Abstract
Chlamydiae are a large group of obligate endosymbionts of eukaryotes that includes the Chlamydiaceae family, comprising several animal pathogens. Among Chlamydiaceae, Chlamydia trachomatis causes widespread ocular and urogenital infections in humans. Like many bacterial pathogens, all Chlamydiae manipulate host cells by injecting them with type III secretion effector proteins. We previously characterized the C. trachomatis effector CteG, which localizes at the host cell Golgi and plasma membrane during distinct phases of the chlamydial infectious cycle. Here, we show that CteG is a Chlamydiaceae-specific effector with over 60 homologs phylogenetically categorized into two distinct clades (CteG I and CteG II) and exhibiting several inparalogs and outparalogs. Notably, cteG I homologs are syntenic to C. trachomatis cteG, whereas cteG II homologs are syntenic among themselves but not with C. trachomatis cteG. This indicates a complex evolution of cteG homologs, which is unique among C. trachomatis effectors, marked by numerous events of gene duplication and loss. Despite relatively modest sequence conservation, nearly all tested CteG I and CteG II proteins were identified as type III secretion substrates using Yersinia as a heterologous bacterial host. Moreover, most of the type III secreted CteG I and CteG II homologs were delivered by C. trachomatis into host cells, where they localized at the Golgi region and cell periphery. Overall, this provided insights into the evolution of bacterial effectors and revealed a Chlamydiaceae family of type III secreted proteins that underwent substantial divergence during evolution while conserving the capacity to localize at specific host cell compartments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlamydiae is a bacterial phylum containing obligate endosymbionts of eukaryotes, including unicellular protozoa and diverse animals [1, 2]. It contains species that have been isolated from various environments, the so-called environmental chlamydiae, which can be phylogenetically grouped into eight distinct families [2, 3]. In addition, the Chlamydiaceae family comprises the Chlamydia genus that includes human pathogens (C. trachomatis and C. pneumoniae) and several other established or proposed species that are pathogens of diverse animals and in some cases have been shown to have zoonotic potential [4]. Recently, among the Chlamydiaceae, the Chlamydiifrater genus was described, comprising two species isolated from flamingos [5]. Among Chlamydiae, C. trachomatis is the most studied species as it is a model obligate intracellular bacterial pathogen and causes trachoma and sexually transmitted diseases that affect millions of people worldwide [6].
Chlamydiae share a characteristic developmental cycle including the infectious, but non-replicative, elementary bodies (EBs) and the non-infectious, but replicative, reticulate bodies (RBs) [6, 7]. EBs invade host cells while forming a membrane-bound vacuolar compartment. Within this compartment, EBs differentiate into RBs, which multiply leading to the formation of a large vacuole, known as the inclusion. Eventually, RBs re-differentiate back into EBs. The cycle is completed by host cell egress of the infectious EBs, either by extrusion of the entire inclusion or by host cell lysis.
Another unifying feature of Chlamydiae is that they utilize a type III secretion (T3S) system to deliver effector proteins into host cells [6, 8,9,10,11,12]. Collectively, Chlamydiae effectors act on a wide range of eukaryotic cell processes at different stages of the developmental cycle to promote host cell invasion, immune evasion, chlamydial survival, proliferation, and egress [6, 8]. Inclusion membrane proteins (Incs) are a large group of Chlamydiae T3S effectors that share characteristic hydrophobic domains and insert in the chlamydial vacuolar membrane [12, 13]. Other Chlamydiae effectors have been shown to be dispersed in the host cell cytosol [14, 15], or to localize at the nucleus [15, 16], Golgi [17], lipid droplets [18], or plasma membrane [17, 19]. In C. trachomatis, about 60 effectors have been identified and many more are expected to exist [8]. Some of these effectors have been shown to be conserved among Chlamydiaceae [14,15,16, 20, 21], while others have been described to be specific to C. trachomatis or to a restricted number of Chlamydia species [21]. However, these searches for the presence of homologous effector genes in Chlamydiaceae only considered a few genes, and many were done before the disclosure of the currently known genome sequences.
We previously identified a C. trachomatis effector, named CteG, that does not show significant similarity to other proteins except putative homologues within Chlamydiaceae and localizes at the host cell Golgi and plasma membrane at distinct stages of infection [17]. More recently, we showed that CteG promotes host cell lytic exit of C. trachomatis by a yet unknown mechanism [22]. Furthermore, CteG binds the host cell centrosomal protein centrin-2 and promotes centrosomal duplication [23]. In a previous work, we identified putative CteG homologs among Chlamydiaceae by performing preliminary Position-Specific Iterated (PSI)-protein BLAST (PSI-BLAST) analyses [17]. In this work, we show that cteG gene has over 60 homologs in Chlamydiaceae, including several inparalogs and outparalogs, indicating a complex evolution unique among C. trachomatis effectors that is marked by several gene duplication and gene loss events. Although the vast majority of the CteG homologs show modest amino acid sequence similarity to C. trachomatis CteG, many of them are also T3S substrates, and can be delivered by C. trachomatis into host cells where they also localize at the Golgi region and cell periphery.
Materials and methods
Genome data
Publicly available assembled genomes and respective annotations for each Chlamydia or Chlamydiifrater species analysed (as of June 2023) were retrieved from the National Center for Biotechnology Information (NCBI).
The reference strains used for the analyses and their respective GenBank genome assembly accession numbers are the following: C. trachomatis serovar L2 strain 434/Bu (L2/434; GCA_000068585.1); C. abortus strain S26/3 (GCA_000026025.1); C. avium strain 10DC88 (GCA_000583875.1); C. buteonis str. IDL17-4553 (GCA_019056495.1); C. caviae strain GPIC (GCA_000007605.1); Candidatus C. corallus (referred as C. corallus) strain G3/2742 − 324 (GCA_002817655.1); C. crocodili str. 12 (GCA_018343815.1); C. felis strain Fe/C-56 (GCA_000009945.1); C. gallinacea strain 08-1274/3 (GCA_000471025.2); Candidatus C. ibidis [referred as C. ibidis] strain 10-1398 (GCA_000454725.1); C. muridarum strain Nigg (GCA_000006685.1); C. pecorum strain E58 (GCA_000204135.1); C. poikilothermis strain S15-834 K (GCA_900239975.1); C. pneumoniae strain CWL029 (GCA_000008745.1); C. psittaci strain 6BC (GCA_000204255.1); Candidatus C. sanzinia [referred as C. sanzinia] strain 2742 − 308 (GCA_001653975.1); C. serpentis strain H15-1957–10 C (GCA_900239945.1); C. suis strain MD56 (GCA_000493885.1); C. abortus strain 15-70d24 (GCA_900416725.2); Chlamydiifrater volucris sp. nov. (referred as C. volucris) strain 15-2067_O50 (GCA_902806995.1); Chlamydiifrater phoenicopteri sp. nov. (referred as C. phoenicopteri) strain 14-2711_R47 (GCA_902807005.1). Species of the Chlamydiaceae sister clade Chlamydiae Clade IV (CC-IV) [24] were also included: Chlamydiae bacterium isolate K940_chlam_9 (GCA_011064985.1), Chlamydiae bacterium isolate KR126_chlam_2 (GCA_011064935.1), Chlamydiae bacterium isolate K1000_chlam_4 (GCA_011065205.1). Closest related species Candidatus Protochlamydia naegleriophila strain KNic (GCA_001499655.1), Estrella lausannensis strain CRIB 30 (GCA_900000175.1) and Simkania negevensis strain Z (GCA_000237205.1) were used as outgroups.
Reciprocal tBLASTx screen
Putative cteG homologs were preliminarily searched by tBLASTx searches using the nucleotide sequence of cteG (ctl0360) from C. trachomatis L2/434 as query against the genome databases created for each chlamydial species. Genomic regions of each species encoding proteins showing similarities with proteins encoded by cteG were considered potential hits when e-values < 0.01. Then, the nucleotide sequence corresponding to 2 kbp upstream from the start and 2 kbp downstream from the end of these genomic regions was used for ab initio gene prediction with AUGUSTUS [25], using Staphylococcus aureus as the bacterial reference organism. The amino acid sequence of proteins potentially encoded by genes predicted at these regions were used in protein BLASTp searches against the NCBI non-redundant standard database of the corresponding Chlamydia or Chlamydiifrater species. Whenever the best hit protein in NCBI corresponded to the identity of the protein encoded by the query gene, it was assumed that the gene was present.
