Abstract
Oral epithelial dysplasia (OED) is diagnosed and graded using a range of histological features, making grading subjective and challenging. Mitotic counting and phosphohistone-H3 (PHH3) staining have been used for the prognostication of various malignancies; however, their importance in OED remains unexplored. This study conducts a quantitative analysis of mitotic activity in OED using both haematoxylin and eosin (H&E)-stained slides and immunohistochemical (IHC) staining for PHH3. Specifically, the diagnostic and prognostic importance of mitotic number, mitotic type and intra-epithelial location is evaluated. Whole slide images (WSI) of OED (n = 60) and non-dysplastic tissue (n = 8) were prepared for analysis. Five-year follow-up data was collected. The total number of mitosis (TNOM), mitosis type and intra-epithelial location was manually evaluated on H&E images and a digital mitotic count performed on PHH3-stained WSI. Statistical associations between these features and OED grade, malignant transformation and OED recurrence were determined. Mitosis count increased with grade severity (H&E: p < 0.005; IHC: p < 0.05), and grade-based differences were seen for mitosis type and location (p < 0.05). The ratio of normal-to-abnormal mitoses was higher in OED (1.61) than control (1.25) and reduced with grade severity. TNOM, type and location were better predictors when combined with histological grading, with the most prognostic models demonstrating an AUROC of 0.81 for transformation and 0.78 for recurrence, exceeding conventional grading. Mitosis quantification and PHH3 staining can be an adjunct to conventional H&E assessment and grading for the prediction of OED prognosis. Validation on larger multicentre cohorts is needed to establish these findings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oral epithelial dysplasia (OED) describes a spectrum of histologically identified architectural and cytological disturbances involving the oral epithelium [1]. These lesions may progress to oral squamous cell carcinoma (OSCC) [2]. Higher grade lesions have higher risk of transformation, highlighting the need for an early and accurate diagnosis [1]. OSCC is the most common malignant neoplasm of the oral cavity associated with a myriad of environmental aetiologies and genetic alterations [3,4,5].
Because of the direct relationship between OED and malignant transformation, the dysplasia grade is considered the most important prognosticator for malignant transformation [5]. However, the current grading system (WHO, 2017) is associated with poor reproducibility, which can result in an inconsistent and unreliable diagnosis [6]. Suggestions to mitigate these shortcomings include the use of clinical determinants and molecular markers [7]. The binary grading system is an alternative criteria proposed to improve observer reproducibility by quantifying the minimum number of cytological and architectural features required for a diagnosis [8]. However, this classification uses the same histological features listed in the WHO Classification, and there remains a lack of high-quality evidence to support the prognostic importance of many of these features [2]. The recent update from the 5th Edition of the WHO Classification includes additional features, such as apoptotic mitoses and single cell keratinisation. However, the clinical relevance for inclusion of these features is unclear [9]. A recent study explored histological feature-specific associations in OED with clinical outcomes. The predictive performance of the proposed models for OED progression exceeded conventional grading [10]. However, a more detailed and prospective analysis of individual histological features is still needed to establish a more objective predictive/grading system.
Mitotic figure counting is used for diagnosis and prognostication of various malignancies [11,12,13,14] including breast, gastric and neuroendocrine carcinomas [13, 15,16,17]. However, its importance in precancer diagnosis and progression is yet to be explored. The main limitation of mitosis counting is the tediousness of the manual approach, in addition to interpretation differences due to variations in chromatin arrangements in the different mitotic stages, and the resemblance of apoptotic bodies and pyknotic nuclei with mitotic bodies (Fig. 1) [18]. Many of these limitations can now be overcome by the increasing number of digital/computational tools which allow for automated quantification, providing more objective, efficient and reliable outputs [19]. However, in the case of mitotic cell counting, attention also needs to be given to the presence of abnormal mitotic forms, characterised by mitotic asymmetry or an abnormal segregation of chromosomes [20].
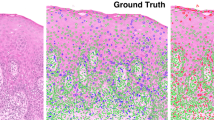
Photomicrographs (40 ×) demonstrating the different mitotic stages observed in OED (black arrows) based on H&E (A) and PHH3-IHC staining (B). Photomicrographs (20 ×) demonstrating the different OED grades (WHO, 2017) on H&E (C) and PHH3-IHC-stained images (D). H&E photomicrographs (40 ×) demonstrating ‘normal’ appearance of mitosis (E) and ‘abnormal’ appearance of mitosis (F) highlighted by black arrow
Various biomarkers have been implicated in OED progression, but the evidence to support their routine use is still lacking [21]. Phosphohistone-H3 (PHH3) is a specific protein phosphorylated during chromatin condensation in mitosis [22]. It stains positively during the late G2 phase and M phase. Phosphorylation of the histone H3 starts to occur just before prophase which is not identifiable on haematoxylin and eosin (H&E) examination [18], lending to the role of PHH3 a useful marker.
The aims of this study were threefold: first, to conduct a quantitative analysis of mitotic activity in OED (including number, type and intra-epithelial location of mitoses) using digitised H&E sections and immunohistochemical (IHC)-stained tissue with PHH3; second, to evaluate changes in mitotic activity relative to OED progression; and third, to develop and explore multivariable models using mitotic features for prediction of OED recurrence and malignant transformation, with comparison to conventional grading.
