Abstract
Significant advances have been observed in the field of cell biology, with numerous studies exploring the molecular genetic pathways that have contributed to species evolution and disease development. The current study adds to the existing body of research evidence by reviewing information related to the role of leftover viruses and/or viral remnants in human physiology. To explore leftover viruses, their incorporation, and their roles in human physiology. The study entailed conducting a systematic search in the PsycINFO, PubMed, Web of Science, and CINAHL databases to locate articles related to the topic of investigation. The search terms included “leftovers,” “viruses,” “genome sequences,” “transposable elements,” “immune response,” and “evolution.” Additional articles were selected from the references of the studies identified in the electronic databases. Evidence showed that both retroviruses and nonretroviruses can be integrated into the human germline via various mechanisms. The role of leftover viruses in human physiology has been explored by studying the activation of human retroviral genes in the human placenta, RNA transfer between neurons through virus-like particles, and RNA transfer through extracellular vesicles. Research evidence suggested that leftover viruses play key roles in human physiology. A more complete understanding of the underlying pathways may provide an avenue for studying human evolution and allow researchers to determine the pathogenesis of some viral infections. Evidence obtained in this review shows that leftover viruses may be incorporated into the human genome. Retroviral genes are critical for the development of different parts of the body, such as the placenta in mammals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, researchers have confirmed the existence of over 5000 species of viruses, and efforts are ongoing to verify the discovery of approximately 320,000 other species (Katsura and Asai 2019). Research evidence shows that RNA viruses pose a major threat to human health by causing serious complications such as severe acute respiratory syndrome and Middle East respiratory syndrome (Noble 2012; Sharma 2013; Parrish and Tomonaga 2016). When humans are infected by viruses, the host immune system works to eliminate both the viruses and infected cells. In some instances, viruses enter the human genome. Apparently, 45% of the human genome consists of long interspersed nuclear elements and transposons (Katsura and Asai 2019). Approximately 5–8% of the transposable elements in the human genome are derived from viral sequences that resemble those of infective retroviruses (Katsura and Asai 2019).
Retroviruses and nonretroviral RNA and DNA form viral sequences that affect the genomes of various vertebrates. Retroviral genetic elements comprise sequences from human immune deficiency viruses (HIV), human T-lymphotropic viruses (HTLV), and simian foamy viruses (SFV) that can be integrated into the human germline and become heritable (Brochier-Armanet et al. 2012; Feschotte and Gilbert 2012; Papke and Gogarten 2012; Forterre 2012). On the other hand, nonretroviral RNA viruses include Bornavirus—a nonsegmented, negative-strand RNA virus with different designated endogenous elements, such as endogenous Borna-like N (EBLN) elements (Honda and Tomonaga 2016). Understanding retroviruses and nonretroviruses is clinically significant since the accumulation of endogenous viral sequences has influenced the current human genome (Katsura and Asai 2019; Miska and Ferguson-Smith 2016). This systematic review aims to examine the current knowledge of viral remnants in human physiology in terms of their function in the immune and nervous systems and the role of extracellular vesicles (EVs) in human physiology.
Methodology
The current study aimed to explore the topic of leftover viruses in human physiology and provide evidence on how different viruses and transposable features contribute to the development and function of the immune system. The study entailed analyzing the results of investigations related to the topic of interest as published in the PsycINFO, PubMed, Web of Science, and CINAHL databases. The search terms and phrases were “leftovers,” “viruses,” “genome sequences,” “transposable elements,” “immune response,” and “evolution,” and Boolean operators (AND/OR) were used to combine search terms to identify additional sources for inclusion in the systematic review. The search was limited to articles that were published in the four electronic databases between 2012 and 2019. The abstracts of the available articles were carefully reviewed to assess their quality and appropriateness (Fig. 1). The initial search yielded approximately 350 articles. However, after applying other parameters, including the study type, publication year, text options, search field tags, and language, the search was limited to 105 articles from the databases and 10 obtained through cross-referencing. These articles were carefully examined following the process illustrated in Fig. 1 to determine their suitability for this review, and after careful scrutinization, 90 articles were selected.
Results
After the search and review process, the final list of articles that were used in the systematic review consisted of prospective clinical trials, experimental studies, and clinical reviews. The 90 studies that met the inclusion criteria provided vital insights into the role of mobile genetic elements in the development and evolution of cellular immune pathways. A review of previous studies showed that virus-derived sequences and transposable elements constitute substantial portions of different cellular genomes (Griffith and Wagner 2017; Mess 2014; Carter 2012). Various investigations in the reviewed studies showed a strong evolutionary link between cellular genome sequences and diverse cellular immune pathways (Zhang et al. 2019; Grow et al. 2015; Dupressoir et al. 2012). At the basic level, some studies showed superinfection exclusion as a prototypical virus-mediated immune mechanism described for both prokaryotes and eukaryotes (Imakawa et al. 2015; Enders and Carter 2012; Franco de Oliveira et al. 2012). Complex immune response mechanisms may be partially or fully derived from diverse mobile genetic elements, such as CRISPR-Cas in prokaryotes and the RAG1/2 system in vertebrates (Carter et al. 2013; Ferner et al. 2014). These genetic features enable the creation of immunological memory of foreign genetics and facilitate the generation of antibodies and development of cell receptor diversity.
