Abstract
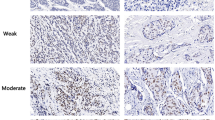
About 30% of patients with hormone receptor (HR)-positive breast cancers and up to 50% of human epidermal growth factor receptor 2 (HER2)-positive patients develop progression due to treatment resistance, highlighting the need for more differentiated tumor classifications within the breast cancer molecular subtype to optimize the therapies. We aim to examine the roles of histone modification markers. The levels of common repressive histone markers, histone H3 lysine 9 trimethylation (H3K9me3), histone H3 lysine 27 trimethylation (H3K27me3), and histone H4 lysine 20 trimethylation (H4K20me3), in tumors were evaluated by immunohistochemistry for 914 breast cancer patients. The subjects were followed up until December 2021. Hazard ratios (HRs) for overall survival (OS) and progression-free survival (PFS) were estimated using Cox regression models. For H3K27me3, patients with the high level had a longer PFS rate (81.3%) than that with the low level (73.9%) within HR-positive/HER2-negative subtype during a follow-up of 85 months only in univariate analysis (P < 0.05). For H3K9me3, the significant association between the high level of it and the longer OS [HR = 0.57, P < 0.05] was found within HR-positive/HER2-negative subtype in multivariate analysis. For H4K20me3, patients with the high level had a longer both OS [HR = 0.38] and PFS [HR = 0.46] within HR-positive/HER2-negative subtype, while had a shorter OS [HR = 3.28] in triple-negative breast cancer (TNBC) in multivariate analysis (all P < 0.05). H3K9me3 and H3K27me3 were the potential prognostic markers for breast cancer patients with HR-positive/HER2-negative subtype. Importantly, H4K20me3 was a robust prognostic marker for both HR-positive/HER2-negative and TNBC patients.

Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
The R code used during the current study is available from the corresponding author on reasonable request.
References
van Maaren MC, de Munck L, Strobbe LJA, Sonke GS, Westenend PJ, Smidt ML, Poortmans PMP, Siesling S (2019) Ten-year recurrence rates for breast cancer subtypes in the Netherlands: a large population-based study. Int J Cancer 144:263–272. https://doi.org/10.1002/ijc.31914
Razvi H, Tsang JY, Poon IK, Chan SK, Cheung SY, Shea KH, Tse GM (2021) INSM1 is a novel prognostic neuroendocrine marker for luminal B breast cancer. Pathology 53:170–178. https://doi.org/10.1016/j.pathol.2020.07.004
Lian J, Ma HX, Xu EW, Bu P, Yun KM, Xi YF (2022) Subclassifying triple-negative breast cancers and its potential clinical utility. Virchows Arch 481:13–21. https://doi.org/10.1007/s00428-022-03329-0
Dowsett M, Cuzick J, Wale C, Howell T, Houghton J, Baum M (2005) Retrospective analysis of time to recurrence in the ATAC trial according to hormone receptor status: an hypothesis-generating study. J Clin Oncol 23:7512–7517. https://doi.org/10.1200/JCO.2005.01.4829
Dowsett M, Houghton J, Iden C, Salter J, Farndon J, A’Hern R, Sainsbury R, Baum M (2006) Benefit from adjuvant tamoxifen therapy in primary breast cancer patients according oestrogen receptor, progesterone receptor, EGF receptor and HER2 status. Ann Oncol 17:818–826. https://doi.org/10.1093/annonc/mdl016
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 25:118–145. https://doi.org/10.1200/JCO.2006.09.2775
Rakha EA, Reis-Filho JS, Ellis IO (2010) Combinatorial biomarker expression in breast cancer. Breast Cancer Res Treat 120:293–308. https://doi.org/10.1007/s10549-010-0746-x
Haque MM, Desai KV (2019) Pathways to endocrine therapy resistance in breast cancer. Front Endocrinol (Lausanne) 10:573. https://doi.org/10.3389/fendo.2019.00573
Ramadan WS, Talaat IM, Hachim MY, Lischka A, Gemoll T, El-Awady R (2021) The impact of CBP expression in estrogen receptor-positive breast cancer. Clin Epigenetics 13:72. https://doi.org/10.1186/s13148-021-01060-2
Yang Y, Li J, Liu Y, Zhong Y, Ren W, Tan Y, He Z, Li C, Ouyang J, Hu Q, Yu Y, Yao H (2021) Magnetic resonance imaging radiomics signatures for predicting endocrine resistance in hormone receptor-positive non-metastatic breast cancer. Breast 60:90–97. https://doi.org/10.1016/j.breast.2021.09.005
Dimitrakopoulos FI, Kottorou A, Tzezou A (2021) Endocrine resistance and epigenetic reprogramming in estrogen receptor positive breast cancer. Cancer Lett 517:55–65. https://doi.org/10.1016/j.canlet.2021.05.030