To verify putative homology, the nucleotide sequence of the putative cteG homolog genes found in the first search was used in a reciprocal tBLASTx against the genome sequence of C. trachomatis L2/434. If cteG appeared as the top hit with an e-value < 0.01, then the gene was considered a putative cteG homolog. A similar procedure was used to identify putative homologs of genes encoding other C. trachomatis effectors.
Phylogenetic analyses
To reconstruct the species tree, single copy orthologs (SCO) were retrieved using Orthofinder 2 [26] (-M msa -S blast -A mafft) from the predicted proteomes of the studied species. The resulting concatenated alignment produced by Orthofinder contained 198,493 amino acid positions that were subsequently used to infer a ML tree with IQ-TREE v1.6.11 [27] using an automatic detection of the best-fitting model of amino acid evolution and an ultrafast bootstrap method (-bb 1,000) [28]. Five independent tree searches were performed in total (--runs 5) and the tree with the highest likelihood score was considered the one representing the most likely phylogenetic relationships between species.
For the CteG phylogeny, putative CteG homologs were obtained through BLASTp searches against the local proteome databases using the CteG amino acid sequence from C. trachomatis L2/434 as query (AGJ64459.1; identical to CAP03800.1). A preliminary phylogenetic tree was constructed in IQ-TREE v. 1.6.11 [27] using all top BLASTp hits (e-value < 0.01). As putative CteG sequences were identified in Chlamydia species distantly related to C. trachomatis, the amino acid sequence of a putative CteG homolog from C. caviae (AAP05046.1) was subsequently used in a second BLASTp search. All top BLASTp hits obtained using both blast searches were analysed (e-value < 0.1), except for one sequence from Chlamydiifrater volucris (WP_213319067.1) which did not return cteG as top hit by reciprocal blast. Redundant sequences were removed with CD-HIT v4.6.7 [29]. The resulting sequences were aligned with MAFFT v. 7.407 [30] using the iterative refinement method L-INS-i (--localpair). Poorly aligned regions were removed with trimAl v1.2 using the “gappyout” option [31]. Phylogenies were constructed for both trimmed and not trimmed alignments. The Maximum Likelihood (ML) trees were inferred with IQ-TREE v1.6.11 [27] using an automatic detection of the best-fitting model of amino acid evolution and ultrafast bootstraps (-bb 1,000) [28]. A total of five runs (-runs 5) were conducted and the tree with the highest likelihood was selected.
Synteny analyses
Synteny conservation across Chlamydia and Chlamydiifrater species was done by first identifying genes in the neighbourhood of cteG homologs within Chlamydiaceae. This was done either manually using the available Genbank (NCBI) genome assemblies or by ab initio prediction using AUGUSTUS [25]. For each protein potentially encoded by the predicted genes, its identity to a putative C. trachomatis homolog was verified by top hit BLASTp and e-value < 0.001.
Escherichia coli and Yersinia enterocolitica strains and growth conditions
Escherichia coli NEB® 10β (New England Biolabs) was used for plasmid construction and purification, and E. coli ER2925 (New England Biolabs) was used to replicate and purify plasmids for transformation of C. trachomatis. Yersinia enterocolitica ΔHOPEMT (MRS40 pIML421 [yopHΔ1− 352, yopOΔ65− 558, yopP23, yopE21, yopM23, yopT135]), deficient for the Yersinia Yop T3S effectors H, O, P, E, M, and T, but T3S-proficient [32] and T3S-deficient Y. enterocolitica ΔHOPEMT ΔYscU (MRS40 pFA1001 yopHΔ1− 352, yopOΔ65− 558, yopP23, yopE21, yopM23, yopT135], yscUΔ1− 354) [33] were used for T3S assays. The yscU gene encodes an essential component of the Y. enterocolitica T3S system, and the yscUΔ1− 354 mutation is non-polar [34]. E. coli and Y. enterocolitica strains were grown in liquid or agar lysogeny broth (LB; NZYTech) with the appropriate selective antibiotics and supplements. E. coli and Y. enterocolitica cells were transformed with the plasmids by electroporation. T3S assays using Y. enterocolitica were done as previously described [34, 35].
Plasmids, DNA oligonucleotides, and DNA manipulations
The plasmids used in this work are listed in Table S1. The DNA oligonucleotides used in plasmid construction and in other molecular biology procedures are listed in Table S2. In general, plasmids were generated by cloning with restriction enzymes using standard molecular biology procedures. Briefly, DNA sequences were amplified with proof-reading Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific). DNA sequences and backbone plasmids were then digested with FastDigest restriction enzymes (Thermo Fisher Scientific) and ligated with T4 DNA ligase (Thermo Fisher Scientific). NZYTaq II DNA polymerase (NZYTech) was used for screening of positive clones. DNA fragments and plasmids were purified with DNA Clean & Concentrator-5TM kit (Zymo Research), ZymocleanTM Gel DNA recovery kit (Zymo Research), NZYMiniprep kit (NZYTech) or NZYMidiprep kit (NZYTech) following the manufacturer’s instructions. In general, pLJM3 [36] was used as vector to generate plasmids for protein production and analysis of T3S in Y. enterocolitica; plasmids pSVP246 [17] and pMC114 (generated in this work), both derivatives of p2TK2-SW2 [37], were used as vectors to generate plasmids for protein production and analysis of delivery into host cells by C. trachomatis. To generate plasmids encoding cteG homologs we used genomic DNA, kindly provided by Agathe Subtil (C. caviae and C. pneumoniae), Ian Clarke (C. muridarum), María Rosa Vergara (C. abortus), and Nicole Borel (C. pecorum and C. suis), as template for the PCR reactions. The accuracy of the nucleotide sequence of all the inserts in the constructed plasmids was checked by DNA sequencing (done at STAB VIDA).
Mammalian cell lines
HeLa 229 cells (from the European Collection of Authenticated Cell Culture; ECACC) were passaged in 4.5 g/L glucose, L-glutamine Dulbecco’s Modified Eagle’s Medium (DMEM; Corning) supplemented with heat-inactivated 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Scientific) at 37ºC in a humidified atmosphere of 5% (v/v) CO2 and detached from culture plates or flasks with TrypLE™ Express (Thermo Fisher Scientific). Cell cultures were regularly tested for Mycoplasma by conventional PCR as described [38].
C. trachomatis strains and growth conditions
The C. trachomatis strains used and generated in this work are listed in Table S3. They were propagated in HeLa 229 cells using standard procedures [39], and as described in our previous studies [17, 22]. C. trachomatis transformants were generated essentially as described by Agaisse and Derré (66), and as described in our previous studies [17, 40], except that newly generated C. trachomatis strains used for analysis of protein delivery into HeLa 229 cells were not plaque purified. Chlamydia stocks were tested for Mycoplasma by conventional PCR [38] and DNA sequencing techniques.
Infection of HeLa cells with C. trachomatis
Infection of HeLa cells with C. trachomatis for subsequent immunofluorescence and immunoblotting analyses were done as described previously [17, 22]. In experiments where HeLa 229 cells were infected with C. trachomatis strains carrying plasmids encoding genes under the control of the tetA tetracycline-inducible promoter (Table S3), anhydrotetracycline was added to 50 ng/ml at time zero of infection to process cells at 24 h post-infection (p.i), or at time zero and at 24 h p.i. to process cells at 46 h p.i.
Immunoblotting and immunofluorescence microscopy
Immunoblotting and immunofluorescence microscopy were performed as previously described [17, 22]. For immunoblotting the following antibodies were used: rat anti-HA (3F10; Roche; 1:1,000), mouse anti-chlamydial Hsp60 (A57-B9; Thermo Fisher Scientific; 1:1,000), mouse anti-α-tubulin (clone B-5-1-2; Sigma-Aldrich; 1:1,000), followed by anti-rat or anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare and Jackson ImmunoResearch; 1:10,000). For immunofluorescence microscopy the following antibodies and dyes were used: rat anti-HA (3F10; Roche; 1:200), rabbit anti-GM130 (Sigma Aldrich; 1:200), rabbit anti-Cap1 (kindly provided by Agathe Subtil; 1:200) [41], goat anti-Chlamydia major outer membrane protein (MOMP) (Abcam; 1:200), and goat anti-C. trachomatis FITC-conjugated antibody (Sigma-Aldrich, 1:150), followed by fluorophore-conjugated Rhodamine Red-X-conjugated anti-rat, AF488-conjugated anti-rabbit, DyLight 405-conjugated anti-goat antibodies (Jackson ImmunoResearch; 1:200), and staining with 4′,6-Diamidino-2-phenylindole (DAPI; 1:30,000).