Viral leftovers and the activation of human retroviral genes in the placenta
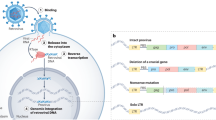
Some of the reviewed studies reported a link between leftover viruses and the activation of human retroviral genes in the placenta (Cornelis et al. 2017; Rezende et al. 2012). Other studies noted that the incorporation of retroviral genes in the placenta is important for its development, particularly in placental organisms (Chuong 2018; Gundling and Wildman 2015). Research showed that retroviral elements are usually expressed in the placenta. As a result, the expression of retroviruses in the placenta has evolved over time to allow viral exchange between the mother and fetus (Roberts et al. 2016; Armstrong et al. 2017). Based on these reports, some authors found that the placenta is a crucial site for the domestication of retroviral genes to help these genes serve an essential adaptive function in the host (Garratt et al. 2013; Schmidt et al. 2015; Chuong 2013; Jacques et al. 2013). Some of the effects linked to domesticated viruses include an enhanced immune response and changes in maternal physiology that benefit the fetus (Simonti et al. 2017; Pavlicev et al. 2015; Chuong et al. 2013; Ferreira et al. 2016). Some studies also reported evidence of the interplay and interaction between host defenses and retroviruses, which has been noted to contribute substantially to diversity in the mammalian placenta while facilitating different mechanisms of genomic imprinting (Katsura and Asai 2019; Parrish and Tomonaga 2016; Schroeder et al. 2013; Lavialle et al. 2013). Of particular interest for this review was the findings of some authors showing that infectious retroviruses have ribonucleic acid (RNA) genomes that can be reverse transcribed into double-stranded deoxyribonucleic acid (DNA) (Küry et al. 2018; Mager and Stoye 2015; Göke et al. 2015; Malik 2012). These viruses insert themselves into the host genome in the form of a provirus (Denner 2016; Furukawa et al. 2014), and once within the host genome, the retrovirus can replicate rapidly by transcribing new RNAs.
Retroviruses have been observed to possess a particular characteristic unlike that of any other virus. Rather than entering a host, replicating, and then leaving, these viruses penetrate the genome of the host, undergo replication, and gain heritable attributes, implying that they can be passed from one generation to the next (Dupressoir et al. 2011). With the help of viral genes, leftover viral DNA is copied and inserted at different locations in the host genome. Notably, these copies can be expressed in diverse parts of the body, where they use the available resources to develop new functions (Chamley et al. 2014; Esnault et al. 2013). Since viral proteins already have unique functions, it is easier for them to influence the development and evolution of different body parts, including the placenta. Oxygen and nutrient delivery are required for the fetus to mature inside the uterus of the mother. Furthermore, the fetus can survive only if waste is successfully removed and the fetal and maternal blood supplies remain separate. Some of the reviewed studies revealed that retroviruses in the genome are crucial for placental development (Malik 2012; Esnault et al. 2013; Zhuang et al. 2014; Vernochet et al. 2014). Additionally, the leftover viruses can help the embryo develop into a ball of cells in the womb, which is essential in the development of an infant. As a result, scientists have noted that leftover viruses and junk DNA are potent and important forces in the evolution of new species.
Malik stated that approximately 8% of the human genome comprises retroviruses at different phases of fossilization (Malik 2012). These retroviruses have directly infected the genome in germline tissue in a manner that facilitates the creation of imprints to be passed to genomes in other cells and subsequent generations. The majority of insertions into the host genome usually occur instantly or slowly through the accumulation of mutations (Malik 2012; Denner 2016; Vernochet et al. 2014). Although a significant portion of retroviral imprints degenerate as a result of evolution, some are preserved and can be activated through mutations. In such cases, the host genome takes up retroviral genes for its own use. The best-known genes linked to the domesticated retroviral envelope are syncytin genes, which are associated with embryo development in eutherian mammals. Cornelis et al. stated that the domestication of syncytin genes is an example of convergent evolution due to the cooption of retroviral genes for critical biological functions in reproductive systems and biology (Cornelis et al. 2017; Redelsperger et al. 2014). The domestication of syncytin genes may have occurred at least seven times during the evolution of mammals.
Other studies showed that leftover retroviral envelopes have been associated with immunosuppressive properties that protect fetal tissues from attack by the maternal immune system (Cornelis et al. 2017; Redelsperger et al. 2014). Nonetheless, the requirement for this functionality is unclear, although there is a link with placental development. Researchers have found three main types of placenta in eutherian mammals (Cornelis et al. 2017; Garcia-Etxebarria and Jugo 2013). The first is the hemochorial type, which develops in simian primates, rodents, and lagomorphs. Syncytin genes are thought to play a critical role in the development of the hemochorial aspects of the placenta. In addition, this type of placenta is characterized by direct exposure of the fetal chorion to maternal blood. The second is the endotheliochorial type that can be found in different mammals. These types of placentas have direct contact between the fetal chorion and the endothelial walls of the maternal blood vessels. Although researchers have linked placental development to seven different classes of syncytin genes, it is not clear whether these genes play a role in the endotheliochorial placenta (Malik 2012; Nakagawa et al. 2013; Cornelis et al. 2014). In other studies, a complete analysis of the genome sequence of animal samples led to the identification of Syncytin-Car1 as one gene that retained the features of retroviral envelopes and preserved them in orthologous locations (Malik 2012).
Some studies showed that although the biological requirements for endotheliochorial and hemochorial placentation differ, both processes involve syncytins (Malik 2012; Denner et al. 2012; Hummel et al. 2015). The last placentation type, known as epitheliochorial placentation, is found in other eutherian mammals, such as horses and pigs. This kind of placentation is different from the other types in the sense that it lacks syncytiotrophoblasts or syncytin-like genes. The results from phylogenetic reconstruction projects show that hemochorial placentation is the most invasive form and is ancestral to all mammalian species (Malik 2012). In this case, the placentation process is believed to have involved the domestication of syncytin genes linked to leftover retroviruses and the conservation of proviruses linked to the HERV-W and HERV-FRD groups (Malik 2012). The expression of proteins associated with conserved proviruses in placental cells is demonstrated by their fusogenic properties. Based on the results, researchers concluded that retroviruses that encode proteins such as syncytin-1 and 2 may have existed in the placental germline for 30–40 million years (Malik 2012; Hummel et al. 2015; Cornelis et al. 2015). Furthermore, the fact that the two genes linked to retroviruses have maintained a unique open reading frame, competing with the group-specific antigen (gag) and polymerase (pol) genes, may be an indication of the selection process that led to the retention of functional proteins in the placenta.
