidation was also greatly improved by using these three steps, although with a lower CRP cut-off (40 mg/L) and using WB-Ply PCR, which was more sensitive than WB-LytA for the diagnosis of non-complicated pneumonia.
Post-test probability of typical lobar pneumonia or complicated pneumonia with the three-step assessment: (1) clinical model, (2) C-reactive protein >80 mg/L, and (3) whole-blood pneumococcal PCRs (PLY specific for typical lobar pneumonia, LYT-A specific for complicated pneumonia) based on the prevalence (pre-test probability). Abbreviations: LR likelihood ratio for a positive test (LR+) and a negative test (LR−). Impact of LR+ is shown by straight black lines; impact of LR− by dotted lines. Clinical model: positive = 2 out of 3 features are present; PLY+ and LYT-A+ positive pneumococcal whole-blood PCRs (Pneumolysin or Autolysin). *post-test probabilities are calculated for patients with complete laboratory workup
Discussion
In this study, we showed that a model combining negative clinical signs with low levels of inflammatory markers could identify children at a low risk of pneumonia with consolidation. Having two out of three clinical signs—unilateral hypoventilation, grunting, and an absence of wheezing—showed good specificity for identifying CAP with consolidation but was not very sensitive.
The lack of sensitivity of clinical signs has been emphasized previously [6, 9]. Thus, we evaluated a model combining clinical signs and inflammatory markers.
Recent studies demonstrated that these markers were useful for predicting the bacterial etiology of CAP [1, 18, 21, 25] and that PCT and CRP values were better than WBC count for differentiating bacterial from viral CAP [28]. Furthermore, in the present study, children with positive blood pneumococcal PCR or negative NP viral PCR had higher levels of inflammatory markers. We subsequently showed that adding CRP (cut-off 40 mg/L) as a second step to the model allowed us to rule out pneumonia with excellent sensitivity (92%) and with a negative predictive value of 87% for pneumonia with consolidation and 94% for complicated pneumonia.
Children with negative clinical signs and low CRP were at a low risk of pneumonia. The challenge in the setting of a consultation in an emergency department is the identification and treatment of children at a high risk of developing pneumonia with consolidation while minimizing unnecessary investigations, treatment, or hospital admissions for children with self-limiting viral respiratory illnesses. The negative predictive value of the rules developed here allowed us to identify which children, from among all those with fever and tachypnea, were at a low risk of pneumonia with consolidation and thus required neither a chest radiograph nor immediate antibiotic treatment. This rule is applicable within the time frame allotted for an emergency consultation as a clinical evaluation and CRP value are rapidly available. The present study confirmed that rules associating clinical signs with inflammatory markers could identify children with a low risk of pneumonia [40].
In order to more reliably rule in the diagnosis of pneumonia with consolidation, we combined clinical signs and CRP with a third step using blood pneumococcal PCR markers. The post-test probability of pneumonia with consolidation for a child with positive clinical signs, high CRP, and positive blood pneumococcal PLY PCR reached 77%, and the PPV was 72%. Different models for predicting pneumonia have been studied recently. Erdman et al. developed a model associating CRP and Chitinase 3-like-1 which could rule in radiological pneumonia with a lower PPV of 54% [19]. Elemraid et al. constructed a model based on age, CRP, and a neutrophil count with a 75% sensitivity and a 91% PPV for the diagnosis of radiologically confirmed pneumonia [18]. Finally, we previously developed a model using CRP or PCT, associated with a positive pneumococcal urinary antigen, which could predict pneumococcal pneumonia with a similar (75–83%) PPV [24]. These different models re-emphasize the central role that CRP or PCT have in the prediction of pneumonia.
Although identifying children at a high risk of pneumonia with consolidation is important, rapidly identifying children at risk of pneumonia with complications such as bacteremia, pleural effusion, or empyema is also necessary. These children require immediate management with investigation and often intravenous antibiotics and hospital admission. The three-step model presented here, involving clinical signs, high CRP, and positive blood P-PCR increases the post-test probability of complicated pneumonia from 52 to 87%. It is worth noting that the blood pneumococcal PCR was most useful in the group of children with high levels of inflammatory markers. This might be explained by the fact that the group of children with high levels of inflammatory markers probably had a high prevalence of pneumococcal pneumonia and, therefore, the influence of false-positive PCR results, due to nasopharyngeal colonization, may be reduced. Associating a negative NP virus PCR further increased the positive likelihood ratio to 22, allowing the identification of children at a high risk of complications.