Results
Preliminary screening of putative homologs of CteG and of other C. trachomatis effectors within Chlamydiaceae
To reanalyse the distribution of putative CteG homologs across Chlamydiaceae, we started by performing a preliminary tBLASTx analysis against a local genome database. The top hits were subsequently used in a reciprocal tBLASTx against C. trachomatis, aiming to further verify putative homology to cteG, as illustrated in Fig. 1A. This revealed the presence of 36 putative homologs of CteG (including C. trachomatis CteG) encoded within Chlamydiaceae genomes with some species presenting several copies (four in C. caviae, C. felis, and C. poikilothermis; three in C. pecorum; and two in C. buteonis, C. crocodili, C. ibidis, C. pneumoniae, C. psittaci, C. serpentis, and C. suis) and only two species (C. avium and C. gallinacea) showing no putative CteG homologs (Table S4). Using this strategy, putative homologs of CteG were not found in species from a sister lineage of the Chlamydiaceae (CC-IV; [24]) or in closely related species belonging to the Chlamydiales/Parachlamydiales orders (E. lausannensis, S. negevensis, Ca. P. naegleriophila), suggesting that the ancestral cteG gene appeared after the divergence of Chlamydiaceae from other Chlamydiae families and from sister lineage CC-IV.
CteG is the C. trachomatis effector with the higher relative number of putative homologs within Chlamydiaceae. (A) Graphical summary of the reciprocal tBLASTx approach to identify putative homologs of CteG within Chlamydiaceae. A similar procedure was used to identify putative homologs of non-Inc effectors and Incs. (B) The number of putative homologs found for each protein and in each Chlamydia or Chlamydiifrater species is coloured as indicated in the legend above the graph and depicted relative to the number of paralogs in C. trachomatis. The length of the coloured rectangles is proportional to the number of putative homologs in each species (e.g., only one homolog in CT222). See Table S4 for details
As previously described [17], amino acid similarity along the full-length of C. trachomatis CteG and its putative homologs was only observed in C. muridarum and C. suis (Fig. S1). All the other putative homologs of CteG only show sequence similarity to specific regions of CteG, ranging from amino acid residue 143 to the C-termini of CteG (Fig. S1). Furthermore, although C. buteonis HBN95_03995, C. caviae CCA_00297 and CCA_00298, C. crocodili H9Q19_03970, C. felis CF_0705 and CF_0706, C. poikilothermis C834K_0321 and C834K_0322, C. serpentis C10C_1043 were identified in the reciprocal BLAST analysis as putative homologs of CteG, they show a limited range of amino acid similarity (20 to 151 residues) and mostly display low sequence similarity (20–27% of identity; e-values from 0.013 to 3e− 6) to CteG (Fig. S1).
A procedure similar to what is illustrated in Fig. 1A was used to search for putative homologs within Chlamydiaceae of 54 additional C. trachomatis effectors, including 35 Incs experimentally shown to localize at the inclusion membrane (reviewed in [8] and references therein). Among the chlamydial effectors, most of the DUF582 proteins (CT619, CT620, CT621, CT711, CT712) [15] and the deubiquitinases Cdu1 and Cdu2 [42] led to the detection of different numbers of putative paralogs within C. trachomatis (Table S4). Therefore, for a visualization of how putative homologs of CteG and other C. trachomatis effectors are spread across Chlamydiaceae, the total number of putative homologs found in each case were plotted relative to the number of corresponding putative paralogs identified in C. trachomatis (Fig. 1B). Among the C. trachomatis effectors analysed, CteG displayed the higher relative number of putative homologs within Chlamydiaceae (Fig. 1B). By comparison to C. trachomatis, this is because of the existence of multiple CteG putative homologs in several Chlamydiaceae species, which is not seen to such an extent for any other C. trachomatis effector (Fig. 1B). This was intriguing and prompted us to analyse in further detail the evolutionary history of CteG, and if some of the putative CteG homologs are also T3S substrates delivered into host cells during infection.
Evolution of CteG in Chlamydiaceae
To deepen the study of the evolution of cteG among Chlamydiaceae species and closest relatives, and recover the highest number of homologs possible, we next inferred a species tree of the Chlamydiaceae (Fig. 2) and searched for all putative CteG homologs in a local proteome database, lowering the stringency of the BLASTp search (e-value < 0.1). The species tree was constructed using a phylogenomic approach, consisting of an alignment of 214 concatenated proteins obtained from Orthofinder (Fig. 2). The phylogenetic relationships among the Chlamydiaceae members is either consistent [43] or slightly differs from that described in previous studies [3, 24, 44]. Such variations could be attributed to the inclusion of more chlamydial species or to the use of different strategies for phylogenetic inference.
Phylogenomic tree of the Chlamydiaceae and closest related species of the Chlamydiae Clade IV (CC-IV) clade. The phylogeny was constructed with 214 single copy orthologs and rooted with S. negevensis, E. lausannensis and Ca. P. naegleriophila. Only bootstrap values below 100 are shown next to the respective nodes. Presence and absence of CteG homologs is illustrated as indicated next to each species name according to the local BLASTp analyses using the respective whole proteomes (see Fig. 3). Blue represents CteG belonging to clade I, red CteG belonging to clade II, and white means that CteG was absent, as also shown in Fig. 3. Original tree files and alignments can be found in Figshare: https://figshare.com/s/b5ee1bbacc3d9ec347fb
We next inferred the phylogenetic tree of CteG using all putative homologs recovered by the local BLASTp search (total of 63 sequences, including C. trachomatis CteG; Fig. 3 and Table S5). The phylogeny separated the CteG homologs in two main clades: CteG I (C. trachomatis CteG and 25 other proteins) and CteG II (37 proteins). Clade CteG I includes CteG from C. trachomatis and putative orthologs from closely related species, grouping essentially according to the species phylogeny (Figs. 2 and 3). However, and in line with our initial screen for CteG homologs (Fig. 1), several paralogs could be identified. These include inparalogs (e.g., in C. pecorum) and outparalogs (e.g., in C. cavie, C. crocodili or C. poikilothermis). This suggests that several cteG duplication events occurred at different time points during the evolution of Chlamydiaceae. Clade CteG II includes more distantly related homologs from Chlamydiaceae species, but not from C. trachomatis and closest relatives C. suis and C. muridarum. Interestingly, this clade also includes CteG homologs from distantly related Chlamydiifrater species, which are seemingly absent from CteG I clade. As in CteG I, in CteG II, the global relationships between species/clades follow those from the species tree (Figs. 2 and 3). However, inparalogs (C. volucris) and several outparalogs (e.g., in C. abortus, C. caviae, or C. poikilothermis) could also be inferred (Fig. 3).
Evolutionary history of CteG across Chlamydiaceae and closest relatives. CteG phylogeny comprising all top CteG BLASTp hits (e-value < 0.1). The two sequences used as queries for the BLASTp searches are highlighted in bold. Proteins selected for further analyses of protein secretion and delivery into host cells are highlighted by black filled squares. Phylogeny was midpoint rooted using iTOL v5 [71]. Only bootstrap values higher than 95 are shown as black dots next to the respective nodes. Phylogenies were visualized and rooted using iTOL [71]. Original tree files and alignments can be found in Figshare: https://figshare.com/s/b5ee1bbacc3d9ec347fb
There are differences in the number and identity of putative CteG homologs retrieved from the preliminary reciprocal tBLASTx approach (Fig. 1A and Table S4) and the homologs identified by the BLASTp and subsequent CteG phylogeny (Fig. 3 and Table S5). However, the expansion of cteG-related genes among Chlamydiaceae suggested by the initial screen revealed to be more significative with the more robust analysis of CteG phylogeny: 63 CteG homologs (including C. trachomatis CteG) instead of the 35 putative homologs initially found (Figs. 1 and 3). Only C. trachomatis and C. muridarum do not possess CteG paralogs encoded in their genomes. All other Chlamydiaceae species encode 2 (C. suis, C. corallus, C. avium, C. gallinacea, C. phoenicopteri), 3 (C. serpentis, C. sanzinia, C. abortus, C. ibidis, C. volucris), 4 (C. pneumoniae, C. pecorum, C. buteonis, C. psittaci), or 5 (C. crocodili, C. poikilotermis, C. caviae, C. felis) proteins evolutionarily related to C. trachomatis CteG. As cteG homologs show significant divergence among each other it is plausible that other putative cteG homologs were overlooked using our approach. However, we constructed an HMM profile for CteG using the 62 homologs (recovered by BLASTp), used a phmmer-based search [45] and did not find additional homologs. Overall, when considering the distribution (Fig. 2) and phylogenetic relationships (Fig. 3) of CteG proteins, it seems plausible that an ancient duplication in the most recent common ancestor (MRCA) of the Chlamydiaceae took place originating two distinct cteG ‘alleles’, followed by multiple gene duplication and differential loss events.