Activation of human retroviruses such as HIV and impact on the brain
Retroviruses have been observed in relation to the functionality of the placenta. In addition, other researchers have reported the role they play in the physiology of the nervous system, particularly the brain. HIV-associated central nervous system (CNS) disease has been reported to cause challenges among various patients, leading to opportunistic infections, neurocognitive disorders, immune reconstitution inflammatory syndrome (IRIS), collateral damage to the brain from combination antiretroviral therapy (cART), accelerated aging, and the associated impact of cerebrovascular disease (Clifford and Ances 2013; Cohen et al. 2015). Most of the disorders are caused by the induction of immune suppression arising from the pathogenic influence of HIV on the CNS (Ene 2018). This effect contributes to the reactivation of latent brain infection.
Evidence points out that the entry of HIV into the brain occurs in the early stages of acute infection with the virus (Ene 2018). The virus uses a “Trojan horse” model to reach brain cells, where it triggers the activation of astrocytes with the help of neurotoxic proteins, leading to increased production of glutamate. The effect on neurons occurs indirectly through elevated levels of extracellular glutamate, which cause neuronal bioenergetic disturbances and subsequent injury to neurons (Williams et al. 2014). Additionally, neuronal injury can be caused by the production of chemokines and cytokines due to the replication of the virus accompanied by systemic inflammation and microbial translocation.
HIV can seed in the brain and undergo evolution independent of the systemic compartment. Soon after infection, immune activation occurs despite the reduction in cerebrospinal fluid (CSF), HIV load, and inflammation. This impact is known as compartmentalization of the CSF, which occurs during primary infection with HIV and subsequent replication (Sturdevant et al. 2015). As a result, evidence has shown that numerous individuals who are HIV-positive have a particular degree of noncognitive impairment (Ene 2018). This indicates that the virus is found in CSF among patients with neurological diseases. However, there is still a dearth of research regarding patients with cognitive impairment due to opportunistic infections.
Nonretroviral RNA viral elements: bornaviruses (BDV/BoDV)
Conventionally, eukaryotic genomes comprise numerous virus-affiliated sequences primarily obtained from retroviruses. Research scientists have discovered that in addition to retroviruses, several vertebrate genomes encompass endogenous sequences obtained from DNA and RNA viruses of nonretroviral nature (Honda and Tomonaga 2016). A typical example is bornavirus, a negative-strand RNA virus that is nonsegmented (Kuhn et al. 2015). The virus has a monopartite single-stranded RNA genome with negative polarity that resembles that of the members of the order Mononegavirales (Kuhn et al. 2015). The virus is unique, as it is the only nonretroviral RNA virus with the ability to splice genes, and as a result, it cannot replicate the nucleus. Additionally, the alignment of the BDV gene and the sequences of proteins, as well as their regulatory signals in relation to the other mononegaviruses, show that BDV has never belonged to any of the established families of the order Mononegavirales, which includes Filoviridae, Paramyxoviridae, and Rhabdoviridae (Kuhn et al. 2015). The classification and nomenclature have remained unchanged throughout the editions of the naming reports.
Bornavirus is found in the human genome with endogenous elements. Most of the endogenous fragments of bornavirus in the human genome appear to have stemmed from reverse transcription and incorporation of nucleoprotein mRNA in ancient bornavirus from fossils that date back approximately 40 million years (Horie et al. 2010). As a result, the sequences noted have been coined endogenous bornavirus-like nucleoproteins (EBLNs), which signify evidence of an RNA-to-DNA flow of information between the host and virus via a novel mechanism or retrotransposon mediation. However, some viruses, such as Ebola virus (Filoviridae), leave endogenous genetic elements in mammalian genomes but not in the human genome (Belyi et al. 2010).
Notably, infections arising from bornavirus in humans have been the center of controversial scientific debates over the last three decades. The discourses emanate from the notion that Borna disease virus 1 (BoDV-1) was initially responsible for a fatal encephalitis and behavioral alterations observed in infected animals (Rubbenstroth et al. 2019; Schlottau et al. 2018). As a result, and because some viruses are zoonotic, early virologists and clinicians pointed out the possibility of bornavirus causing behavioral and psychiatric disorders among humans (Lipkin et al. 2011). A study by Rott et al. (1985) was the first to report that signs of persistent infection by BDV were identified in the sera of psychiatric patients suffering from major depressive disorders in the mid-1980s. With time, more sporadic cases of acute, nonpurulent, lethal BDV encephalitides have been observed in various parts of the world, such as Germany (Schlottau et al. 2018). Recently, Niller et al. (2020) continued to support the notion that BoDV-1 infection is a fatal zoonosis transmitted in endemic areas by the reservoir host known as the bicolored white-toothed shrew. However, the hypothesis is far-fetched, as the view has dismissed previous epidemiological studies, including the association of BoDV-1 with psychiatric disorders. On the other hand, Guo et al. (2020) compared sequences of human and animal BoDV-1 strains and through phylogenetic analysis, found that close sequence similarities occurred between different hosts and distant geographical regions, thus arguing against zoonotic concepts in narrowed endemic regions in Germany. BoDV-1 strains that infect human and mammalian hosts appeared to be very closely related. As a result, there are still many debates surrounding the zoonotic nature of BDV, thereby underscoring the need for more research to shed light on the human incidences of BDV infection, pathogenicity, and route of transmission, whether direct or indirect.