Marsolier J, Prompsy P, Durand A, Lyne AM, Landragin C, Trouchet A, Bento ST, Eisele A, Foulon S, Baudre L, Grosselin K, Bohec M, Baulande S, Dahmani A, Sourd L, Letouze E, Salomon AV, Marangoni E, Perie L, Vallot C (2022) H3K27me3 conditions chemotolerance in triple-negative breast cancer. Nat Genet 54:459–468. https://doi.org/10.1038/s41588-022-01047-6
Ong LT, Lee WC, Ma S, Oguz G, Niu Z, Bao Y, Yusuf M, Lee PL, Goh JY, Wang P, Yong KSM, Chen Q, Wang W, Ramasamy A, Hoon DSB, Ditzel HJ, Tan EY, Lee SC, Yu Q (2022) IFI16-dependent STING signaling is a crucial regulator of anti-HER2 immune response in HER2+ breast cancer. Proc Natl Acad Sci U S A 119:e2093591177. https://doi.org/10.1073/pnas.2201376119
Zucchetti B, Shimada AK, Katz A, Curigliano G (2019) The role of histone deacetylase inhibitors in metastatic breast cancer. Breast 43:130–134. https://doi.org/10.1016/j.breast.2018.12.001
Zhang Q, Thakur C, Fu Y, Bi Z, Wadgaonkar P, Xu L, Liu Z, Liu W, Wang J, Kidder BL, Chen F (2020) Mdig promotes oncogenic gene expression through antagonizing repressive histone methylation markers. Theranostics 10:602–614. https://doi.org/10.7150/thno.36220
Yu-Rice Y, Jin Y, Han B, Qu Y, Johnson J, Watanabe T, Cheng L, Deng N, Tanaka H, Gao B, Liu Z, Sun Z, Bose S, Giuliano AE, Cui X (2016) FOXC1 is involved in ERalpha silencing by counteracting GATA3 binding and is implicated in endocrine resistance. Oncogene 35:5400–5411. https://doi.org/10.1038/onc.2016.78
Ye S, Ding YF, Jia WH, Liu XL, Feng JY, Zhu Q, Cai SL, Yang YS, Lu QY, Huang XT, Yang JS, Jia SN, Ding GP, Wang YH, Zhou JJ, Chen YD, Yang WJ (2019) SET domain-containing protein 4 epigenetically controls breast cancer stem cell quiescence. Cancer Res 79:4729–4743. https://doi.org/10.1158/0008-5472.CAN-19-1084
Chen QX, Yang YZ, Liang ZZ, Chen JL, Li YL, Huang ZY, Weng ZJ, Zhang XF, Guan JX, Tang LY, Yun JP, Ren ZF (2021) Time-varying effects of FOXA1 on breast cancer prognosis. Breast Cancer Res Treat 187:867–875. https://doi.org/10.1007/s10549-021-06125-7
Zhao QY, Lei PJ, Zhang X, Zheng JY, Wang HY, Zhao J, Li YM, Ye M, Li L, Wei G, Wu M (2016) Global histone modification profiling reveals the epigenomic dynamics during malignant transformation in a four-stage breast cancer model. Clin Epigenetics 8:34. https://doi.org/10.1186/s13148-016-0201-x
Li QL, Lei PJ, Zhao QY, Li L, Wei G, Wu M (2017) Epigenomic analysis in a cell-based model reveals the roles of H3K9me3 in breast cancer transformation. Epigenomics-UK 9:1077–1092. https://doi.org/10.2217/epi-2016-0183
Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, Albarracin C, Yu D, Abbruzzese JL, Mills GB, Jr Bast RC, Hortobagyi GN, Hung MC (2008) Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog 47:701–706. https://doi.org/10.1002/mc.20413
Yokoyama Y, Matsumoto A, Hieda M, Shinchi Y, Ogihara E, Hamada M, Nishioka Y, Kimura H, Yoshidome K, Tsujimoto M, Matsuura N (2014) Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res 16:R66. https://doi.org/10.1186/bcr3681
Berry WL, Janknecht R (2013) KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res 73:2936–2942. https://doi.org/10.1158/0008-5472.CAN-12-4300
Yang J, Jubb AM, Pike L, Buffa FM, Turley H, Baban D, Leek R, Gatter KC, Ragoussis J, Harris AL (2010) The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res 70:6456–6466. https://doi.org/10.1158/0008-5472.CAN-10-0413
Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, Yang X, Li Y, Han X, Zhang Y, Xuan C, Yao Z, Shang Y (2011) Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A 108:7541–7546. https://doi.org/10.1073/pnas.1017374108
Healey MA, Hu R, Beck AH, Collins LC, Schnitt SJ, Tamimi RM, Hazra A (2014) Association of H3K9me3 and H3K27me3 repressive histone marks with breast cancer subtypes in the Nurses’ Health Study. Breast Cancer Res Treat 147:639–651. https://doi.org/10.1007/s10549-014-3089-1
Sasidharan NV, El SH, Taha RZ, John A, Ali BR, Elkord E (2018) DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin Epigenetics 10:78. https://doi.org/10.1186/s13148-018-0512-1
Varghese B, Del Gaudio N, Cobellis G, Altucci L, Nebbioso A (2021) KDM4 involvement in breast cancer and possible therapeutic approaches. Front Oncol 11:750315. https://doi.org/10.3389/fonc.2021.750315