Synteny of cteG homologs
We next analysed gene organization in the vicinity of the cteG homologs. While the analysis was performed based on the genomic sequence of the C. trachomatis L2/434 strain, we used the nomenclature of the C. trachomatis serovar D UW3 strain (D/UW3). The genomic nucleotide sequences of the two strains are syntenic and 99.55% identical [46], and as strain D/UW3 corresponds to the first sequenced chlamydial genome [11] its gene nomenclature is often used as reference. The analysis revealed the existence of homologs that are syntenic to cteG (ct105 in C. trachomatis D/UW3) and that almost all localize in a locus encompassing C. trachomatis ct102 to ct107 homologous genes (Fig. 4). They all correspond to the CteG I clade in the phylogenetic tree of CteG, suggesting they are all cteG orthologs (Fig. 3). In addition, the genes encoding the CteG homologs corresponding to the CteG II clade in the phylogenetic tree of CteG (Fig. 3) are non-syntenic to C. trachomatis cteG (Fig. S2). However, they are all syntenic with each other occupying a locus flanked by homologs of C. trachomatis ct009-ct008-ct007 on one side (except C. corallus, C. pneumoniae, and C. serpentis where only homologs of ct009-ct008 were found), and tRNA-Ser and ct356-ct355-ct354 (most Chlamydia species) or tRNA-Ser and ct003-ct002-ct001 (C. serpentis, C. corallus, and C. pneumoniae) on the other side (Fig. S2). In the Chlamydiifrater species, the locus containing the CteG homologs also includes several pmp (polymorphic membrane protein) genes [47] and homologs of ct009, ct008, and ct007 with altered localization relative to C. trachomatis (Fig. S2). This locus is flanked by homologs of ct356-ct355-ct354 on one side and of ct004-ct003-ct002-ct001 on the other side (Fig. S2), thus showing further rearrangements relative to the syntenic locus in most Chlamydia species.
Genomic region of cteG and of each of its syntenic homologs in other Chlamydia and Chlamydiifrater species. cteG homologs identified by the analysis of CteG phylogeny depicted in Fig. 2 are coloured in red. Our preliminary search for putative CteG homologs (Fig. 1 and Table S4) retrieved a C. ibidis protein (H359_0450) that was not retrieved by the CteG phylogeny. The corresponding gene is indicated coloured in red and white stripes. Other genes are coloured as indicated within the figure. Genes for which no putative homologs were found are coloured in white. C. abortus cab377 was not identified as a putative homolog of cteG and is annotated in NCBI databases as a fragmented pseudogene. Genomic regions are depicted according to the species tree in Fig. 2A. The nomenclature in the legend for each group of putative homologs is from C. trachomatis D/UW3 strain [11]
In summary, the synteny of cteG homologs (from the CteG I and CteG II clades) within Chlamydiaceae further supports that they are evolutionarily related.
Most CteG homologs within Chlamydiaceae are T3S substrates
To analyse if the CteG homologs identified within Chlamydiaceae are also T3S substrates, we selected both CteG I and CteG II proteins and tested if they can be secreted as full-length proteins by Yersinia enterocolitica. Bacteria such as Shigella flexneri or Yersinia spp. with well-characterized T3S systems were used as heterologous systems to show that Chlamydia encodes T3S substrates [10, 12], and have been used to screen for putative chlamydial effectors [35, 48, 49] and to test if specific chlamydial proteins are T3S substrates [41, 50,51,52,53].
The CteG homologs analysed were from C. muridarum (AAF39239.1/TC_0381), C. suis (ESN89684.1/Q499_0113, ESN89662.1/Q499_0114), C. pneumoniae (AAD18548.1/Cpn_0404, AAD18549.1/Cpn_0405), C. abortus (CAH63829.1/CAB376), C. pecorum (AEB41680.1/G5S_0729, AEB41682.1/G5S_0731, AEB41684.1/G5S_0733) and C. caviae (AAP05046.1/CCA_00297, AAP05047.1/CCA_00298, AAP05136.1/CCA_00389, AAP05137.1/CCA_00390). This includes CteG I proteins (TC_0381, Q499_0113, which are closely related to CteG; and Q499_0114, Cpn_0404, Cpn0405, CAB376, G5S_0729, G5S_0731, G5S_0733, CCA_00389, CCA_00390, which by comparison to TC_0381 and Q499_0113 show less similarity to CteG) and CteG II proteins (CCA_00297, CCA_00298) (Fig. 3 and Fig. S1).
We first generated plasmids enabling gene expression from the Yersinia yopE T3S effector gene promoter of CteG homologs with a C-terminal haemagglutinin (HA) epitope tag. While generating the plasmids encoding C. suis Q499_0114-HA and C. pecorum G5S_0729-HA, we noted by DNA sequencing that the genes were truncated relative to what is annotated in the NCBI database. For C. suis Q499_0114 we generated two plasmids encoding two putative proteins that we named Q499_0114A (residues 1 to 283 of the annotated Q499_0114 sequence) and Q499_0114B (residues 314 to 529 of the annotated Q499_0114 sequence) while for C. pecorum G5S_0729 we only analysed the truncated protein (residues 1 to 120 and 274 to 337 of the annotated sequence).
The plasmids encoding HA-tagged CteG homologs were then separately introduced into T3S-proficient Y. enterocolitica (ΔHOPEMT), which lacks most Yersinia effectors [32, 33, 35]. As positive and negative controls in the secretion assays, we used Y. enterocolitica strains encoding CteG-HA and a HA-tagged chlamydial ribosomal protein (RplJ-HA), respectively. In general, the proteins migrated according to their predicted molecular mass (Fig. 5A and B). Many CteG homologs revealed multiple bands besides the band corresponding to the predicted molecular mass (Fig. 5A and B), a feature of unknown relevance that we consistently found in our studies involving CteG [17, 22, 35]. This revealed that 11 out of the 14 (∼ 80%) CteG homologs tested were secreted except for C. suis Q499_0114B and C. pecorum G5S_0731, which were not secreted, and for C. pecorum G5S_0733, where the results were unclear (Fig. 5A and B; summarized in Table 1). Then, the plasmids encoding the HA-tagged CteG homologs that were secreted (including the plasmid encoding G5S_0733) were individually introduced into T3S-deficient Y. enterocolitica ΔHOPEMT ΔYscU. Assays with the resulting strains confirmed that secretion of these CteG homologs was T3S-dependent (Fig. S3). In summary, these data suggest that, overall, the CteG homologs within Chlamydiaceae are also T3S substrates, even those more distantly related such as C. caviae CCA_00297 and CCA_00298.
Analysis of Type III secretion (T3S) of CteG homologs within Chlamydiaceae using Y. enterocolitica as heterologous host. (A) Type III secretion (T3S)-proficient Y. enterocolitica ΔHOPEMT was used to analyse secretion of CteG homologs within Chlamydiaceae with a C-terminal HA epitope tag. Immunoblots show the result of T3S assays in which proteins in culture supernatants (S, secreted proteins) and in bacterial pellets (P, non-secreted proteins) from ∼ 5 × 108 and ∼ 5 × 107 bacteria, respectively, were loaded per lane. CteG, a known C. trachomatis T3S substrate [35], was used as positive control, and the C. trachomatis ribosomal protein RplJ was used as a negative control [35]. The bands corresponding to the predicted molecular mass of the proteins analysed are indicated with a white asterisk (RplJ-HA, ∼ 19 kDa; CteG-HA, ∼ 68 kDa; Cpn_0404-HA, ∼ 37 kDa; Cpn_0405-HA, ∼ 26 kDa; TC_0381-HA, ∼ 71 kDa; CAB376-HA, ∼ 80 kDa; CCA_00389-HA, ∼ 79 kDa; CCA_00390-HA, ∼ 94 kDa; CCA_00297-HA, ∼ 49 kDa; CCA_00298-HA, ∼ 60 kDa; G5S_0729-HA, ∼ 19 kDa; G5S_0733-HA, ∼ 63 kDa; G5S_0731-HA, ∼ 38 kDa; Q499_0113-HA, ∼ 68 kDa; Q499_0114A-HA, ∼ 29 kDa; Q499_0114A-HA, ∼ 32 kDa). SycO is a strictly cytosolic Yersinia T3S chaperone [72] and its immunodetection ensured that the presence of HA-tagged proteins in the culture supernatants was not a result of bacterial lysis or contamination. (B) The percentage (%) of secretion of each protein by Y. enterocolitica ΔHOPEMT was calculated by densitometry of the bands in the immunoblots, as the ratio between the amount of secreted and total protein. Data are the mean ± standard error of the mean from at least 3 independent experiments
Many CteG homologs can be delivered into host cells by C. trachomatis
Next, we sought to test if the CteG homologs that are type III secreted by Y. enterocolitica, including G5S_0733, could be delivered by C. trachomatis into infected host cells. To avoid possible toxic effects in C. trachomatis, we initially generated plasmids where the expression of the genes encoding the CteG homologs was under control of the tetracycline-inducible tetA promoter. In all cases, the constructs were designed for the CteG homologs to have a C-terminal double (2HA) epitope tag. After individual introduction of the plasmids into CteG-deficient C. trachomatis (cteG::aadA), we successfully obtained transformants in eleven of the twelve cases. The exception was for the strain that should encode CCA_00390-2HA, and this protein was not further analysed.