In addition to bornaviruses, other nonretroviral viruses from different families have contributed considerably to the evolutionary gene flow from virus to animal genomes and thus have a considerably yet underestimated role in coevolution (Katzourakis and Gifford 2010). Nonetheless, the mechanism of integration has remained unclear, although some researchers have mentioned infection of the host germ cells and integration into the host nuclear chromosome as the mechanisms of integration (Horie and Tomonaga 2011). Conversely, the integration of human EBLNs follows that of typical structures known as long interspersed element 1 (LINE-1). Coufal et al. (2010) showed evidence for de novo retrotransposition events through LINE-1 retrotransposons with increased copy numbers in the human brain (hippocampus). LINE-1 conversion of viral RNA into DNA is thought to be the likely mechanism by which bornaviruses replicate in the nucleus to integrate EBLNs into the genomes of humans and their primate ancestors (Honda and Tomonaga 2016; Horie et al. 2010). Consequently, the importance of long-term conservation, particularly of EBLNs, has been addressed based on the fact that 4 out of 7 human EBLNs encode intact open reading frames (ORFs) (Belyi et al. 2010). hsEBLN-1 encodes an almost complete ORF of the bornavirus N protein that has 41% identity with the viral N protein (Belyi et al. 2010). Additionally, transcribed and translated EBLNs likely play a protective role against exogenous BoDV-1 infection.
RNA transfer between neurons through virus-like particles
The role of leftover viruses in biological processes has also been explored by studying RNA transfer between neurons through virus-like particles. This mechanism is important because it provides a basis for understanding the concept of long-term learning (Moelling and Broecker 2019; Flores et al. 2014). Researchers have argued that neurons and immune systems during early evolution may have comprised viroids and viroid-like RNA (Flores et al. 2014; Legendre et al. 2014). In this context, viroids are considered virus-linked protein-free agents consisting of circular and highly structured noncoding RNAs that cause infections. In several viroids, RNA becomes catalytically active through the ribozyme activity pathway (Feschotte and Gilbert 2012; Legendre et al. 2014). Moreover, viroids contain remnants of ancient RNAs that are believed to have evolved prior to the evolution of both protein and DNA (Flores et al. 2014). Importantly, these exact types of viroids have been identified only in plants, indicating that their noncoding capability made them an important and universal cellular ancestor (Feschotte and Gilbert 2012; Legendre et al. 2014). Furthermore, viroids reveal the unique elements of ancient RNAs that eventually evolved to facilitate the development of immune systems and neuron networks (Feschotte and Gilbert 2012; Legendre et al. 2014). The realization that transcleaving derivatives and transcleaving ribozymes may exist in neuron networks and transfer processes highlights the possible role of leftover viruses and viroids in human development.
The brain has evolved to allow the transfer and storage of information via synaptic connections between neuron networks. Although the fundamental importance of the brain in regard to information storage has been highlighted, researchers do not yet understand the cellular and molecular processes related to its evolutionary origin (Feschotte and Gilbert 2012; Moelling 2012, 2013; Hayward 2017). Recent investigations have shown that eukaryotic genomes include DNA of transposon or retroviral origin (Moelling and Broecker 2019). An increasing volume of research evidence reveals that the coding sequences in eukaryotic genomic DNA provide important avenues for elucidating the development of new neuron functions and the evolution of brain regulatory elements (Moelling 2012; Hayward 2017). In vertebrates, researchers have identified virus-like elements and protein-coding genes that are linked to transposons and retroviruses (Moelling 2012, 2013; Hayward 2017). Most of these viral-derived genes are expressed in the brain, but their cellular and molecular functions remain largely unknown. One such element that has attracted research attention is the neuronal gene Arc, which has structural components that are also found in Gag polyproteins in retroviruses. Furthermore, polyproteins are believed to have evolved from the Ty3/gypsy retrotransposon family (Moelling and Broecker 2019; Pastuzyn and Shepherd 2017). Gag polyproteins and the Arc gene are important regulators that affect synaptic plasticity and are required for protein synthesis-linked long-term potentiation (LTP) (Cruz et al. 2018; Dodonova et al. 2019). Pastuzyn and Shepherd (2017) stated that the Arc gene can control synaptic plasticity via AMPA-type glutamate receptors (AMPARs). This particular endocytic pathway helps control neural activities and homeostasis through synaptic scaling.
RNA transfer through EVs
The role of leftover viruses in the development and evolution of biological systems was also highlighted by studies in which researchers noted similar RNA transfer from one cell to another through EVs, leading to the movement of messenger RNA (mRNA) and small interfering RNA (siRNA) (Di Liegro et al. 2017; Biller et al. 2014). The function of EVs in development highlights their important role in this process. According to Di Liegro et al. (2017), the release of EVs by various cells and their fusion with surrounding cells are ancient processes that form the basis for studying the role of leftover viruses in the development and evolution of biological systems. The release of EVs and their fusion with recipient cells are highly conserved processes in the evolution from bacteria to humans (Di Liegro et al. 2017). There is research evidence for the interspecies transfer of EVs from microorganisms to mammalian cells (Di Liegro et al. 2017; Biller et al. 2014; Mateescu et al. 2017). Moreover, EVs can carry various proteins, RNAs, and lipids to facilitate the horizontal transfer of biochemicals from one cell to another.
RNAs are the central protagonist of EV-mediated exchanges in the body. In particular, RNA–protein complexes have been linked to EV exchanges (Muraca et al. 2015; Iraci et al. 2016). Similar to retroviruses that bud from infected cells and infect neighboring cell populations, RNA–protein complexes can reach cells other than those in which they are initially produced. Once present in recipient cells, RNA–protein complexes modify gene expression in a number of ways. In recent years, researchers have strived to explore the function of the RNA components in such complexes (Muraca et al. 2015; Iraci et al. 2016; Fatima and Nawaz 2017; Li et al. 2015). The results have shown that EVs can transfer different classes of RNAs, such as mRNA, noncoding RNA (ncRNA), long noncoding RNA (lncRNA), circular RNA (circRNA), and microRNA (miRNA) (Iraci et al. 2016; Au Yeung et al. 2016). EVs are implicated in mRNA translation in recipient cells, thus facilitating the introduction of new proteins (Au Yeung et al. 2016; Morris and Mattick 2014; Nolte-’t Hoen et al. 2016). miRNAs, on the other hand, can target mRNA in recipient cells, decreasing their degradation or evoking the depletion of specific host proteins (Nolte-’t Hoen et al. 2016). circRNAs are considered sponges that can bind miRNAs and reduce their binding capacity. Furthermore, these RNAs act as scaffolds during the formation and localization of protein complexes that regulate gene transcription. The most important major findings are summarized in Table 1.
