Abstract
The use of autopsies in medicine has been declining. The COVID-19 pandemic has documented and rejuvenated the importance of autopsies as a tool of modern medicine. In this review, we discuss the various autopsy techniques, the applicability of modern analytical methods to understand the pathophysiology of COVID-19, the major pathological organ findings, limitations or current studies, and open questions. This article summarizes published literature and the consented experience of the nationwide network of clinical, neuro-, and forensic pathologists from 27 German autopsy centers with more than 1200 COVID-19 autopsies. The autopsy tissues revealed that SARS-CoV-2 can be found in virtually all human organs and tissues, and the majority of cells. Autopsies have revealed the organ and tissue tropism of SARS-CoV-2, and the morphological features of COVID-19. This is characterized by diffuse alveolar damage, combined with angiocentric disease, which in turn is characterized by endothelial dysfunction, vascular inflammation, (micro-) thrombosis, vasoconstriction, and intussusceptive angiogenesis. These findings explained the increased pulmonary resistance in COVID-19 and supported the recommendations for antithrombotic treatment in COVID-19. In contrast, in extra-respiratory organs, pathological changes are often nonspecific and unclear to which extent these changes are due to direct infection vs. indirect/secondary mechanisms of organ injury, or a combination thereof. Ongoing research using autopsies aims at answering questions on disease mechanisms, e.g., focusing on variants of concern, and future challenges, such as post-COVID conditions. Autopsies are an invaluable tool in medicine and national and international interdisciplinary collaborative autopsy-based research initiatives are essential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The COVID-19 pandemic has clearly highlighted the high importance of autopsies as an integral part of modern medicine. COVID-19 autopsies have provided insights into cellular and molecular pathomechanisms [8] by allowing direct analyses of the affected organs, thereby providing profound and viable data supporting concepts for therapeutic approaches, e.g., the striking absence of lymphocytic myocarditis despite suggestive clinical presentation [59]. Autopsies also documented the role of pre-existing medical conditions and vulnerable patients, provided the ultimate causes of death, and provided feedback on the effectiveness of treatment strategies [43, 106]. At the beginning of this global health crisis, post-mortem examinations revealed important histopathological findings on the nature of SARS-CoV-2 infection, such as the pronounced thrombotic angiopathy in the lungs [162] and the high incidence of thromboembolic events [157]. Despite this, some countries still discourage, or have even forbidden, COVID-19 autopsies [66, 133].
To facilitate the systematic evaluation of autopsy findings, research endeavors and multicentric trials, surgical pathologists, neuropathologists, and forensic pathologists, supported by virologists, scientific, and professional societies and healthcare authorities, combined forces to build the German Research Network for Autopsies in Pandemics (DEFEAT PANDEMIcs). DEFEAT PANDEMIcs established a highly organized nationwide network to collect and share data, materials, and findings as swiftly as possible. This has strengthened the comprehensive medical research of COVID-19, with over 90 papers published by the parties involved so far. DEFEAT PANDEMIcs also serves as a foundation for potential future pandemics, by providing standard operating procedures and emergency plans for autopsies, validating innovative techniques for sample collection and tissue analysis, and creating structures for systematic reporting in outbreak scenarios. Within this framework, a key building block is the nationwide registry of COVID-19 autopsies (DeRegCOVID; www.DeRegCOVID.ukaachen.de), which has been operational since early April 2020. The DeRegCOVID provides an electronic backbone for centralized and coordinated support for autopsy centers and researchers, for data reporting, biomaterial availability, and structured data coordination [148, 151] with the benefit of decentralized sample storage.
This article summarizes autopsy techniques and major COVID-19 associated pathological organ findings, based on literature review and a synoptic contemplation from over 1200 autopsies within DeRegCOVID performed and analyzed by the DEFEAT PANDEMIcs centers (for details, see Supplementary Table 1). Each specific organ pathology section was primarily prepared by national reference centers for the particular organ and consented by the consortium.