Material and methods
Case selection and tissue processing
Following ethical approval (reference 18/WM/0335), a retrospective sample of 68 H&E-stained tissue sections were retrieved from the department archive. The sample comprised OED sections (n = 60) of varying grades (mild, moderate, severe) with 5-year post-diagnosis data, in addition to non-dysplastic control samples (n = 8) which included cases of benign hyperplasia, scar tissue and inflammatory oral lichen planus. Verrucous and HPV-related OED lesions were excluded based on morphological features, as they are distinct entities with reportedly different behaviours.
Prior to the inclusion, cases were independently reviewed by a consultant oral and maxillofacial pathologist (SAK) to ensure there was sufficient epithelial tissue for analysis. Cases with insufficient tissue, gross artefact or tangentially cut sections were excluded. All cases were then blindly re-evaluated by SAK, HM (clinician with extensive expertise and specialist interest in OED analysis) and PH (trainee oral and maxillofacial pathologist) to confirm the original diagnosis and where necessary assign an updated OED grade (using WHO and binary systems). Grading variability was measured by a Cohen’s kappa score, which resulted in a value of 0.900, demonstrating good interobserver agreement.
New 5-μm-thick formalin-fixed paraffin-embedded sections of the selected cases were obtained for H&E and IHC staining. The sections were scanned at 40 × magnification using an Aperio-CS2 scanner (Leica Biosystems, Milton Keynes, UK) to obtain high-resolution whole slide images (WSI) producing 68 H&E slides and 67 IHC slides for analysis. The IHC sample had one less case due to technical scanning/imaging difficulties, resulting in its exclusion at the final stage.
Clinical data collection included patient age at diagnosis, sex, biopsy site, original histological grade (WHO, 2017), status of malignant transformation and recurrence (lesion that progressed to OSCC or recurred at the same clinical site following treatment within 5 years).
Immunohistochemical staining for PHH3
IHC staining was carried out for the mitosis marker PHH3 (Ser10) using a previously described protocol [23]. A primary rabbit anti-human PHH3 polyclonal antibody (#9701; Cell Signalling Technology, 1:100 dilution) and a secondary goat anti-rabbit antibody was used. Following IHC, counterstaining with haematoxylin and mounting in DPX was done for further analysis.
Analysis of mitosis activity in OED
QuPath software (v.0.3.2) was used for identification of regions of interest (ROI) and subsequent mitotic feature analysis [24]. For all slides, five rectangular-shaped ROIs of a consistent size (area≈165,000 mm2) corresponding to representative dysplastic and non-dysplastic regions were selected at 20 × magnification and verified by two experienced clinicians (HM, SAK).
For the H&E sample (n = 68), two observers (HS, SAK), blinded to clinical outcomes, were asked to independently count and record (i) the total number of mitoses (TNOM), (ii) the number of ‘normal’ and ‘abnormal’ mitoses and (iii) the intra-epithelial mitosis location (‘basal’ or ‘suprabasal’) in each field. An agreement between the observers was made on how to qualify a ‘normal’ and ‘abnormal’ mitosis. An equational bipartition of the chromosomal material was used as standard for ‘normal’ mitosis [25], whereas the presence of abnormalities like binucleation, pyknotic nuclei, micronuclei and broken-egg appearances qualified the mitoses to be ‘abnormal’ [26]. A kappa score of 0.646 was obtained between the two observers for independent mitosis counting. In cases of wide disagreement, a consensus score was agreed/used for the downstream analyses. The means and standard deviation for the mitosis variables (TNOM, type and location) from the five ROIs were recorded and an average obtained for each case.
For the PHH3-IHC sample (n = 67), QuPath’s inbuilt ‘positive cell detection’ algorithm was applied for automated quantification of positively stained mitoses, and intra-epithelial mitosis location recorded through manual assessment (by HS, SAK). Due to the nature of the automated detection, the mitosis type could not be confirmed in the IHC sample. All data were exported onto a pre-structured spreadsheet in Microsoft Excel® (v.2206).
Statistical analyses
Statistical analyses were conducted in GraphPad Prism (v9) and IBM SPSS Statistics (v29.0.1.0). Data was tested for normality following which appropriate statistical tests were selected. Unpaired Student’s t-tests and one-way ANOVA were performed to compare differences in the TNOM, mitosis type and intra-epithelial location between OED grades and relative to control. Where relevant, an appropriate post hoc analysis (Tukey’s/Dunnett’s) was performed for pairwise comparisons. For the H&E analysis, the mean mitosis number and ratio of normal-to-abnormal mitoses were measured and compared between grades. Paired sample t-tests were conducted to compare the number of normal and abnormal mitoses across OED grades.
Multivariable logistic regression models were explored separately for H&E and PHH3-IHC samples, to assess statistical relationships between individual and combined mitotic variables (TNOM, mitosis type, intra-epithelial location) with clinical outcomes (malignant transformation and OED recurrence). The effect of adding clinical variables (age, sex, intraoral site) and histological grade (WHO, binary) on model performance was assessed. The area under the receiver operator characteristic (ROC) curve was used to assess model accuracy and visualise performance. A p value of < 0.05 was considered statistically significant. Figure 2 depicts the workflow methodology for this study.