Discussion
Researchers have noted that mammalian genomes usually harbor diverse types of retrovirus sequences because of mutational decay and evolutionary processes. However, research indicates an important distinction between exogenous and endogenous retroviruses (Simonti et al. 2017; Pavlicev et al. 2015). Exogenous retroviruses are usually incorporated into the host germline and thus are found in all daughter cells within the infected host (Nakagawa et al. 2013). Endogenous retroviruses, in contrast, are transmitted from the parent to the child through gametes, and this transmission process does not adversely infect the new host cell (Katsura and Asai 2019; Parrish and Tomonaga 2016; Lavialle et al. 2013). Instead, these retroviruses jump from the mother to the offspring and vice versa to maintain somata that are distinct from infected germ cells. For instance, bornaviruses replicate in the nucleus to integrate EBLNs into the genomes of humans and their primate ancestors (Honda and Tomonaga 2016; Horie et al. 2010). Interestingly, there are some lineages of retroviruses with both endogenous and exogenous capabilities (Küry et al. 2018; Mager and Stoye 2015). In such instances, exogenous viruses may evolve into endogenous viruses by changing their properties and sequences once they enter germ cells (Göke et al. 2015; Malik 2012; Denner 2016). Nonetheless, in both exogenous and endogenous subpopulations, successful insertion into the host germline will result in the selection of elements that can replicate and move to other sites within the host (Furukawa et al. 2014). Thus, viruses undergo adaptation to improve transposition and facilitate survival in the new host environment, which makes them harmful organisms.
Leftover retroviruses have had a significant and persistent effect on animal genomes. In particular, approximately 8% of the human genome is believed to consist of retroviruses at diverse stages of fossilization. Furthermore, retroviruses directly affect genomes and germline tissues, and their imprints can be passed from one generation to the next (Biller et al. 2014; Mateescu et al. 2017). In most instances, genome insertions in the host tend to be deleterious, but in some cases, retroviruses have minimal effects on host biology, especially in the absence of an accumulation of mutations. The current review further reveals that a fraction of virus imprints show elements of mutational degeneration, which can be preserved in different generations when there is no mutational activation (Iraci et al. 2016; Au Yeung et al. 2016; Morris and Mattick 2014). In other studies, researchers have stated that retroviruses can use the microvesicle transport system to survive. Microvesicles can suppress or activate cellular responses to protect retroviruses (Hayward 2017; Biller et al. 2014). Understanding the effects of leftover viruses on the evolution and development of different biological systems will provide strategies for the development of strong antiviral medications. Furthermore, knowledge of the phenotypes and functions of different biological systems and tissues that have developed in response to viral domestication and inoculation may assist researchers in generating novel methods for enhancing the safety and efficacy of virus vaccines.
Conclusion
This review focused on exploring emerging evidence for the link between biological systems and specific genome sequences, transposable elements, and viruses. The data showed that retroviruses are an important part of the human genome. The link between leftover viruses and human physiology has been explored through the study of human nonretroviral and retroviral gene activation in the human placenta, RNA transfer between neurons through virus-like particles, and RNA transfer through EVs. Although these two forms of RNA viruses impact human physiology to varying degrees, research has shown that their mechanisms of action are balanced in terms of the aspects they address. Overall, a more complete understanding of these processes may help researchers and practitioners develop novel antiviral therapies. Further studies are required to gather additional information on the integration pathways to examine the link between retroviruses and the human genome.