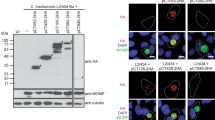
To confirm that the generated C. trachomatis strains were producing the desired proteins, HeLa cells were individually infected for 24 and 46 h with each of the eleven strains, and with a derivative of the cteG::aadA mutant harbouring a plasmid encoding CteG-2HA, also expressed from the tetA promoter [CteG-2HA(PtetA)]. Immunoblotting of whole cell extracts confirmed the production of Q499_0113-2HA, Cpn0404-2HA, Cpn0405-2HA, CAB376-2HA, G5S0729-2HA, G5S0733-2HA, CCA_00297-2HA, CCA_00298-2HA, and CCA_00389-2HA (Fig. S4). However, we could not detect production of TC_0381-2HA and Q499_0114A-2HA. Alternatively, we generated strains derived from the cteG::aadA mutant harbouring plasmids encoding these two proteins expressed from the cteG promoter. Immunoblotting from whole cell extracts of HeLa cells infected for 24 and 46 h with these two strains, and with a derivative of the cteG::aadA mutant harbouring a plasmid encoding CteG-2HA expressed from its own promoter [CteG-2HA(PcteG)], confirmed the production of both TC_0381-2HA and Q499_0114A-2HA (Fig. S5). As previously observed in Fig. 5A and B and in previous studies of C. trachomatis CteG [17, 22, 35], the detection by immunoblotting of the production of many of its homologs in Chlamydiaceae revealed multiple bands besides the band corresponding to the predicted molecular mass (Figs. S4 and S5).
To analyse if the various proteins were delivered by C. trachomatis into host cells, HeLa cells were infected for 24 and 46 h with each of the newly generated strains encoding homologs of CteG and also with the strains producing CteG-2HA(PtetA) and CteG-2HA(PcteG). We then immunolabelled the infected cells with antibodies against HA and Cap1 [a C. trachomatis effector known to localize at the inclusion membrane; [54]], followed by fluorophore-conjugated secondary antibodies and DAPI (to stain the DNA in the host cell nucleus and within chlamydiae). Analysis by fluorescence microscopy revealed that among the 11 proteins tested, ∼ 65%/7 (C. muridarum TC_0381, C. suis Q499_0113, C. abortus CAB376, C. pecorum G5S_0733, and C. caviae CCA_00297, CCA_00298, and CCA_00389) were delivered by C. trachomatis into host cells, while for ∼ 35%/4 (C. suis Q499_0114A, C. pneumoniae Cpn_0404 and Cpn_0405, and C. pecorum G5S_0729) their delivery into host cells by C. trachomatis was not observed (Fig. 6 and Fig. S6; summarized in Table 1). In a few cases (C. abortus CAB376, C. pecorum G5S_0733, and C. caviae CCA_00389), the proteins were only found to be delivered into host cells at 46 h p.i., but not at 24 h p.i. (Fig. 6 and Fig. S6; summarized in Table 1). The C. trachomatis strain producing C. caviae CCA_00298 showed a clear growth defect and at 46 h p.i. almost only small inclusions were observed (Fig. 6).
Delivery into host cells by C. trachomatis of CteG homologs in Chlamydiaceae. HeLa cells were infected for 46 h with C. trachomatis cteG::aadA harboring plasmids encoding CteG or CteG homologs within Chlamydiaceae (Q499_0113 and Q499_0114A, from C. suis; Cpn0404 and Cpn0405, from C. pneumoniae; CAB376 from C. abortus; CCA00389, CCA00297 and CCA00298, from C. caviae; G5S_0733 and G5S_0729, from C. pecorum; TC_0381, from C. muridarum) with a 2HA C-terminal epitope tag. The gene encoding CteG was expressed from its own promoter (PcteG) or from the tetA promoter (PtetA)) and the genes encoding its homologs within Chlamydiaceae were mostly expressed from PtetA, except for the genes encoding TC_0381 and Q499_0114A that were expressed from PcteG. Infected cells were fixed with 4% (w/v) paraformaldehyde and immunolabelled with antibodies against HA (red) and the inclusion membrane-localized protein Cap1 (green) and appropriate fluorophore-conjugated secondary antibodies. The host and chlamydial were also stained with DAPI (blue). The immunolabeled and stained cells were analysed by fluorescence microscopy. Scale bars, 5 μm
To compare the subcellular localization of the CteG homologs that were delivered into HeLa cells by C. trachomatis to the known localization of CteG at the Golgi (at about 24 h p.i.) and at the plasma membrane (at late infection times such as 46 h p.i.), HeLa cells infected for 24 and 46 h were immunolabelled with antibodies against HA, C. trachomatis MOMP, and the cis-Golgi protein GM130, followed by fluorophore-conjugated secondary antibodies. Analysis by fluorescence microscopy of cells infected for 24 h showed that CteG homologs that were delivered by C. trachomatis into host cells also localized at the Golgi region (C. muridarum TC_0381, C. suis Q499_0113, C. caviae CCA_00297, and C. caviae CCA_00298; Fig. 7) and partially at the cell periphery (C. suis Q499_0113, caviae CCA_00297, and C. caviae CCA_00298; Fig. 7 and Fig. S6), which is suggestive of a plasma membrane localization. At 46 h p.i., several CteG homologs clearly localized at the cell periphery (C. suis Q499_0113, C. caviae and CCA_00389, and C. abortus CAB376; Fig. 8A), at the Golgi region and partially at the cell periphery (C. muridarum TC_0381, C. caviae CCA_00297, and C. caviae CCA_00298; Fig. 8B), or only at the Golgi region (C. pecorum G5S_0733; Fig. 8C). The data on the subcellular localization of CteG homologs is summarized in Table 1.
Subcellular localization of CteG homologs in Chlamydiaceae delivered by C. trachomatis into host cells infected for 24 h. HeLa cells were infected for 24 h with C. trachomatis cteG::aadA harboring plasmids encoding CteG or CteG homologs within Chlamydiaceae (Q499_0133 from C. suis; CAB376 from C. abortus; CCA00389, CCA00297 and CCA00298, from C. caviae; G5S_0733 from C. pecorum; TC_0381, from C. muridarum) with a 2HA C-terminal epitope tag. The gene encoding CteG was expressed from its own promoter or from the tetA promoter (as indicated) and the genes encoding its homologs within Chlamydiaceae were mostly expressed from the tetA promoter, except for the gene encoding TC_0381 that was expressed from the cteG promoter. Infected cells were fixed with 4% (w/v) paraformaldehyde and immunolabelled with antibodies against HA (red), cis-Golgi protein GM130 (green) and C. trachomatis Major Outer Membrane Protein (MOMP; blue), and appropriate fluorophore-conjugated secondary antibodies. The immunolabeled cells were analysed by immunofluorescence microscopy. The indicated regions of overlap between the GM130/Golgi and HA/chlamydial protein immunofluorescence signals were magnified and are shown as zoomed images. Scale bars, 5 μm
Subcellular localization of CteG homologs in Chlamydiaceae delivered by C. trachomatis into host cells infected for 46 h. HeLa cells were infected for 46 h, as detailed in the legend of Fig. 7. The images illustrate localization of CteG and its homologs at the cell periphery (A), at the cell periphery and Golgi region (B), or solely at the Golgi region (C). In (B) and (C), the indicated regions of overlap between the GM130/Golgi and HA/chlamydial protein immunofluorescence signals were magnified and are shown as zoomed images
In summary, and in general, the experiments analysing secretion of CteG homologs by Y. enterocolitica and their delivery into host cells by C. trachomatis indicate that they are also T3S substrates, and many can be delivered into host cells by C. trachomatis and localize at the Golgi region and periphery of infected cells.