Overall workflow methodology of the study. A Identification, retrieval and preparation of H&E sample (n = 68). B Preparation of PHH3-IHC sample (n = 67). Conversion of tissue sections to digital WSI and identification of ROI for H&E (C) and PHH3-IHC analysis (D). E Manual assessment of mitosis activity (number, type, location) on H&E. F Automated mitosis quantification for PHH3-IHC sample. G Statistical analysis to assess mitotic activity in OED with correlation to clinical outcomes
Results
Characteristics of the OED cohort
Amongst the 60 OED cases, 39 (65%) were male, and 21 (35%) were female, with a mean age of 61.73 years (IQR 18.5). The clinical intraoral site distribution was the tongue n = 28 (46.67%), floor of mouth n = 15 (25%), buccal mucosa n = 8 (13.33%), gingivae n = 5 (8.33%) and palate n = 4 (6.67%). The WHO histological grade distribution (following blind re-analysis) was mild OED = 20 (33.33%), moderate OED = 17 (28.33%) and severe OED = 23 (38.33%). Binary grade distribution was low-grade OED = 25 (41.7%) and high-grade OED = 35 (58.3%). A total of 14 cases (23.33%) transformed to OSCC, amongst which 8 (57.1%) were moderately dysplastic and 6 (42.9%) were severely dysplastic. Of the 19 (31.67%) cases that recurred after treatment, 8 were moderately dysplastic (42.1%), and 11 were severely dysplastic (57.9%).
Analysis of H&E and PHH3 mitotic count
Both the H&E and IHC analyses yielded a statistically significant difference in the TNOM between WHO grades (H&E: p = 0.0005; IHC: p = 0.0073) and binary OED grades (H&E: p = 0.0012; IHC: p = 0.0403) (Fig. 3). A significant difference was also seen when comparing TNOM between the following groups: mild OED vs severe OED (H&E: p = 0.0006; IHC: p = 0.0197), moderate OED vs severe OED (H&E: p = 0.0113; IHC: p = 0.0181), severe OED vs control (H&E: p = 0.0004; IHC: p = 0.0009) and high-grade OED vs control (H&E: p = 0.0022; IHC: p = 0.0064) (Fig. 3).The remaining pairwise comparisons (mild OED vs moderate OED, mild OED vs control, moderate OED vs control and low-grade OED vs control) were not statistically significant. The mean mitosis number increased with grade severity (H&E: mild OED 1.32, moderate OED 2.09, severe OED 4.93, low-grade OED 1.32, high-grade OED 4.07) and relative to control (0.20). A similar trend was seen in IHC analysis (mild OED 3.47, moderate OED 3.26, severe OED 7.16, low-grade OED 3.36, high-grade OED 5.84, control 0.825).
Analysis of the TNOM based on H&E sections (A, B) and PHH3-IHC sections (C, D) with comparisons between histological grades and relative to control. Analysis of intra-epithelial mitosis location based on H&E sections (E) and PHH3-IHC sections (F) with comparisons between histological grade and relative to control. Asterisk indicates a statistically significant finding (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, **** p ≤ 0.0001)
H&E analysis of mitosis type
Normal mitotic figures
There was a significant difference in the average number of ‘normal’ mitoses between WHO grades (p = 0.0016) and binary grades (p = 0.0040) (Table 1). Significant differences were also seen between the following groups: control vs severe OED (p = 0.0004), control vs high-grade OED (p = 0.0023), mild OED vs severe OED (p = 0.0026) and moderate OED vs severe OED (p = 0.0143) (Table 1).
Abnormal mitotic figures
Similar trends were seen for the presence of ‘abnormal’ mitoses between WHO grades (p = 0.0010) and binary grades (p = 0.0016) (Table 1) in addition to comparisons between control vs severe OED (p = 0.0032), control vs high-grade OED (p = 0.0116), mild OED vs severe OED (p = 0.0010) and moderate OED vs severe OED (p = 0.0322) (Table 1).
Normal-to-abnormal mitosis ratio
The ratio of normal-to-abnormal mitoses was higher in OED (1.61) compared to control (1.25). This ratio was found to reduce with increasing grade severity. The ratios for mild, moderate and severe grades were 3.26, 1.49 and 1.43, and for low and high grades, 2.75 and 1.44, respectively. Statistically significant differences were observed when comparing the ratio of normal/abnormal mitoses across different grades (p = 0.0001 mild OED, p = 0.0289 moderate OED, p = 0.0470 severe OED, p < 0.0001 low-grade OED, p = 0.0137 high-grade OED).