The role of blood and NP PCR in patient management will probably evolve in the future. Previously, viral or pneumococcal PCR was a standard method for etiological diagnosis in research studies [13, 20, 36, 47]. However, since the development of rapid multiplex PCR systems that integrate sample preparation, amplification, detection, and analysis, such as FilmArray®, PCR may come to have a more significant impact on patient management than previously anticipated. With the need for just a few minutes of hands-on-time and results available in less than 1 h, multiplex PCR can work within the time frame of an emergency consultation, something that was impossible before. The results of pneumococcal and viral PCR examinations could thus be used rapidly in a decision-making model for patient therapy.
With regard to etiology, Mycoplasma pneumoniae PCR was positive in fewer than 6% of cases and only in children >5 years old. CRP did not differentiate between mycoplasma pneumonia and other types of pneumonia, as previously underscored by Medjo et al. [34]. With regard to PCT, mycoplasma pneumonia was associated with lower PCT values than non-mycoplasma pneumonia, as previously reported by Moulin et al. [37].
One of the main limitations of the present study, common to all bacterial pneumonia studies, is the lack of a gold standard etiological diagnosis. Radiologically confirmed pneumonia has been used as a proxy for bacterial pneumonia in numerous studies on epidemiological investigations [3], clinical trials of antimicrobial agents [26], and conjugate pneumococcal vaccine efficacy [44]. Recently, Nascimento-Carvalho et al. determined that pneumonia with alveolar consolidation had a 93% sensitivity for pneumococcal infection [38]. We therefore validated our diagnosis of pneumonia by radiologically confirming pneumonia with consolidation even though the limits to this method’s reliability are well known [16].
A second limitation is the relatively low median age of the study population, which could be a bias toward the viral etiology of infections as lower age is linked to viral etiology [18]. However, this age group does represent the typical pneumonia population encountered in a pediatric emergency department [16, 21].
Furthermore, the positive results of this three-step method, using clinical and laboratory markers, need to be externally validated with a different population before being proposed more widely for the management of children with pneumonia.
In conclusion, the risk of pneumonia can be assessed using the three-step workup described herein. Step one is to identify children with “positive clinical signs,” such as unilateral hypoventilation, grunting, and absence of wheezing. Step two is to measure CRP. For children with positive clinical signs and high inflammatory markers; step three could be measuring blood pneumococcal PCR in order to more effectively identifying children with pneumonia with consolidation or a complication. For children with negative markers, future studies proposing safety-netting with a clinical follow-up at 48 h, but no immediate antibiotic treatment or chest radiograph, should be scheduled.
This study adds information that may be useful in implementing current guidelines for the management of pneumonia. This could reduce consumption of antibiotics, decrease resistance, and save unnecessary investigations.
Abbreviations
- CAP:
-
Community-acquired pneumonia
- CI:
-
Confidence interval
- CRP:
-
C-reactive protein
- IQR:
-
Interquartile range
- LytA:
-
Autolysin A
- NP viral PCR:
-
Nasopharyngeal viral PCR
- OR:
-
Odds ratio
- PCR:
-
Polymerase chain reaction
- PCT:
-
Procalcitonin
- PCV-7:
-
Seven-valent pneumococcal conjugate vaccine
- Ply:
-
Pneumolysin
- P-PCR:
-
Pneumococcal (Streptococcus pneumoniae) PCR
- PPV:
-
Positive predictive value
- NPV:
-
Negative predictive value
- SP:
-
Streptococcus pneumoniae = S. pneumoniae = pneumococcus
- WBC:
-
White blood cell
- WB-LytA:
-
Whole-blood Autolysin A pneumococcal PCR
- WB-Ply:
-
Whole-blood Pneumolysin pneumococcal PCR
References
Berg AS, Inchley CS, Fjaerli HO, Leegaard TM, Lindbaek M, Nakstad B (2017) Clinical features and inflammatory markers in pediatric pneumonia: a prospective study. Eur J Pediatr. doi:10.1007/s00431-017-2887-y
Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, Jha P, Campbell H, Walker CF, Cibulskis R et al (2010) Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 375(9730):1969–1987
Cantais A, Mory O, Pillet S, Verhoeven PO, Bonneau J, Patural H, Pozzetto B (2014) Epidemiology and microbiological investigations of community-acquired pneumonia in children admitted at the emergency department of a university hospital. J Clin Virol 60(4):402–407
Cevey-Macherel M, Galetto-Lacour A, Gervaix A, Siegrist CA, Bille J, Bescher-Ninet B, Kaiser L, Krahenbuhl JD, Gehri M (2009) Etiology of community-acquired pneumonia in hospitalized children based on WHO clinical guidelines. Eur J Pediatr 168(12):1429–1436
Cherian T, Mulholland EK, Carlin JB, Ostensen H, Amin R, de Campo M, Greenberg D, Lagos R, Lucero M, Madhi SA et al (2005) Standardized interpretation of paediatric chest radiographs for the diagnosis of pneumonia in epidemiological studies. Bull World Health Organ 83(5):353–359
Clark JE (2015) Determining the microbiological cause of a chest infection. Arch Dis Child 100(2):193–197
Coon ER, Maloney CG, Shen MW (2015) Antibiotic and diagnostic discordance between ED physicians and hospitalists for pediatric respiratory illness. Hosp Pediatr 5(3):111–118
Corless CE, Guiver M, Borrow R, Edwards-Jones V, Fox AJ, Kaczmarski EB (2001) Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J Clin Microbiol 39(4):1553–1558
Craig JC, Williams GJ, Jones M, Codarini M, Macaskill P, Hayen A, Irwig L, Fitzgerald DA, Isaacs D, McCaskill M (2010) The accuracy of clinical symptoms and signs for the diagnosis of serious bacterial infection in young febrile children: prospective cohort study of 15 781 febrile illnesses. BMJ 340:c1594
Dagan R, Shriker O, Hazan I, Leibovitz E, Greenberg D, Schlaeffer F, Levy R (1998) Prospective study to determine clinical relevance of detection of pneumococcal DNA in sera of children by PCR. J Clin Microbiol 36(3):669–673
Dagan R, Bhutta ZA, de Quadros CA, Garau J, Klugman KP, Khuri-Bulos N, Levine O, Saha SK, Sow S, Were F et al (2010a) The remaining challenge of pneumonia: the leading killer of children. Pediatr Infect Dis J
Dagan R, Quadros CA, Garau J, Klugman KP, Khuri-Bulos N, Levine O, Sow S, Yang Y (2010b) Marking November 12, 2010—World Pneumonia Day: where are we, where are vaccines? Hum Vaccin 6(11):922–925
De Schutter I, Vergison A, Tuerlinckx D, Raes M, Smet J, Smeesters PR, Verhaegen J, Mascart F, Surmont F, Malfroot A (2014) Pneumococcal aetiology and serotype distribution in paediatric community-acquired pneumonia. PLoS One 9(2):e89013
Dominguez J, Gali N, Blanco S, Pedroso P, Prat C, Matas L, Ausina V (2001) Detection of Streptococcus pneumoniae antigen by a rapid immunochromatographic assay in urine samples. Chest 119(1):243–249
Dominguez J, Blanco S, Rodrigo C, Azuara M, Gali N, Mainou A, Esteve A, Castellvi A, Prat C, Matas L et al (2003) Usefulness of urinary antigen detection by an immunochromatographic test for diagnosis of pneumococcal pneumonia in children. J Clin Microbiol 41(5):2161–2163
Don M, Valent F, Korppi M, Canciani M (2009) Differentiation of bacterial and viral community-acquired pneumonia in children. Pediatrics international : official journal of the Japan Pediatric Society 51(1):91–96
Dowell SF, Garman RL, Liu G, Levine OS, Yang YH (2001) Evaluation of Binax NOW, an assay for the detection of pneumococcal antigen in urine samples, performed among pediatric patients. Clin Infect Dis 32(5):824–825
Elemraid MA, Rushton SP, Thomas MF, Spencer DA, Gennery AR, Clark JE (2014) Utility of inflammatory markers in predicting the aetiology of pneumonia in children. Diagn Microbiol Infect Dis 79(4):458–462
Erdman LK, D'Acremont V, Hayford K, Rajwans N, Kilowoko M, Kyungu E, Hongoa P, Alamo L, Streiner DL, Genton B et al (2015) Biomarkers of host response predict primary end-point radiological pneumonia in Tanzanian children with clinical pneumonia: a prospective cohort study. PLoS One 10(9). doi:10.1371/journal.pone.0137592