Discussion
In this work, we revealed that CteG shows a unique expansion within Chlamydiaceae, characterized by a total of at least 62 homologs, including several inparalogs and outparalogs in various Chlamydia and Chlamydiifrater species. Phylogenetically, the homologs of C. trachomatis CteG defined two clades that correlate with synteny of their encoding genes to C. trachomatis cteG (clade CteG I) or synteny between the encoding genes but not with C. trachomatis cteG (clade CteG II). In contrast, regardless of being CteG I or CteG II, almost all CteG homologs tested were type III secreted by Y. enterocolitica. Furthermore, most of the homologs that were type III secreted were also delivered by C. trachomatis into host cells where they localized at the Golgi region and/or at the cell periphery, suggesting a plasma membrane localization. Future studies should clarify if the identified homologs are functionally resemblant to C. trachomatis CteG regarding its known capacities to promote lytic exit from host cells [22] and to induce centrosome amplification [23], or even its putative ability to interfere with eukaryotic vesicular trafficking [17]. Interestingly, C. trachomatis CteG may have additional functions early in the chlamydial infectious cycle as cteG gene expression peaks between 1 and 2 h p.i. [17, 35, 55].
Based on the distribution, phylogeny and synteny analyses, a duplication event probably took place in an ancestor of Chlamydia species that originated two cteG-related paralogous genes in two separate loci. This is supported by the presence of CteG homologs from clade CteG II in Chlamydiifrater species. One of the paralogs was eventually lost in C. muridarum, C. trachomatis, and C. suis (CteG II, loci non-sytenic to cteG) and the other paralog in C. avium and C. gallinacea (CteG I, loci syntenic to cteG). There is also evidence for more recent within locus duplication events as several Chlamydia species show more than one cteG-related gene within a single locus (e.g. C. pecorum and Chlamydiifrater).
Gene duplication is a major mechanism in the evolution of eukaryotes [56], but it is thought to be less common in prokaryotes, where horizontal gene transfer has a more important role in driving protein diversification [57,58,59]. There are, however, examples of gene duplication in bacterial genomes [60, 61]. In general, if there is no selective advantage in maintaining a duplicated gene, then the gene is inactivated by mutation and is eventually deleted from the genome. When the duplicated gene is maintained, this is normally associated to a novel function (neofunctionalization), segregation of the functions of the ancestral gene (subfunctionalization), or to conservation of functions in both duplicates to provide redundancy and robustness to environmental challenges or if increased gene dosage is favourable [56]. At present, when considering the multiple functions (host cell lytic exit and centrosome duplication) described for C. trachomatis CteG [22, 23], as well as possible additional functions earlier in the infectious cycle [17, 35, 55], and the wide diversity of hosts of the Chlamydiaceae [43], all hypothetical scenarios that normally explain the maintenance of duplicated genes seem possible to explain the apparent importance of effective gene duplication in the evolution of CteG homologs.
There are two families of effectors that show paralogs in C. trachomatis: the DUF582 proteins (CT619, CT620, CT621, C711, and CT712) [15], which are transported to the host cell nucleus [15, 62] and target the eukaryotic ESCRT (endosomal sorting complexes required for transport) machinery [63], and the deubiquitinases Cdu1 and Cdu2 [42], which have been shown to limit the host response [64], to mediate Golgi fragmentation [65], and to control chlamydial exit by stabilizing other effectors [66]. It should be noted that in our preliminary tBLASTx search for putative homologs of different C. trachomatis effectors, the paralogs of CT619 in C. trachomatis were not detected, despite having been so in other Chlamydia and Chlamydiifrater species. Regardless of this lack of robustness of the initial reciprocal tBLASTx approach, it is very unlikely that the expansion of C. trachomatis effectors other than CteG within Chlamydiaceae would have not been detected. Differently from CteG, the duplication events that likely led to the five or four paralogs of DUF582 proteins present in Chlamydiaceae species probably occurred earlier in evolution, as they are present in all Chlamydia and Chlamydiifrater species. As for the deubiquitinases, it is known that there is a significant expansion of ubiquitination-related gene families in Chlamydiae [67]. However, homologs of Cdu1 are present in many, but not all, Chlamydiaceae [66], and the presence of both Cdu1 and Cdu2 is exclusive of C. trachomatis, C. muridarum and C. suis [66]. Overall, this suggests a recent duplication event that led to the emergence of Cdu2, and not a generalized expansion in Chlamydiaceae as observed for CteG.
Most CteG homologs are type III secreted by Y. enterocolitica and are delivered into host cells by C. trachomatis. The signals that direct proteins for T3S are normally located in their first 20–30 amino acids [68], and this seems to be the case of CteG [35]. Therefore, it could be puzzling that most C. trachomatis CteG homologs do not show significant sequence similarity in their N-terminal regions. However, while the exact nature of the T3S signal is still elusive, the amino sequence of the T3S signal can tolerate multiple changes, and a flexible and non-structured N-terminus region may play an important role in targeting substrates to the T3S machinery [69]. All this may explain why CteG homologs are almost all secreted by Yersinia and many delivered into host cells by C. trachomatis despite the lack of sequence similarity in their N-termini. [17]Not considering the cases of C. suis Q499-0114 and C. pecorum G5S_0729, whose annotated open-reading frames were disrupted in the strains used to amplify their gene DNA, for the putative homologs of CteG that were not type III secreted by Y. enterocolitica (C. pecorum G5S_0731) and/or delivered into host cells by C. trachomatis (C. pneumoniae Cpn_0404 and Cpn_0405), they may have evolved as cytosolic proteins of chlamydiae. Alternatively, their secretion by Y. enterocolitica and/or delivery into host cells by C. trachomatis could require specific targeting signals or uncharacterized endogenous factors, such as T3S chaperones [70], hypothetically present in the original species but not in Y. enterocolitica and/or C. trachomatis.
We previously showed that the first 100 amino acids residues of CteG function as a Golgi targeting region after ectopic expression in uninfected cells [17], However, the exact Golgi and plasma membrane targeting signals of C. trachomatis CteG in infected cells are presently unknown. As discussed above for the T3S signal, there is no significant sequence similarity in the N-terminal region of CteG (including the described Golgi targeting region; [17]) with its homologs and only modest similarity over the entire polypeptide sequence. Despite this, when CteG homologs are delivered by C. trachomatis into host cells, generally, they are also directed to the Golgi region and to the cell periphery of infected cells. This suggests that the Golgi and plasma membrane targeting signals of CteG should be conserved among its homologs, and further supports that they should also be effectors.
For most C. trachomatis effectors, the paralogy detected by the tBLASTx approach in C. trachomatis (i.e., no paralogs, or presence of one or more paralogous pairs) is either maintained across Chlamydiaceae species or putative homologs are only found in a few Chlamydiaceae species. This is particularly striking for the Incs analysed, with some seemingly only present in C. trachomatis and C. suis (CT222, CT224, CT225), in C. trachomatis and C. muridarum (CT227), or in C. trachomatis, C. suis and C. muridarum (IncA, IncD-G, CT134, CT135, CT192, CT226, CT228, CpoS, CT249, CT345, and InaC), while most of the others appear to be present in all (IncS, IncB, CT440, CT565, and CT850) or almost all Chlamydiaceae species (IncV, CT006, MrcA, IncC, IncM, CT383, CT442, CT483, and CT618). This contrasts with a previous study analysing the distribution of Incs in Chlamydiaceae that led to the identification of 23 “core Incs” present in all Chlamydia species [21]. However, the genomic sequences available at the time were limited to C. trachomatis, C. muridarum, C. caviae, and C. pneumoniae. From our analysis, such “core Incs” are limited to IncV, CT006, MrcA, IncS, IncB, CT383, CT440, CT442, CT565, and CT850, when considering those that have been experimentally detected in the inclusion membrane.
In conclusion, we showed that CteG homologs are present in all Chlamydiaceae and their evolution was likely driven by gene duplication, which is uncommon in bacteria. Despite modest sequence conservation, most CteG homologs are delivered into host cells where they localize in the Golgi region and cell periphery, thus suggesting that they are also effectors. Future additional functional studies on the CteG effector family may provide important insights on the molecular functions of C. trachomatis CteG, on how bacterial effector proteins evolved, and on the relevance of gene duplication in the evolution of obligate intracellular bacterial pathogens.