Analysis of H&E and PHH3 mitosis location
Basal mitoses
A higher number of basal mitoses were observed with increasing grade severity, for WHO (mild OED = 1.3, moderate OED = 1.905882353, severe OED = 3.269565217) and binary grading (low-grade OED = 1.296, high-grade OED = 2.89) and relative to control (0.2) on H&E assessment (p < 0.0001). A similar trend was also seen on PHH3-IHC assessment between WHO grades and relative to control (p = 0.0287) (Fig. 3). Further comparisons demonstrated significance differences between mild OED vs. severe OED (H&E: p < 0.0001), moderate OED vs. severe OED (H&E: p = 0.0076; IHC: p = 0.0383), moderate OED vs. control (H&E: p = 0.0163), severe OED vs. control (H&E: p < 0.0001; IHC: p = 0.0005), low-grade OED vs. control (IHC: p = 0.0495) and high-grade OED vs. control (H&E: p < 0.0001; IHC: p = 0.0024) (Fig. 3). The remaining pairwise comparisons were not statistically significant.
Suprabasal mitoses
An increasing number of suprabasal mitoses were also observed with grade severity. Significant differences were shown between WHO grades (H&E: p = 0.0174; IHC: p = 0.0076) and binary grades (H&E: p = 0.0364; IHC: p = 0.0202) as well as between the following groups: mild OED vs. severe OED (H&E: p = 0.0302; IHC: p = 0.0123), moderate OED vs. severe OED (only IHC: p = 0.0446) and severe OED vs. control (only IHC: p = 0.0435) (Fig. 3). The remaining pairwise comparisons were not statistically significant.
Multivariable model development exploration
The association between mitosis variables, clinical characteristics, histological grades and clinical outcomes was assessed (for H&E and PHH3-IHC analysis) using multiple logistic regression. For comparative purposes, the prognostic strength of conventional grading systems (WHO and binary) was also evaluated (Tables 2 and 3).
Prognostic potential of TNOM on H&E and PHH3-IHC sections
The TNOM alone had a modest association with malignant transformation (H&E: AUROC 0.5753; IHC: 0.5468) and OED recurrence (H&E: AUROC 0.6297; IHC: 0.5197), though the strength of association increased when combined with WHO grading (H&E: AUROC 0.7065 for transformation, 0.7401 for recurrence; IHC: AUROC 0.7460 for transformation, 0.7783 for recurrence) and binary grading (H&E: AUROC 0.722 for transformation, AUROC 0.6926 for recurrence; IHC: AUROC 0.7484 for transformation, AUROC 0.7184 for recurrence). The addition of clinical variables to TNOM had little or no effect on model performance (Table 2).
Prognostic potential of mitosis location on H&E and IHC-PHH3 sections
‘Basal’ mitosis was modestly associated with malignant transformation (H&E: AUROC 0.5815; IHC: AUROC 0.6381) and recurrence (H&E: AUROC 0.6175; IHC: 0.5411). In comparison, ‘suprabasal’ mitosis had a marginally weaker prognostic association (H&E: AUROC 0.5388 for transformation, AUROC 0.5854 for recurrence; IHC: 0.6278 for transformation, AUROC 0.6217 for recurrence). Whilst the addition of clinical variables had little overall effect on the prognostic strength of mitosis location, the incorporation of histological grading improved predictive strength, particularly for ‘suprabasal’ mitoses on H&E (‘suprabasal mitoses’ + ‘WHO grade’ = AUROC of 0.736 for transformation and 0.7458 for recurrence) (Table 2).
Prognostic potential of mitosis type on H&E sections
‘Abnormal’ mitoses alone had a greater predictive strength than ‘normal’ mitoses on H&E for transformation (AUROC 0.6856 vs 0.5016, respectively) and recurrence (AUROC 0.7022 vs 0.5552, respectively). However, incorporation of histological grading improved the predictive strength for ‘normal’ mitoses to a greater extent than for ‘abnormal’ mitoses (‘normal mitoses’ + ‘WHO grade’ = AUROC 0.7469, p = 0.0055 vs ‘abnormal mitoses’ + ‘WHO grade’ = AUROC 0.6537, p = 0.0836). The addition of clinical variables had little or no effect on model performance (Table 3).
Prognostic models using combined mitosis features
Combining the different mitosis variables with histological grading produced the most predictive models. The most superior model for prediction of transformation (‘abnormal mitoses’ + ‘suprabasal mitoses’ + ‘TNOM’ + ‘WHO grade’) produced an AUROC of 0.8113 (p = 0.0005, 95% CI 0.6987 to 0.9239), and the most superior model for prediction of recurrence (‘abnormal mitosis’ + ‘basal mitoses’ + ‘TNOM’ + ‘WHO grade’) achieved an AUROC of 0.7895 (p = 0.0003, 95% CI 0.6777 to 0.9013). Both these models outperformed conventional grading systems (Table 3).
Discussion
This study highlights the potential importance of mitosis assessment and quantification in OED diagnosis and prognostication. Mitosis counting has been effectively implemented in the diagnosis of various malignancies [13, 17, 27,28,29], but its diagnostic importance in oral precancers remains largely unexplored. Due to the limitations of manual mitotic figure counting, PHH3 was explored to evaluate its role as a diagnostic and prognostic adjunct to conventional H&E assessment.