Abbreviations
- AMPAR:
-
α-Amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor
- BDV/BoDV:
-
Borna disease virus
- BoDV-1:
-
Borna disease virus 1
- cART:
-
Combination antiretroviral therapy
- CINAHL:
-
Cumulative Index of Nursing and Allied Health Literature
- circRNA:
-
Circular RNA
- CNS:
-
Central nervous system
- CRISPR-Cas:
-
Clustered regularly interspaced short palindromic repeats
- CSF:
-
Cerebrospinal fluid
- DNA:
-
Deoxyribonucleic acid
- EBLNs:
-
Endogenous bornavirus-like nucleoproteins
- EVs:
-
Extracellular vesicles
- HERV-FRD:
-
Family of human endogenous retroviruses
- HERV-W:
-
Human endogenous retrovirus-W
- HIV:
-
Human immunodeficiency virus
- HTLV:
-
Human T-lymphotropic virus
- IRIS:
-
Immune reconstruction inflammatory syndrome
- lncRNA:
-
Long noncoding RNA
- LINE-1:
-
Long interspersed element 1
- LTP:
-
Long-term potentiation
- miRNA:
-
MicroRNA
- mRNA:
-
Messenger RNA
- ncRNA:
-
Noncoding RNA
- RAG 1/2:
-
Recombination-activating gene 1 and 2
- RNA:
-
Ribonucleic acid
- SFV:
-
Simian foamy virus
- siRNA:
-
Small interfering RNA
References
Armstrong DL, McGowen MR, Weckle A, Pantham P, Caravas J, Agnew D, Benirschke K, Savage-Rumbaugh S, Nevo E, Kim CJ, Wagner GP, Romero R, Wildman DE (2017) The core transcriptome of mammalian placentas and the divergence of expression with placental shape. Placenta 57:71–78
Au Yeung CL, Co N-N, Tsuruga T, Yeung T-L, Kwan S-Y, Leung CS, Li Y, Lu ES, Kwan K, Wong K-K, Schmandt R, Lu KH, Mok SC (2016) Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun 29(7):11150
Belyi VA, Levine AJ, Skalka AM (2010) Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog 6(7):e1001030
Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW (2014) Bacterial vesicles in marine ecosystems. Science 343(6167):183–186
Brochier-Armanet C, Gribaldo S, Forterre P (2012) Spotlight on the Thaumarchaeota. ISME J 6(2):227–230
Carter AM (2012) Evolution of placental function in mammals: the molecular basis of gas and nutrient transfer, hormone secretion, and immune responses. Physiol Rev 92(4):1543–1576
Carter AM, Enders AC, Jones CJP, Keovichit PK, Hugot JP (2013) A new form of rodent placentation in the relict species, Laonastes aenigmamus (Rodentia: Diatomyidae). Placenta 34(7):548–558
Chamley LW, Holland OJ, Chen Q, Viall CA, Stone PR, Abumaree M (2014) Review: where is the maternofetal interface? Placenta 35(Suppl):S74-80
Chuong EB (2013) Retroviruses facilitate the rapid evolution of the mammalian placenta. BioEssays News Rev Mol Cell Dev Biol 35(10):853–861
Chuong EB (2018) The placenta goes viral: retroviruses control gene expression in pregnancy. PLoS Biol 16(10):e3000028
Chuong EB, Rumi MAK, Soares MJ, Baker JC (2013) Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet 45(3):325–329
Clifford DB, Ances BM (2013) HIV-associated neurocognitive disorder. Lancet Infect Dis 13(11):976–986
Cohen RA, Seider TR, Navia B (2015) HIV effects on age-associated neurocognitive dysfunction: premature cognitive aging or neurodegenerative disease? Alzheimers Res Ther 7(1):37
Cornelis G, Vernochet C, Malicorne S, Souquere S, Tzika AC, Goodman SM, Catzeflis F, Robinson TJ, Milinkovitch MC, Pierron G, Heidmann O, Dupressoir A, Heidmann T (2014) Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc Natl Acad Sci USA 111(41):E4332-4341
Cornelis G, Vernochet C, Carradec Q, Souquere S, Mulot B, Catzeflis F, Nilsson MA, Menzies BR, Renfree MB, Pierron G, Zeller U, Heidmann O, Dupressoir A, Heidmann T (2015) Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci USA 112(5):E487-496
Cornelis G, Funk M, Vernochet C, Leal F, Tarazona OA, Meurice G, Heidmann O, Dupressoir A, Miralles A, Ramirez-Pinilla MP, Heidmann T (2017) An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc Natl Acad Sci USA 114(51):E10991–E11000
Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH (2010) L1 retrotransposition in human neural progenitor cells. Nature 460(7259):1127–1131
Cruz L, Romero JAA, Iglesia RP, Lopes MH (2018) Extracellular vesicles: decoding a new language for cellular communication in early embryonic development. Front Cell Dev Biol 6:94
Denner J (2016) Expression and function of endogenous retroviruses in the placenta. APMIS Acta Pathol Microbiol Immunol Scand 124(1–2):31–43
Denner J, Mihica D, Kaulitz D, Schmidt C-M (2012) Increased titers of neutralizing antibodies after immunization with both envelope proteins of the porcine endogenous retroviruses (PERVs). Virol J 5(9):260
Di Liegro CM, Schiera G, Di Liegro I (2017) Extracellular vesicle-associated RNA as a carrier of epigenetic information. Genes 8(10):240. https://doi.org/10.3390/genes8100240
Dodonova SO, Prinz S, Bilanchone V, Sandmeyer S, Briggs JAG (2019) Structure of the Ty3/Gypsy retrotransposon capsid and the evolution of retroviruses. Proc Natl Acad Sci USA 116(20):10048–10057
Dupressoir A, Vernochet C, Harper F, Guégan J, Dessen P, Pierron G, Heidmann T (2011) A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci USA 108(46):E1164-1173
Dupressoir A, Lavialle C, Heidmann T (2012) From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33(9):663–671
Enders AC, Carter AM (2012) Review: the evolving placenta: different developmental paths to a hemochorial relationship. Placenta 33(Suppl):S92-98
Ene L (2018) Human immunodeficiency virus in the brain-culprit or facilitator? Infect Dis 11:1178633717752687
Esnault C, Cornelis G, Heidmann O, Heidmann T (2013) Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet 9(3):e1003400
Fatima F, Nawaz M (2017) Vesiculated long non-coding RNAs: offshore packages deciphering trans-regulation between cells, cancer progression and resistance to therapies. Non-Coding RNA 3(1):10