Data availability
Data is provided within the manuscript or supplementary information files. Additionally, original tree files and alignments can be found in Figshare: https://figshare.com/s/b5ee1bbacc3d9ec347fb.
References
Collingro A, Kostlbacher S, Horn M (2020) Chlamydiae in the Environment. Trends Microbiol. https://doi.org/10.1016/j.tim.2020.05.020. 28:877 – 88
Horn M (2008) Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol. https://doi.org/10.1146/annurev.micro.62.081307.162818. 62:113 – 31
Dharamshi JE, Kostlbacher S, Schon ME, Collingro A, Ettema TJG, Horn M (2023) Gene gain facilitated endosymbiotic evolution of Chlamydiae. Nat Microbiol 8:40–54. https://doi.org/10.1038/s41564-022-01284-9
Luu LDW, Kasimov V, Phillips S, Myers GSA, Jelocnik M (2023) Genome organization and genomics in Chlamydia: whole genome sequencing increases understanding of chlamydial virulence, evolution, and phylogeny. Front Cell Infect Microbiol 13:1178736. https://doi.org/10.3389/fcimb.2023.1178736
Vorimore F, Holzer M, Liebler-Tenorio EM, Barf LM, Delannoy S, Vittecoq M, Wedlarski R et al (2021) Evidence for the existence of a new genus Chlamydiifrater gen. nov. inside the family Chlamydiaceae with two new species isolated from flamingo (Phoenicopterus roseus): Chlamydiifrater phoenicopteri sp. nov. and chlamydiifrater volucris sp. nov. Syst Appl Microbiol 44:126200. https://doi.org/10.1016/j.syapm.2021.126200
Elwell C, Mirrashidi K, Engel J (2016) Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. https://doi.org/10.1038/nrmicro.2016.30
Abdelrahman YM, Belland RJ (2005) The chlamydial developmental cycle. FEMS Microbiol Rev. https://doi.org/10.1016/j.femsre.2005.03.002. 29:949 – 59
Bugalhão JN, Mota LJ (2019) The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg. Microb cell. https://doi.org/10.15698/mic2019.09.691. 6:414 – 49
Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P et al (2004) Illuminating the evolutionary history of chlamydiae. Science 304:728–730. https://doi.org/10.1126/science.1096330
Fields KA, Hackstadt T (2000) Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol 38:1048–1060. https://doi.org/10.1046/j.1365-2958.2000.02212.x
Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W et al (1998) Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. https://doi.org/10.1126/science.282.5389.754
Subtil A, Parsot C, Dautry-Varsat A (2001) Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol Microbiol 39:792–800. https://doi.org/10.1046/j.1365-2958.2001.02272.x
Rockey DD, Scidmore MA, Bannantine JP, Brown WJ (2002) Proteins in the chlamydial inclusion membrane. Microbes Infect. https://doi.org/10.1016/s1286-4579(02)01546-0. 4:333 – 40
Cosse MM, Barta ML, Fisher DJ, Oesterlin LK, Niragire B, Perrinet S, Millot GA, Hefty PS, Subtil A (2018) The loss of expression of a single type 3 Effector (CT622) strongly reduces Chlamydia trachomatis Infectivity and Growth. Frontiers in cellular and infection microbiology. 8:145. https://doi.org/10.3389/fcimb.2018.00145
Muschiol S, Boncompain G, Vromman F, Dehoux P, Normark S, Henriques-Normark B, Subtil A (2011) Identification of a family of effectors secreted by the type III secretion system that are conserved in pathogenic Chlamydiae. Infect Immun 79:571–580. https://doi.org/10.1128/IAI.00825-10
Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A (2010) Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog 6:e1000995. https://doi.org/10.1371/journal.ppat.1000995
Pais SV, Key CE, Borges V, Pereira IS, Gomes JP, Fisher DJ, Mota LJ (2019) CteG is a Chlamydia trachomatis effector protein that associates with the golgi complex of infected host cells. Sci Rep 9:6133. https://doi.org/10.1038/s41598-019-42647-3
Kumar Y, Cocchiaro J, Valdivia RH (2006) The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol 16:1646–1651. https://doi.org/10.1016/j.cub.2006.06.060
Wang X, Hybiske K, Stephens RS (2018) Direct visualization of the expression and localization of chlamydial effector proteins within infected host cells. Pathogens Disease 76. https://doi.org/10.1093/femspd/fty011
Lutter EI, Bonner C, Holland MJ, Suchland RJ, Stamm WE, Jewett TJ, McClarty G, Hackstadt T (2010) Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect Immun 78:3678–3688. https://doi.org/10.1128/IAI.00515-10
Lutter EI, Martens C, Hackstadt T (2012) Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp Funct Genomics 2012:362104. https://doi.org/10.1155/2012/362104
Pereira IS, Pais SV, Borges V, Borrego MJ, Gomes JP, Mota LJ (2022) The type III secretion Effector CteG mediates host cell lytic exit of Chlamydia trachomatis. Front Cell Infect Microbiol 12:902210. https://doi.org/10.3389/fcimb.2022.902210
Steiert B, Icardi CM, Faris R, McCaslin PN, Smith P, Klingelhutz AJ, Yau PM, Weber MM (2023) The Chlamydia trachomatis type III-secreted effector protein CteG induces centrosome amplification through interactions with centrin-2. Proc Natl Acad Sci USA 120:e2303487120. https://doi.org/10.1073/pnas.2303487120
Dharamshi JE, Tamarit D, Eme L, Stairs CW, Martijn J, Homa F, Jorgensen SL, Spang A, Ettema TJG (2020) Marine sediments illuminate Chlamydiae Diversity and Evolution. Curr Biol 30:1032–48e7. https://doi.org/10.1016/j.cub.2020.02.016
Keller O, Kollmar M, Stanke M, Waack S (2011) A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27:757–763. https://doi.org/10.1093/bioinformatics/btr010
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. https://doi.org/10.1186/s13059-019-1832-y
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular biology and evolution. 35:518 – 22. https://doi.org/10.1093/molbev/msx281
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Katoh K, Standley DM (2014) MAFFT: iterative refinement and additional methods. Methods Mol Biol. https://doi.org/10.1007/978-1-62703-646-7_8. 1079:131 – 46
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. https://doi.org/10.1093/bioinformatics/btp348. 25:1972-3
Iriarte M, Cornelis GR (1998) YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol Microbiol. https://doi.org/10.1046/j.1365-2958.1998.00992.x. 29:915 – 29
Almeida F, Borges V, Ferreira R, Borrego MJ, Gomes JP, Mota LJ (2012) Polymorphisms in Inc Proteins and Differential expression of inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of Lymphogranuloma Venereum isolates. J Bacteriol 194:6574–6585. https://doi.org/10.1128/JB.01428-12
Sorg I, Wagner S, Amstutz M, Muller SA, Broz P, Lussi Y, Engel A, Cornelis GR (2007) YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J 26:3015–3024. https://doi.org/10.1038/sj.emboj.7601731
da Cunha M, Milho C, Almeida F, Pais SV, Borges V, Mauricio R, Borrego MJ, Gomes JP, Mota LJ (2014) Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol 14:40. https://doi.org/10.1186/1471-2180-14-40
Marenne MN, Journet L, Mota LJ, Cornelis GR (2003) Genetic analysis of the formation of the Ysc-Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, YscF and YopN. Microb Pathog 35:243–258. https://doi.org/10.1016/s0882-4010(03)00154-2
Agaisse H, Derre I (2013) A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS ONE 8:e57090. https://doi.org/10.1371/journal.pone.0057090
Uphoff CC, Drexler HG (2011) Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Biol 731:93–103. https://doi.org/10.1007/978-1-61779-080-5_8
Scidmore MA (2005) Cultivation and laboratory maintenance of Chlamydia trachomatis. Curr Protoc Microbiol Chapter 11. https://doi.org/10.1002/9780471729259.mc11a01s00. Unit 11A 1
da Cunha M, Pais SV, Bugalhão JN, Mota LJ (2017) The Chlamydia trachomatis type III secretion substrates CT142, CT143, and CT144 are secreted into the lumen of the inclusion. PLoS ONE 12:e0178856. https://doi.org/10.1371/journal.pone.0178856