The role of various oncogenes in OED progression to cancer still remains unvalidated [30]. Ki-67 being a cell cycle marker, rather than a specific marker of mitosis, has shown conflicting results. In one study, the value of PHH3 and Ki-67 for measuring mitotic activity in OSCC demonstrated a significant association between expression of PHH3 (p = 0.016) and mitotic activity (p = 0.031) with survival time; however, no similar relationship was found with Ki-67 (p = 0.295) [31]. In another study, the presence, location and pattern of Ki-67 positivity demonstrated variable results for differentiation between normal tissue, OED and OSCC [32]. The unreliability of Ki-67 [32, 33] and the successful use of PHH3 as an independent biomarker in various different malignancies [13, 15, 17, 22, 34] led us to explore this marker further.
The TNOM was shown to increase proportionally with grade severity on both H&E and PHH3-IHC analyses, supporting findings in the existing literature [35–38. This could be explained by the increased stem cell turnover and quantity of abnormal mutations [39]. Overall, PHH3 mitotic count was greater than H&E, likely due to the inclusion of early prophase stage, which cannot be reliably distinguished on H&E-stained sections. In a previous study, a comparison in mitotic count between H&E and crystal violet-stained sections demonstrated significant differences between non-dysplastic oral mucosa, OED and OSCC [39]. Whilst our findings revealed a greater difference between mild and severe OED, control and high-grade/severe OED, promising differences were also observed between the more ‘demanding’ groups (moderate vs severe OED) in terms of mitosis number, mitosis type and mitosis location.
H&E analysis of mitosis type demonstrated a higher ratio of normal-to-abnormal mitoses in OED than control, which decreased with grade severity. Mitosis location assessment on H&E and IHC analysis demonstrated significant differences in the number of ‘basal’ and ‘suprabasal’ mitoses between grades. ‘Suprabasal’ mitoses were shown to be more predictive than ‘basal’ mitoses on PHH3-IHC. A study on meningioma demonstrated that PHH3 mitotic counts had a better interobserver correlation than H&E mitotic counts (Rm = 0.83 vs 0.77, respectively) [40], with good discrimination between grades (AUROC 0.91). Our study suggested similar findings, with better generally performance for PHH3-IHC models than H&E models, particularly for TNOM and mitosis location (Table 2). This is likely to be related to greater objectivity of mitosis assessment with PHH3 staining.
Prognostic models combining TNOM, mitosis type, location and histological grading showed better prediction for transformation and recurrence. Generally, the addition of clinical variables had minimal impact on model performances, whereas histological grading boosted predictive potential. Such a trend was also observed in a study by Mahmood et al. where inclusion of grades improved prognostic strength of histological OED models [10].
The most predictive H&E models for malignant transformation (‘abnormal mitoses’ + ‘suprabasal mitoses’ + ‘TNOM’ + ‘WHO grade’ = AUROC 0.8113) and OED recurrence (‘abnormal mitosis’ + ‘basal mitoses’ + ‘TNOM’ + ‘WHO grade’ = AUROC 0.7895) (AUROC 0.65) incorporated multiple mitotic features and outperformed conventional WHO grading on its own. In the case of PHH3-IHC models, the most superior models utilised fewer mitotic features for prediction of transformation (‘basal mitoses’ + ‘binary grading’ = AUROC 0.7714) and recurrence (‘TNOM’ + ‘WHO grading’ = AUROC 0.7783). These findings indicate that PHH3-IHC may be important for prognostication of OED, complementing H&E analysis.
The authors acknowledge a few limitations. First, the follow-up period comprised 5 years. Whilst transformation may occur later [41], a number of studies have shown transformation incidence to be highest during the first 5 years. [5, 41,42,43,44] A study by Hankinson et al. (2021) reported a median transformation time of 22 months (IQR 46.0) for a cohort of OED cases (n = 150) retrieved from the same centre as that used for this study [45]. Second, cases were from a single-centre, and the sample size could be regarded as small [46, 47]. However, the unit in question is a national tertiary centre providing service to a large geographical region, thereby increasing the biological diversity of the sample. Furthermore, the sample has an equitable distribution of dysplasia grades with inclusion of transformed and non-transformed cases. For an early exploratory study that serves as a basis for future work, our sample is similar to many other studies [31, 48, 49] of this kind. The control cases were included for clinical interest and early comparative analysis, hence the small numbers. They did not contribute to the prognostic work, which was the important and novel aspect of this study.
In conclusion, we report increased mitotic activity with OED progression. Mitotic quantification using PHH3-IHC is potentially more reliable than H&E analysis, with typically greater predictive strength, even with inclusion of fewer variables. The addition of histological grading further improved performance of PHH3-IHC models, more so than the H&E models. To the best of our knowledge, this is one of the first studies to utilise mitosis quantification and compare H&E with PHH3-IHC for OED analysis and prognosis prediction. The promising results call for further exploration of H&E and IHC markers to contribute to a more objective grading of OED and reliable prognosis prediction. Further studies with larger multicentre cohorts are required for clinical validation.
Data Availability
All the data derived from this study are included in the manuscript. Further information for reasonable requests (if needed) can be made available, by contacting the last author (s.a.khurram@sheffield.ac.uk).