Esposito S, Marchese A, Tozzi AE, Rossi GA, Da Dalt L, Bona G, Pelucchi C, Schito GC, Principi N, Italian Pneumococcal CAPG (2012) Bacteremic pneumococcal community-acquired pneumonia in children less than 5 years of age in Italy. Pediatr Infect Dis J 31(7):705–710
Esposito S, Di Gangi M, Cardinale F, Baraldi E, Corsini I, Da Dalt L, Tovo PA, Correra A, Villani A, Sacco O et al (2016) Sensitivity and specificity of soluble triggering receptor expressed on myeloid cells-1, midregional proatrial natriuretic peptide and midregional proadrenomedullin for distinguishing etiology and to assess severity in community-acquired pneumonia. PLoS One 11(11):e0163262
Fagan TJ (1975) Letter: nomogram for Bayes theorem. N Engl J Med 293(5):257
Florin TA, Ambroggio L (2014) Biomarkers for community-acquired pneumonia in the emergency department. Curr Infect Dis Rep 16(12):451
Galetto-Lacour A, Alcoba G, Posfay-Barbe KM, Cevey-Macherel M, Gehri M, Ochs MM, Brookes RH, Siegrist CA, Gervaix A (2013a) Elevated inflammatory markers combined with positive pneumococcal urinary antigen are a good predictor of pneumococcal community acquired pneumonia in children. Pediatr Infect Dis J
Galetto-Lacour A, Alcoba G, Posfay-Barbe KM, Cevey-Macherel M, Gehri M, Ochs MM, Brookes RH, Siegrist CA, Gervaix A (2013b) Elevated inflammatory markers combined with positive pneumococcal urinary antigen are a good predictor of pneumococcal community-acquired pneumonia in children. Pediatr Infect Dis J 32(11):1175–1179
Greenberg D, Givon-Lavi N, Sadaka Y, Ben-Shimol S, Bar-Ziv J, Dagan R (2014) Short-course antibiotic treatment for community-acquired alveolar pneumonia in ambulatory children: a double-blind, randomized, placebo-controlled trial. Pediatr Infect Dis J 33(2):136–142
Harris M, Clark J, Coote N, Fletcher P, Harnden A, McKean M, Thomson A, British Thoracic Society Standards of Care C (2011) British Thoracic Society guidelines for the management of community acquired pneumonia in children: update 2011. Thorax 66(Suppl 2):ii1–i23
Hoshina T, Nanishi E, Kanno S, Nishio H, Kusuhara K, Hara T (2014) The utility of biomarkers in differentiating bacterial from non-bacterial lower respiratory tract infection in hospitalized children: difference of the diagnostic performance between acute pneumonia and bronchitis. J Infect Chemother 20(10):616–620
Jokinen C, Heiskanen L, Juvonen H, Kallinen S, Karkola K, Korppi M, Kurki S, Ronnberg PR, Seppa A, Soimakallio S et al (1993) Incidence of community-acquired pneumonia in the population of four municipalities in eastern Finland. Am J Epidemiol 137(9):977–988
Klugman KP, Madhi SA, Albrich WC (2008) Novel approaches to the identification of Streptococcus pneumoniae as the cause of community-acquired pneumonia. Clin Infect Dis 47(Suppl 3):S202–S206
Leyenaar JK, Lagu T, Shieh MS, Pekow PS, Lindenauer PK (2014) Variation in resource utilization for the management of uncomplicated community-acquired pneumonia across community and children’s hospitals. J Pediatr 165(3):585–591
McAvin JC, Reilly PA, Roudabush RM, Barnes WJ, Salmen A, Jackson GW, Beninga KK, Astorga A, McCleskey FK, Huff WB et al (2001) Sensitive and specific method for rapid identification of Streptococcus pneumoniae using real-time fluorescence PCR. J Clin Microbiol 39(10):3446–3451
McIntosh K (2002) Community-acquired pneumonia in children. N Engl J Med 346(6):429–437
Medjo B, Atanaskovic-Markovic M, Radic S, Nikolic D, Lukac M, Djukic S (2014) Mycoplasma pneumoniae as a causative agent of community-acquired pneumonia in children: clinical features and laboratory diagnosis. Ital J Pediatr 40:104
Michelow IC, Olsen K, Lozano J, Rollins NK, Duffy LB, Ziegler T, Kauppila J, Leinonen M, McCracken GH Jr (2004) Epidemiology and clinical characteristics of community-acquired pneumonia in hospitalized children. Pediatrics 113(4):701–707
Moisi JC, Makawa MS, Tall H, Agbenoko K, Njanpop-Lafourcade BM, Tamekloe S, Amidou M, Mueller JE, Gessner BD (2017) Burden of pneumococcal disease in northern Togo before the introduction of pneumococcal conjugate vaccine. PLoS One 12(1):e0170412
Moulin F, Raymond J, Lorrot M, Marc E, Coste J, Iniguez JL, Kalifa G, Bohuon C, Gendrel D (2001) Procalcitonin in children admitted to hospital with community acquired pneumonia. Arch Dis Child 84(4):332–336