Gehre L, Gorgette O, Perrinet S, Prevost MC, Ducatez M, Giebel AM, Nelson DE, Ball SG, Subtil A (2016) Sequestration of host metabolism by an intracellular pathogen. eLife 5:e12552. https://doi.org/10.7554/eLife.12552
Misaghi S, Balsara ZR, Catic A, Spooner E, Ploegh HL, Starnbach MN (2006) Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol 61:142–150. https://doi.org/10.1111/j.1365-2958.2006.05199.x
Nunes A, Gomes JP (2014) Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infect Genet Evol 23:49–64. https://doi.org/10.1016/j.meegid.2014.01.029
Phillips S, Quigley BL, Timms P (2019) Seventy years of Chlamydia Vaccine Research - limitations of the past and directions for the future. Front Microbiol 10:70. https://doi.org/10.3389/fmicb.2019.00070
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763. https://doi.org/10.1093/bioinformatics/14.9.755
Thomson NR, Holden MT, Carder C, Lennard N, Lockey SJ, Marsh P, Skipp P et al (2008) Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res 18:161–171
Vasilevsky S, Stojanov M, Greub G, Baud D (2016) Chlamydial polymorphic membrane proteins: regulation, function and potential vaccine candidates. Virulence 7:11–22. https://doi.org/10.1080/21505594.2015.1111509
Subtil A, Delevoye C, Balana ME, Tastevin L, Perrinet S, Dautry-Varsat A (2005) A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol 56:1636–1647. https://doi.org/10.1111/j.1365-2958.2005.04647.x
Chellas-Gery B, Linton CN, Fields KA (2007) Human GCIP interacts with CT847, a novel Chlamydia trachomatis type III secretion substrate, and is degraded in a tissue-culture infection model. Cell Microbiol 9:2417–2430. https://doi.org/10.1111/j.1462-5822.2007.00970.x
Triboulet S, N’Gadjaga MD, Niragire B, Kostlbacher S, Horn M, Aimanianda V, Subtil A (2022) CT295 Is Chlamydia trachomatis’ Phosphoglucomutase and a Type 3 Secretion Substrate. Frontiers in cellular and infection microbiology. 12:866729. https://doi.org/10.3389/fcimb.2022.866729
Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T (2004) A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci USA 101:10166–10171. https://doi.org/10.1073/pnas.0402829101
Hower S, Wolf K, Fields KA (2009) Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol 72:1423–1437. https://doi.org/10.1111/j.1365-2958.2009.06732.x
Chen YS, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, Valdivia RH (2014) The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog 10:e1003954. https://doi.org/10.1371/journal.ppat.1003954
Fling SP, Sutherland RA, Steele LN, Hess B, D’Orazio SE, Maisonneuve J, Lampe MF, Probst P, Starnbach MN (2001) CD8 + T cells recognize an inclusion membrane-associated protein from the vacuolar pathogen Chlamydia trachomatis. Proc Natl Acad Sci USA 98:1160–1165. https://doi.org/10.1073/pnas.98.3.1160
Wurihan W, Wang Y, Yeung S, Zou Y, Lai Z, Fondell JD, Li WV, Zhong G, Fan H (2024) Expression activation of over 70% of Chlamydia trachomatis genes during the first hour of infection. Infect Immun 92:e0053923. https://doi.org/10.1128/iai.00539-23
Kuzmin E, Taylor JS, Boone C (2022) Retention of duplicated genes in evolution. Trends Genet 38:59–72. https://doi.org/10.1016/j.tig.2021.06.016
Tria FDK, Martin WF (2021) Gene duplications are at least 50 Times less frequent than Gene transfers in Prokaryotic genomes. Genome Biol Evol 13. https://doi.org/10.1093/gbe/evab224
Lerat E, Daubin V, Ochman H, Moran NA (2005) Evolutionary origins of genomic repertoires in bacteria. PLoS Biol 3:e130. https://doi.org/10.1371/journal.pbio.0030130
Treangen TJ, Rocha EP (2011) Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet 7:e1001284. https://doi.org/10.1371/journal.pgen.1001284
Sanchez-Herrero JF, Bernabeu M, Prieto A, Huttener M, Juarez A (2020) Gene Duplications in the genomes of Staphylococci and Enterococci. Front Mol Biosci 7:160. https://doi.org/10.3389/fmolb.2020.00160
Bernabeu M, Sanchez-Herrero JF, Huedo P, Prieto A, Huttener M, Rozas J, Juarez A (2019) Gene duplications in the E. Coli genome: common themes among pathotypes. BMC Genomics 20:313. https://doi.org/10.1186/s12864-019-5683-4
Hobolt-Pedersen AS, Christiansen G, Timmerman E, Gevaert K, Birkelund S (2009) Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol Med Microbiol 57:46–58. https://doi.org/10.1111/j.1574-695X.2009.00581.x
Vromman F, Perrinet S, Gehre L, Subtil A (2016) The DUF582 proteins of Chlamydia trachomatis bind to components of the ESCRT Machinery, which is dispensable for bacterial growth in vitro. Front Cell Infect Microbiol 6:123. https://doi.org/10.3389/fcimb.2016.00123
Fischer A, Harrison KS, Ramirez Y, Auer D, Chowdhury SR, Prusty BK, Sauer F et al (2017) Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. eLife 6. https://doi.org/10.7554/eLife.21465
Pruneda JN, Bastidas RJ, Bertsoulaki E, Swatek KN, Santhanam B, Clague MJ, Valdivia RH, Urbe S, Komander D (2018) A Chlamydia effector combining deubiquitination and acetylation activities induces golgi fragmentation. Nat Microbiol 3:1377–1384. https://doi.org/10.1038/s41564-018-0271-y
Bastidas RJ, Kedzior M, Davidson RK, Walsh SC, Dolat L, Sixt BS, Pruneda JN, Coers J, Valdivia RH (2023) The acetylase activity of Cdu1 regulates bacterial exit from infected cells by protecting Chlamydia effectors from degradation. https://doi.org/10.1101/2023.02.28.530337. bioRxiv
Domman D, Collingro A, Lagkouvardos I, Gehre L, Weinmaier T, Rattei T, Subtil A, Horn M (2014) Massive expansion of Ubiquitination-related gene families within the Chlamydiae. Molecular biology and evolution. 31:2890 – 904. https://doi.org/10.1093/molbev/msu227
Pais SV, Kim E, Wagner S (2023) Virulence-associated type III secretion systems in Gram-negative bacteria. Microbiol (Reading) 169. https://doi.org/10.1099/mic.0.001328
Galán JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature. https://doi.org/10.1038/nature05272. 444:567 – 73
Feldman MF, Cornelis GR (2003) The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol Lett 219:151–158. https://doi.org/10.1016/S0378-1097(03)00042-9
Letunic I, Bork P (2021) Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W6. https://doi.org/10.1093/nar/gkab301
Letzelter M, Sorg I, Mota LJ, Meyer S, Stalder J, Feldman M, Kuhn M, Callebaut I, Cornelis GR (2006) The discovery of SycO highlights a new function for type III secretion effector chaperones. Embo J 25:3223–3233. https://doi.org/10.1038/sj.emboj.7601202
Acknowledgements
We are grateful to Mariana Mendão for the construction of plasmids, and to our colleagues Agathe Subtil, Ian Clarke, María Rosa Vergara, and Nicole Borel for generously providing DNA of several Chlamydia species and antibodies.
Funding
This work was supported by Fundação para a Ciência e Tecnologia (FCT) through grant PTDC/BIA-MIC/28503/2017, and in the scope of the projects UIDP/04378/2020 and UIDB/04378/2020 of the Research Unit on Applied Molecular Biosciences – UCIBIO, and LA/P/0140/2020 of the Associate Laboratory Institute for Health and Bioeconomy - i4HB. ISP and MPL were supported by PhD fellowships SFRH/BD/129756/2017 and SFRH/BD/144284/2019, respectively, also funded by FCT.
Open access funding provided by FCT|FCCN (b-on).
Author information
Authors and Affiliations
Contributions
LJM, CG, and PG contributed to the study conception and design. Material preparation, data collection and analysis were performed by ISP, MdC, IL, MPL, CG, and LJM. The first draft of the manuscript was written by ISP and LJM. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pereira, I.S., da Cunha, M., Leal, I.P. et al. Identification of homologs of the Chlamydia trachomatis effector CteG reveals a family of Chlamydiaceae type III secreted proteins that can be delivered into host cells. Med Microbiol Immunol 213, 15 (2024). https://doi.org/10.1007/s00430-024-00798-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00430-024-00798-9