Abbreviations
- OED:
-
Oral epithelial dysplasia
- OSCC:
-
Oral squamous cell carcinoma
- HPV:
-
Human papillomavirus
- WHO:
-
World Health Organization
- H&E:
-
Haematoxylin and eosin
- PHH3:
-
Phosphohistone-H3
- IHC:
-
Immunohistochemistry
- WSI:
-
Whole slide image
- ROI:
-
Region of interest
- DPX:
-
Dibutyl phthalate polystyrene xylene
- ANOVA:
-
Analysis of variance
- AUROC:
-
Area under receiver operator characteristic
- TNOM:
-
Total number of mitoses
- Ki-67:
-
Kiel-67
References
Warnakulasuriya S, Reibel J, Bouquot J, Dabelsteen E (2008) Oral epithelial dysplasia classification systems: predictive value, utility, weaknesses and scope for improvement. J Oral Pathol Med 37(3):127–133. https://doi.org/10.1111/j.1600-0714.2007.00584.x
Odell E, Kujan O, Warnakulasuriya S, Sloan P (2021) Oral epithelial dysplasia: recognition, grading and clinical significance. Oral Dis 27(8):1947–1976. https://doi.org/10.1111/odi.13993
Pilborough AE, Lambert DW, Khurram SA (2019) Extranodal extension in oral cancer: a role for the nodal microenvironment? J Oral Pathol Med 48(10):863–870. https://doi.org/10.1111/jop.12870
Pires FR, Ramos AB, Coutinho de Oliveira JB, Tavares AS, Ribeiro da Luz PS, Bartholomeu R, dos Santos TC (2013) Oral squamous cell carcinoma: clinicopathological features from 346 cases from a single oral pathology service during an 8-year period. J Appl Oral Sci 21(5):460–467. https://doi.org/10.1590/1679-775720130317
Speight PM, Khurram SA, Kujan O (2018) Oral potentially malignant disorders: risk of progression to malignancy. Oral Surg Oral Med Oral Pathol Oral Radiol 125(6):612–627. https://doi.org/10.1016/j.oooo.2017.12.011
Holmstrup P, Vedtofte P, Reibel J, Stoltze K (2007) Oral premalignant lesions: is a biopsy reliable? J Oral Pathol Med 36(5):262–266. https://doi.org/10.1111/j.1600-0714.2007.00513.x
Ranganathan K, Kavitha L (2019) Oral epithelial dysplasia: classifications and clinical relevance in risk assessment of oral potentially malignant disorders. J Oral Maxillofac Pathol 23(1):19–27. https://doi.org/10.4103/jomfp.JOMFP_13_19
Kujan O, Oliver RJ, Khattab A, Roberts SA, Thakker N, Sloan P (2006) Evaluation of a new binary system of grading oral epithelia dysplasia for prediction of malignant transformation. Oral Oncol 42(10):987–993. https://doi.org/10.1016/j.oraloncology.2005.12.014
Muller S, Tilakaratne WM (2022) Update from the 5th edition of the World Health Organization classification of head and neck tumors: tumours of the oral cavity and mobile tongue. Head Neck Pathol 16(1):54–62
Mahmood H, Bradburn M, Rajpoot N, Islam NM, Kujan O, Khurram SA (2022) Prediction of malignant transformation and recurrence of oral epithelial dysplasia using architectural and cytological feature specific prognostic models. Mod Pathol 35(9):1151–1159. https://doi.org/10.1038/s41379-022-01067-x
Tamori Y, Suzuki E, Deng W-M (2016) Epithelial tumors originate in tumor hotspots, a tissue-intrinsic microenvironment. PLoS Biol 14(9):e1002537. https://doi.org/10.1371/journal.pbio.1002537
Dessauvagie BF, Thomas C, Robinson C, Frost FA, Harvey J, Sterrett GF (2015) Validation of mitosis counting by automated phosphohistone H3 (PHH3) digital image analysis in a breast carcinoma tissue microarray. Pathology 47(4):329–334. https://doi.org/10.1097/pat.0000000000000248
Skaland I, Janssen EAM, Gudlaugsson E et al (2007) Phosphohistone H3 expression has much stronger prognostic value than classical prognosticators in invasive lymph node-negative breast cancer patients less than 55 years of age. Mod Pathol 20(12):1307–1315. https://doi.org/10.1038/modpathol.3800972
Stalhammar G, Robertson S, Wedlund L et al (2018) Digital image analysis of Ki67 in hot spots is superior to both manual Ki67 and mitotic counts in breast cancer. Histopathology 72(6):974–989. https://doi.org/10.1111/his.13452
Takahashi H, Murai Y, Tsuneyama K et al (2006) Overexpression of phosphorylated histone H3 is an indicator of poor prognosis in gastric adenocarcinoma patients. Appl Immunohistochem Mol Morphol 14(3):296–302. https://doi.org/10.1097/00129039-200609000-00007
Colman H, Giannini C, Huang L et al (2006) Assessment and prognostic significance of mitotic index using the mitosis marker phospho-histone H3 in low and intermediate-grade infiltrating astrocytomas. Am J Surg Pathol 30(5):657–664. https://doi.org/10.1097/01.pas.0000202048.28203.25