Ferner K, Siniza S, Zeller U (2014) The placentation of eulipotyphla-reconstructing a morphotype of the Mammalian placenta. J Morphol 275(10):1122–1144
Ferreira LMR, Meissner TB, Mikkelsen TS, Mallard W, O’Donnell CW, Tilburgs T, Gomes HAB, Camahort R, Sherwood RI, Gifford DK, Rinn JL, Cowan CA, Strominger JL (2016) A distant trophoblast-specific enhancer controls HLA-G expression at the maternal-fetal interface. Proc Natl Acad Sci USA 113(19):5364–5369
Feschotte C, Gilbert C (2012) Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13(4):283–296
Flores R, Gago-Zachert S, Serra P, Sanjuán R, Elena SF (2014) Viroids: survivors from the RNA world? Ann Rev Microbiol 68:395–414
Forterre P (2012) Darwin’s goldmine is still open: variation and selection run the world. Front Cell Infect Microbiol 2:106
Franco de Oliveira M, Favaron PO, Ambrósio CE, Miglino MA, Mess AM (2012) Chorioallantoic and yolk sac placentation in Thrichomys laurentinus (Echimyidae) and the evolution of hystricognath rodents. J Exp Zoolog B Mol Dev Evol 318(1):13–25
Furukawa S, Kuroda Y, Sugiyama A (2014) A comparison of the histological structure of the placenta in experimental animals. J Toxicol Pathol 27(1):11–18
Garcia-Etxebarria K, Jugo BM (2013) Evolutionary history of bovine endogenous retroviruses in the Bovidae family. BMC Evol Biol 20(13):256
Garratt M, Gaillard J-M, Brooks RC, Lemaître J-F (2013) Diversification of the eutherian placenta is associated with changes in the pace of life. Proc Natl Acad Sci USA 110(19):7760–7765
Göke J, Lu X, Chan Y-S, Ng H-H, Ly L-H, Sachs F, Szczerbinska I (2015) Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16(2):135–141
Griffith OW, Wagner GP (2017) The placenta as a model for understanding the origin and evolution of vertebrate organs. Nat Ecol Evol 1(4):72
Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Pera RAR, Wysocka J (2015) Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522(7555):221–225
Gundling WE, Wildman DE (2015) A review of inter- and intraspecific variation in the eutherian placenta. Philos Trans R Soc Lond B Biol Sci 370(1663):20140072
Guo Y, He P, Sun L, Zhang X, Xu X, Tang T, Zhou W, Li Q, Zou D, Bode L, Xie P (2020) Full-length genomic sequencing and characterization of Borna disease virus 1 isolates: lessons in epidemiology. J Med Virol. https://doi.org/10.1002/jmv.25951
Hayward A (2017) Origin of the retroviruses: when, where, and how? Curr Opin Virol 25:23–27
Honda T, Tomonaga K (2016) Endogenous non-retroviral RNA virus elements evidence a novel type of antiviral immunity. Mob Genet Elem 6(3):e1165785
Horie M, Tomonaga K (2011) Non-retroviral fossils in vertebrate genomes. Viruses 3(10):1836–1848
Horie M, Honda T, Suzuki Y, Kobayashi Y, Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM, Tomonaga K (2010) Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 463(7277):84–87
Hummel J, Kämmerer U, Müller N, Avota E, Schneider-Schaulies S (2015) Human endogenous retrovirus envelope proteins target dendritic cells to suppress T-cell activation. Eur J Immunol 45(6):1748–1759
Imakawa K, Nakagawa S, Miyazawa T (2015) Baton pass hypothesis: successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes Cells Devoted Mol Cell Mech 20(10):771–788
Iraci N, Leonardi T, Gessler F, Vega B, Pluchino S (2016) Focus on extracellular vesicles: physiological role and signalling properties of extracellular membrane vesicles. Int J Mol Sci 17(2):171
Jacques P-É, Jeyakani J, Bourque G (2013) The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 9(5):e1003504
Katsura Y, Asai S (2019) Evolutionary medicine of retroviruses in the human genome. Am J Med Sci 358(6):384–388
Katzourakis A, Gifford RJ (2010) Endogenous viral elements in animal genomes. PLoS Genet 6(11):e1001191
Kuhn JH, Dürrwald R, Bào Y, Briese T, Carbone K, Clawson AN, deRisi JL, Garten W, Jahrling PB, Kolodziejek J, Rubbenstroth D, Schwemmle M, Stenglein M, Tomonaga K, Weissenböck H, Nowotny N (2015) Taxonomic reorganization of the family Bornaviridae. Arch Virol 160(2):621–632
Küry P, Nath A, Créange A, Dolei A, Marche P, Gold J, Giovannoni G, Hartung H-P, Perron H (2018) Human endogenous retroviruses in neurological diseases. Trends Mol Med 24(4):379–394
Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T (2013) Paleovirology of “syncytins”, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci 368(1626):20120507
Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Couté Y, Rivkina E, Abergel C, Claverie J-M (2014) Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci USA 111(11):4274–4279
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25(8):981–984
Lipkin WI, Briese T, Hornig M (2011) Borna disease virus—fact and fantasy. Virus Res 162(1–2):162–172
Mager DL, Stoye JP (2015) Mammalian endogenous retroviruses. Microbiol Spectr 3(1):MDNA3-0009–2014
Malik HS (2012) Retroviruses push the envelope for mammalian placentation. Proc Natl Acad Sci USA 109(7):2184–2185
Mateescu B, Kowal EJK, van Balkom BWM, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FWN, Das S et al (2017) Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—an ISEV position paper. J Extracell Vesicles 6(1):1286095
Mess A (2014) Placental evolution within the supraordinal clades of eutheria with the perspective of alternative animal models for human placentation. Adv Biol 2014:1–21
Miska EA, Ferguson-Smith AC (2016) Transgenerational inheritance: models and mechanisms of non-DNA sequence-based inheritance. Science 354(6308):59–63
Moelling K (2012) Are viruses our oldest ancestors? EMBO Rep 13(12):1033
Moelling K (2013) What contemporary viruses tell us about evolution: a personal view. Arch Virol 158(9):1833–1848
Moelling K, Broecker F (2019) Viruses and evolution—viruses first? A personal perspective. Front Microbiol 10:523