Zhou M, Chen QX, Yang YZ, Liang ZZ, Li YL, Huang ZY, Weng ZJ, Zhang XF, Guan JX, Tang LY, Ren ZF (2022) Prognostic value of glutaminase 1 in breast cancer depends on H3K27me3 expression and menopausal status. Virchows Arch 480:259–267. https://doi.org/10.1007/s00428-021-03210-6
Kapoor-Vazirani P, Kagey JD, Vertino PM (2011) SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol 31:1594–1609. https://doi.org/10.1128/MCB.00524-10
Dubik D, Dembinski TC, Shiu RP (1987) Stimulation of c-myc oncogene expression associated with estrogen-induced proliferation of human breast cancer cells. Cancer Res 47:6517–6521
Bedi U, Scheel AH, Hennion M, Begus-Nahrmann Y, Ruschoff J, Johnsen SA (2015) SUPT6H controls estrogen receptor activity and cellular differentiation by multiple epigenomic mechanisms. Oncogene 34:465–473. https://doi.org/10.1038/onc.2013.558
Paydar P, Asadikaram G, Nejad HZ, Akbari H, Abolhassani M, Moazed V, Nematollahi MH, Ebrahimi G, Fallah H (2019) Epigenetic modulation of BRCA-1 and MGMT genes, and histones H4 and H3 are associated with breast tumors. J Cell Biochem 120:13726–13736. https://doi.org/10.1002/jcb.28645
Soto-Reyes E, Recillas-Targa F (2010) Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene 29:2217–2227. https://doi.org/10.1038/onc.2009.509
Jones RA, Robinson TJ, Liu JC, Shrestha M, Voisin V, Ju Y, Chung PE, Pellecchia G, Fell VL, Bae S, Muthuswamy L, Datti A, Egan SE, Jiang Z, Leone G, Bader GD, Schimmer A, Zacksenhaus E (2016) RB1 deficiency in triple-negative breast cancer induces mitochondrial protein translation. J Clin Invest 126:3739–3757. https://doi.org/10.1172/JCI81568
Zecchin D, Moore C, Michailidis F, Horswell S, Rana S, Howell M, Downward J (2020) Combined targeting of G protein-coupled receptor and EGF receptor signaling overcomes resistance to PI3K pathway inhibitors in PTEN-null triple negative breast cancer. EMBO Mol Med 12:e11987. https://doi.org/10.15252/emmm.202011987
Sanchez-Munoz A, Perez-Ruiz E, Jurado JM, Ribelles N, Marquez A, Miramon J, Maiz M, Pajares B, Gallego E, Scholtz V, Jimenez B, Soler C, Molina M, Garcia-Rios I, Alba E (2011) Outcome of small invasive breast cancer with no axillary lymph node involvement. Breast J 17:32–38. https://doi.org/10.1111/j.1524-4741.2010.01026.x
Xu H, Jin F, Zhang XJ, Wang DQ, Yu SF, Wang AP (2020) Adherence status to adjuvant endocrine therapy in Chinese women with early breast cancer and its influencing factors: a cross-sectional survey. Cancer Med 9:3703–3713. https://doi.org/10.1002/cam4.3017
Acknowledgements
We sincerely thank the patients who participated in this study, the staff who conducted the baseline and the follow-up data collection, and the medical staff in the breast departments of the Third Affiliated Hospital, and the Cancer Center of Sun Yat-sen University. We also sincerely thank National Natural Science Foundation of China (81973115) and Science and Technology Planning Project of Guangdong Province, China (2019B030316002).
Funding
This study was funded by National Natural Science Foundation of China (81973115) and Science and Technology Planning Project of Guangdong Province, China (2019B030316002). The founders have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Bo Wang, Meng Zhou, and Ze-fang Ren designed and directed the study, wrote, and/or revised the manuscript. Yuan-zhong Yang constructed the TMAs and contributed to the IHC. Yue-xiang Ren and Lu-ying Tang contributed to digital imaging of IHC-stained sections and the assessment of immunohistochemical expression. Bo Wang, Meng Zhou, Yue-yu Shi, and Xing-lei Chen contributed to clinical data collection and curation. Bo Wang, Meng Zhou, Yue-yu Shi, and Xing-lei Chen participated in the statistical analysis plan and interpretation of results. Ze-fang Ren provided administrative support and supervision for the study. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
The study was approved by the Ethics Committee of School of Public Health, Sun Yat-Sen University.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, B., Zhou, M., Shi, Yy. et al. Survival is associated with repressive histone trimethylation markers in both HR-positive HER2-negative and triple-negative breast cancer patients. Virchows Arch 482, 1047–1056 (2023). https://doi.org/10.1007/s00428-023-03534-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-023-03534-5