Tsuta K, Liu DC, Kalhor N, Wistuba II, Moran CA (2011) Using the mitosis-specific marker anti-phosphohistone H3 to assess mitosis in pulmonary neuroendocrine carcinomas. Am J Clin Pathol 136(2):252–259. https://doi.org/10.1309/ajcpdxfopxgef0rp
Veras E, Malpica A, Deavers MT, Silva EG (2009) Mitosis-specific marker phospho-histone H3 in the assessment of mitotic index in uterine smooth muscle tumors: a pilot study. Int J Gynecol Pathol 28(4):316–321. https://doi.org/10.1097/PGP.0b013e318193df97
Veta M, van Diest PJ, Jiwa M, Al-Janabi S, Pluim JPW (2016) Mitosis counting in breast cancer: object-level interobserver agreement and comparison to an automatic method. PLoS One 11(8):e0161286. https://doi.org/10.1371/journal.pone.0161286
Donovan TA, Moore FM, Bertram CA et al (2021) Mitotic figures-normal, atypical, and imposters: a guide to identification. Vet Pathol 58(2):243–257. https://doi.org/10.1177/0300985820980049
Smith J, Rattay T, McConkey C, Helliwell T, Mehanna H (2009) Biomarkers in dysplasia of the oral cavity: a systematic review. Oral Oncol 45(8):647–653. https://doi.org/10.1016/j.oraloncology.2009.02.006
Sawicka A, Seiser C (2012) Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie 94(11):2193–2201. https://doi.org/10.1016/j.biochi.2012.04.018
Abdullah Zubir AZ, Whawell SA, Wong TS, Khurram SA (2020) The chemokine lymphotactin and its recombinant variants in oral cancer cell regulation. Oral Dis 26(8):1668–1676. https://doi.org/10.1111/odi.13500
Bankhead P, Loughrey MB, Fernandez JA et al (2017) QuPath: open source software for digital pathology image analysis. Sci Rep 7(1):16878. https://doi.org/10.1038/s41598-017-17204-5
Steinbeck R (2001) Pathologic mitoses and pathology of mitosis in tumorigenesis. Eur J Histochem 45(4):311–318
Ankle MR, Kale AD, Charantimath S (2007) Comparison of staining of mitotic figures by haematoxylin and eosin-and crystal violet stains, in oral epithelial dysplasia and squamous cell carcinoma. Indian J Dent Res 18(3):101–105
Pidhorecky I, Cheney RT, Kraybill WG, Gibbs JF (2000) Gastrointestinal stromal tumors: current diagnosis, biologic behavior, and management. Ann Surg Oncol 7:705–712
Diaconescu D, Diaconescu S, Chesca A, Toma S (2011) The value of mitotic counting in prostate carcinomas. Int J Math Models Methods Appl Sci 2:379–386
Goodarzi M, Correa AM, Ajani JA et al (2009) Anti-phosphorylated histone H3 expression in Barrett’s esophagus, low-grade dysplasia, high-grade dysplasia, and adenocarcinoma. Mod Pathol 22(12):1612–1621
Pitiyage G, Tilakaratne WM, Tavassoli M, Warnakulasuriya S (2009) Molecular markers in oral epithelial dysplasia: review. J Oral Pathol Med 38(10):737–752. https://doi.org/10.1111/j.1600-0714.2009.00804.x
Tancredi-Cueto N, Vigil-Bastitta G, Bologna-Molina R, Beovide-Cortegoso V (2022) The value of phosphohistone H3 as a cell proliferation marker in oral squamous cell carcinoma. A comparative study with Ki-67 and the mitotic activity index. Med Oral Patol Oral Cir Bucal. 27(5):e444
Birajdar SS, Radhika M, Paremala K, Sudhakara M, Soumya M, Gadivan M (2014) Expression of Ki-67 in normal oral epithelium, leukoplakic oral epithelium and oral squamous cell carcinoma. J Oral Maxillofac Pathol 18(2):169–176. https://doi.org/10.4103/0973-029x.140729
Scholzen T, Gerdes J (2000) The Ki-67 protein: from the known and the unknown. J Cell Physiol 182(3):311–322. https://doi.org/10.1002/(sici)1097-4652(200003)182:3%3c311::aid-jcp1%3e3.0.co;2-9
Zhu P, Zhang C-B, Yang P et al (2016) Phosphohistone H3 (pHH3) is a prognostic and epithelial to mesenchymal transition marker in diffuse gliomas. Oncotarget 7(29):45005–45014. https://doi.org/10.18632/oncotarget.7154
Steinbeck RG (2004) Dysplasia in view of the cell cycle. Eur J Histochem 48(3):203–211
Kovesi G, Szende B (2003) Changes in apoptosis and mitotic index, p53 and Ki67 expression in various types of oral leukoplakia. Oncology 65(4):331–336. https://doi.org/10.1159/000074646
Pilati SFM, Bianco BC, Vieira DSC, Modolo F (2017) Histopathologic features in actinic cheilitis by the comparison of grading dysplasia systems. Oral Dis 23(2):219–224. https://doi.org/10.1111/odi.12597
Camara PR, Dutra SN, Takahama A, Fontes K, Azevedo RS (2016) A comparative study using WHO and binary oral epithelial dysplasia grading systems in actinic cheilitis. Oral Dis 22(6):523–529. https://doi.org/10.1111/odi.12484