Morris KV, Mattick JS (2014) The rise of regulatory RNA. Nat Rev Genet 15(6):423–437
Muraca M, Putignani L, Fierabracci A, Teti A, Perilongo G (2015) Gut microbiota-derived outer membrane vesicles: under-recognized major players in health and disease? Discov Med 19(106):343–348
Nakagawa S, Bai H, Sakurai T, Nakaya Y, Konno T, Miyazawa T, Gojobori T, Imakawa K (2013) Dynamic evolution of endogenous retrovirus-derived genes expressed in bovine conceptuses during the period of placentation. Genome Biol Evol 5(2):296–306
Niller HH, Angstwurm K, Rubbenstroth D, Schlottau K, Ebinger A, Giese S, Wunderlich S, Banas B, Forth LF et al (2020) Zoonotic spillover infections with Borna disease virus 1 leading to fatal human encephalitis, 1999–2019: an epidemiological investigation. Lancet Infect Dis 20(4):467–477
Noble D (2012) A theory of biological relativity: no privileged level of causation. Interface Focus 2(1):55–64
Nolte-‘t Hoen E, Cremer T, Gallo RC, Margolis LB (2016) Extracellular vesicles and viruses: are they close relatives? Proc Natl Acad Sci USA 113(33):9155–9161
Papke RT, Gogarten JP (2012) Ecology. How bacterial lineages emerge. Science 336(6077):45–46
Parrish NF, Tomonaga K (2016) Endogenized viral sequences in mammals. Curr Opin Microbiol 31:176–183
Pastuzyn ED, Shepherd JD (2017) Activity-dependent arc expression and homeostatic synaptic plasticity are altered in neurons from a mouse model of angelman syndrome. Front Mol Neurosci 10:234
Pavlicev M, Hiratsuka K, Swaggart KA, Dunn C, Muglia L (2015) Detecting endogenous retrovirus-driven tissue-specific gene transcription. Genome Biol Evol 7(4):1082–1097
Redelsperger F, Cornelis G, Vernochet C, Tennant BC, Catzeflis F, Mulot B, Heidmann O, Heidmann T, Dupressoir A (2014) Capture of syncytin-Mar1, a fusogenic endogenous retroviral envelope gene involved in placentation in the Rodentia squirrel-related clade. J Virol 88(14):7915–7928
Rezende LC, Barbeito CG, Favaron PO, Mess A, Miglino MA (2012) The fetomaternal interface in the placenta of three species of armadillos (Eutheria, Xenarthra, Dasypodidae). Reprod Biol Endocrinol RBE 4(10):38
Roberts RM, Green JA, Schulz LC (2016) The evolution of the placenta. Reprod Camb Engl 152(5):R179-189
Rott R, Herzog S, Fleischer B, Winokur A, Amsterdam J, Dyson W, Koprowski H (1985) Detection of serum antibodies to Borna disease virus in patients with psychiatric disorders. Science 228(4700):755–756
Rubbenstroth D, Schlottau K, Schwemmle M, Rissland J, Beer M (2019) Human bornavirus research: Back on track! PLoS Pathog 15(8):e1007873
Schlottau K, Forth L, Angstwurm K, Höper D, Zecher D, Liesche F, Hoffmann B, Kegel V, Seehofer D, Platen S, Salzberger B, Liebert UG, Niller H-H, Schmidt B, Matiasek K, Riemenschneider MJ, Brochhausen C, Banas B, Renders L, Moog P, Wunderlich S, Seifert CL, Barreiros A, Rahmel A, Weiss J, Tappe D, Herden C, Schmidt-Chanasit J, Schwemmle M, Rubbenstroth D, Schlegel J, Pietsch C, Hoffmann D, Jantsch J, Beer M (2018) Fatal encephalitic Borna disease virus 1 in solid-organ transplant recipients. N Engl J Med 379(14):1377–1379
Schmidt A, Morales-Prieto DM, Pastuschek J, Fröhlich K, Markert UR (2015) Only humans have human placentas: molecular differences between mice and humans. J Reprod Immunol 108:65–71
Schroeder DI, Blair JD, Lott P, Yu HOK, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, LaSalle JM (2013) The human placenta methylome. Proc Natl Acad Sci USA 110(15):6037–6042
Sharma A (2013) Transgenerational epigenetic inheritance: focus on soma to germline information transfer. Prog Biophys Mol Biol 113(3):439–446
Simonti CN, Pavlicev M, Capra JA (2017) Transposable element exaptation into regulatory regions is rare, influenced by evolutionary age, and subject to pleiotropic constraints. Mol Biol Evol 34(11):2856–2869
Sturdevant CB, Joseph SB, Schnell G, Price RW, Swanstrom R, Spudich S (2015) Compartmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection. PLoS Pathog 11(3):e1004720
Vernochet C, Redelsperger F, Harper F, Souquere S, Catzeflis F, Pierron G, Nevo E, Heidmann T, Dupressoir A (2014) The captured retroviral envelope syncytin-A and syncytin-B genes are conserved in the Spalacidae together with hemotrichorial placentation. Biol Reprod 91(6):148
Williams DW, Veenstra M, Gaskill PJ, Morgello S, Calderon TM, Berman JW (2014) Monocytes mediate HIV neuropathogenesis: mechanisms that contribute to HIV associated neurocognitive disorders. Curr HIV Res 12(2):85–96
Zhang X, Pavlicev M, Jones HN, Muglia LJ (2019) Eutherian-specific gene TRIML2 attenuates inflammation in the evolution of placentation. Mol Biol Evol 37(2):507–523
Zhuang X-W, Li J, Brost BC, Xia X-Y, Chen HB, Wang C-X, Jiang S-W (2014) Decreased expression and altered methylation of syncytin-1 gene in human placentas associated with preeclampsia. Curr Pharm Des 20(11):1796–1802
Funding
No funding was received in connection with this manuscript.
Author information
Authors and Affiliations
Contributions
BA made the conception and design of the work, the acquisition, data analysis, and interpretation of data for the work and drafting of the work, revising it critically for important intellectual content and final approval of the version to be published and takes agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that there are no competing interests in connection with this manuscript.
Ethical approval
Ethics approval was not needed for this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Arneth, B. Leftovers of viruses in human physiology. Brain Struct Funct 226, 1649–1658 (2021). https://doi.org/10.1007/s00429-021-02306-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-021-02306-8