Nascimento-Carvalho CM, Araujo-Neto CA, Ruuskanen O (2015) Association between bacterial infection and radiologically confirmed pneumonia among children. Pediatr Infect Dis J 34(5):490–493
O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T (2009) Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374(9693):893–902
Oostenbrink R, Thompson M, Lakhanpaul M, Steyerberg EW, Coad N, Moll HA (2013) Children with fever and cough at emergency care: diagnostic accuracy of a clinical model to identify children at low risk of pneumonia. Eur J Emerg Med 20(4):273–280
Posfay-Barbe KM, Galetto-Lacour A, Grillet S, Ochs MM, Brookes RH, Kraehenbuhl JD, Cevey-Macherel M, Gehri M, Gervaix A, Siegrist CA 2010 Immunity to pneumococcal surface proteins in children with community-acquired pneumonia: a distinct pattern of responses to pneumococcal choline-binding protein A. Clin Microbiol Infect
Raymond F, Carbonneau J, Boucher N, Robitaille L, Boisvert S, Wu WK, De Serres G, Boivin G, Corbeil J (2009) Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J Clin Microbiol 47(3):743–750
2002) British Thoracic Society guidelines for the management of community acquired pneumonia in childhood. Thorax 57(Suppl 1):i1–24
Tregnaghi MW, Saez-Llorens X, Lopez P, Abate H, Smith E, Posleman A, Calvo A, Wong D, Cortes-Barbosa C, Ceballos A et al (2014) Efficacy of pneumococcal nontypable Haemophilus influenzae protein D conjugate vaccine (PHiD-CV) in young Latin American children: a double-blind randomized controlled trial. PLoS Med 11(6):e1001657
Tsolia MN, Psarras S, Bossios A, Audi H, Paldanius M, Gourgiotis D, Kallergi K, Kafetzis DA, Constantopoulos A, Papadopoulos NG (2004) Etiology of community-acquired pneumonia in hospitalized school-age children: evidence for high prevalence of viral infections. Clin Infect Dis 39(5):681–686
Wallihan R, Ramilo O (2014) Community-acquired pneumonia in children: current challenges and future directions. J Inf Secur 69(Suppl 1):S87–S90
Williams DJ, Zhu Y, Grijalva CG, Self WH, Harrell FE, Jr., Reed C, Stockmann C, Arnold SR, Ampofo KK, Anderson EJ et al (2016) Predicting severe pneumonia outcomes in children. Pediatrics 138(4). doi:10.1542/peds.2016-1019
Acknowledgements
We declare that all the authors have seen and approved the final manuscript, contributed significantly to the work, and that the manuscript has neither been previously published nor is it being considered for publication elsewhere. GA and AGL had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. We would like to thank and acknowledge the extensive help of Ms. Florence Hugon for the organization of files, follow-up, transport and storage of samples, and for her fantastic energy.
Author information
Authors and Affiliations
Contributions
We declare that all the authors have contributed to this study. AG, AGL, KK, MG, RT, and GA participated in the design. AG, AGL, KK, MG, RT, VM, LL, and SM participated actively in implementing the study and collecting data. KK, VM, and GA contributed to the construction and cleaning of the database. GA performed most of the statistical analyses and wrote the initial manuscript. All authors participated in the revision of the manuscript and the design of figure and tables. AGL and AG actively supervised the whole process.
Corresponding author
Ethics declarations
Funding
This study was funded by the Geneva University Hospitals, Division of Pediatric Emergency Medicine (Professor Alain Gervaix).
Ethical approval
All the procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was obtained from all the participants’ parents, and the teenage participants themselves, before enrollment.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Communicated by Nicole Ritz
Rights and permissions
About this article
Cite this article
Alcoba, G., Keitel, K., Maspoli, V. et al. A three-step diagnosis of pediatric pneumonia at the emergency department using clinical predictors, C-reactive protein, and pneumococcal PCR. Eur J Pediatr 176, 815–824 (2017). https://doi.org/10.1007/s00431-017-2913-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00431-017-2913-0