Tandon A, Singh NN, Brave VR, Sreedhar G (2016) Image analysis assisted study of mitotic figures in oral epithelial dysplasia and squamous cell carcinoma using differential stains. J Oral Biol Craniofac Res 6(Suppl 1):S18–S23. https://doi.org/10.1016/j.jobcr.2016.09.003
Duregon E, Cassenti A, Pittaro A et al (2015) Better see to better agree: phosphohistone H3 increases interobserver agreement in mitotic count for meningioma grading and imposes new specific thresholds. Neuro Oncol 17(5):663–669. https://doi.org/10.1093/neuonc/nov002
Nevanpaa TT, Terava AE, Laine HK, Rautava J (2022) Malignant transformation of oral epithelial dysplasia in Southwest Finland. Sci Rep 12(1):8261. https://doi.org/10.1038/s41598-022-12441-9
Ho MW, Risk JM, Woolgar JA et al (2012) The clinical determinants of malignant transformation in oral epithelial dysplasia. Oral Oncol 48(10):969–976. https://doi.org/10.1016/j.oraloncology.2012.04.002
Lumerman H, Freedman P, Kerpel S (1995) Oral epithelial dysplasia and the development of invasive squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endodontol 79(3):321–329
Liu W, Bao Z-X, Shi L-J, Tang G-Y, Zhou Z-T (2011) Malignant transformation of oral epithelial dysplasia: clinicopathological risk factors and outcome analysis in a retrospective cohort of 138 cases. Histopathology 59(4):733–740. https://doi.org/10.1111/j.1365-2559.2011.03938.x
Hankinson PM, Mohammed-Ali RI, Smith AT, Khurram SA (2021) Malignant transformation in a cohort of patients with oral epithelial dysplasia. Br J Oral Maxillofac Surg 59(9):1099–1101. https://doi.org/10.1016/j.bjoms.2021.02.019
Jaber MA, Elameen EM (2021) Long-term follow-up of oral epithelial dysplasia: a hospital based cross-sectional study. J Dent Sci 16(1):304–310. https://doi.org/10.1016/j.jds.2020.04.003
Liu W, Bao ZX, Shi LJ, Tang GY, Zhou ZT (2011) Malignant transformation of oral epithelial dysplasia: clinicopathological risk factors and outcome analysis in a retrospective cohort of 138 cases. Histopathology 59(4):733–740
Ivina AA, Semkin VA, Khabadze ZS, Babichenko II (2019) Immunohistochemical study of Ki-67, PHH3, and CK15 protein expression in oral epithelial malignancy. Arkhiv Patologii 81(5):30–34. https://doi.org/10.17116/patol20198105130
Dragomir L, Simionescu C, Mărgăritescu C, Stepan A, Dragomir IM, Popescu M (2012) P53, p16 and Ki67 immunoexpression in oral squamous carcinomas. Rom J Morphol Embryol 53(1):89–93
Funding
HM is funded by the National Institute for Health Research (reference NIHR300904). PH is partially funded by a pre-doctoral bursary from Cancer Research UK and the Pathological Society for Great Britain and Ireland (reference RCCPSB-Nov21\100001). AZAZ is funded by a British Council Pakistan Grant (reference 20-ICRG-46/RGM/HEC/2020). SAK is partially funded by a Cancer Research UK Project Grant (reference C63489/A29674).
Author information
Authors and Affiliations
Contributions
H.M. and S.A.K. designed the study. H.M. retrieved and prepared the case cohort with clinical data collection. H.M. and P.H. re-analysed the histological grading with S.A.K. H.S. carried out immunohistochemistry with guidance from A.Z.A.Z. A.Z.A.Z. prepared the digital images. H.S. carried out ROI analysis and manual and automated mitosis quantification, and S.A.K. executed the manual recounting. H.S. and H.M. performed statistical analysis. H.S., H.M. and S.A.K. were involved in the analysis and interpretation of results. H.S. and H.M. led on manuscript writing with contributions from A.Z.A.Z., P.H. and S.A.K. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Ethical approval was granted by the West Midlands-Edgbaston Research Ethics Committee (reference 18/WM/0335).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hrishikesh Sathyamoorthy and Hanya Mahmood joint first authorship.
Supplementary Information
Below is the link to the electronic supplementary material.

428_2023_3668_Fig4_ESM.png
Supplementary Figure 1. ROC curves demonstrating the most predictive prognostic models (as highlighted inTable 2) for malignant transformation (A) and OED recurrence (B) based on H&E assessment (n=68, 340 ROI). AUROC= area under receiver operating characteristic. (PNG 174 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sathyamoorthy, H., Mahmood, H., Zubir, A.Z.A. et al. Prognostic importance of mitosis quantification and PHH3 expression in oral epithelial dysplasia. Virchows Arch 484, 47–59 (2024). https://doi.org/10.1007/s00428-023-03668-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-023-03668-6