Autopsy types and techniques
Autopsies of COVID-19 patients, and autopsies in general, can be divided into (a) clinical, (b) ordered by health authorities, or (c) forensic autopsies. A clinical autopsy is most often initiated by clinicians rather than the relatives of the deceased and performed in cases with assumed natural causes of death. Consent is required either from the patient himself (generally in the case of a research project), or from the next of kin. These autopsies are traditionally performed by surgical pathologists, and in the majority of cases, detailed clinical history and data are available; health care authorities can also order autopsies. In Germany, this is done following the Infection Protection Law (Infektionsschutzgesetz) and does not require patient or relative consent; however, this is obtained where possible. These autopsies are performed by both surgical and forensic pathologists [43]. If death occurs at home, clinical data is generally not available prior to autopsy. However, such autopsies provide important insights into causes of death of non-hospitalized patients, and can help to uncover previously unrecognized pathologies, such as asymptomatic infections [83]. Finally, a forensic autopsy is always performed by forensic pathologists and is carried out in cases of (suspected or proven) unnatural causes of death, such as trauma, suicide, homicide, or when the cause of death is unclear.
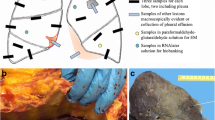
The standard procedure is to undertake a complete autopsy with the opening and assessment of all three body cavities (cranium, thorax, and abdomen), followed by macroscopic and microscopic evaluation (Fig. 1). Alternative autopsy techniques are also being increasingly used, e.g., the so-called rapid autopsy, performed as soon as possible after death [13], or “minimal-invasive” autopsies, which use post-mortem imaging, e.g., computed tomography (CT)- or ultrasound to guide biopsy-based tissue sampling, which can be coupled with robotics [40, 86]. The advantages of these techniques include improved tissue quality and in part higher consent rates with relatives.
Standard biomaterial sampling during autopsy includes formalin-fixed and paraffin-embedded tissues, which can include whole organs, if necessary, such as the brain for neuropathological examination. Further biomaterials required for specific diagnostic purposes can include organ-specific swabs (performed in a sterile way), body fluids (e.g., venous blood, cerebrospinal fluid, urine), native and cryopreserved samples (e.g., for virus and bacteria isolation [20]), or samples for electron microscopy. In a forensic scenario, body fluids and samples are routinely retained for toxicological analyses. As a result, the autopsy examination provides unique access to human material from all organ systems, allowing an otherwise impossible comprehensive assessment of deceased patients. Specific sampling sites, fixation methods, and clinical data available in the German Registry for COVID-19 autopsies (DeRegCOVID), are outlined in detail in a retrospective nationwide multicenter registry study. This study analyzed the electronic data on COVID-19 autopsy cases available in the DeRegCOVID, which mainly included analysis of age and sex distribution, disease duration, postmortal SARS-CoV-2 detection and cause of death. The study also reports on experiences and other achievements of building-up such a registry [150]. In this review, 35 publications with a direct link to the network are included.
Several approaches can be used to detect infectious agents in (autopsy) tissues. The gold standard of SARS-CoV-2 diagnostics is the detection of its ribonucleic acid (RNA) using highly sensitive nucleic acid amplification techniques, mainly quantitative reverse transcription-polymerase chain reaction (RT-qPCR). Since the beginning of the pandemic, several (commercially) available tests have been validated and approved for use with patient swabs and respiratory secretions. These assays can also be adapted for analyses of (postmortem) tissue, either fixed or unfixed, to address challenges of RNA degradation [64, 129, 157]. Currently, however, no widely validated PCR tests for use in tissues are available. Compartment-specific testing proved useful in confirming suspected cases or understanding of viral spread in the human body [129, 160]. Immunohistochemistry and electron microscopy are challenging to interpret in autopsy-derived samples, cannot be recommended for routine nor research detection of SARS-CoV-2, and should be used and interpreted with caution. Specifically, establishment of the staining protocol including adequate positive and negative controls is crucial [89, 149]. In electron microscopy, a variety of cellular structures can easily mimic virus particles; the search for these can be highly time-consuming and the risk of false-positive interpretation is high. Therefore, it can only be recommended for research and diagnostics when applied and interpreted by experts in the field [68, 89].
With the appropriate preanalytical elements for tissue sampling, autopsy material can be analyzed using a broad armamentarium of modern technologies, allowing deeper insights into pathomechanisms and alternative approaches for virus detection alike. These include single cell or single nucleus RNA sequencing [35], in situ hybridization [69, 108, 129, 160], viral genome sequencing [138], gene-expression analysis on the RNA [8] and proteome level [117], innovative tissue imaging, and 3D reconstruction [8, 42] (Fig. 1).
In addition to a large body of data on molecular changes in tissues from COVID-19 patients, application of the above-mentioned techniques in autopsy tissues has also demonstrated protein expression that enable viral infection of the host cell, particularly the viral-entry receptor angiotensin-converting enzyme 2 (ACE2) and transmembrane protease, serine 2 (TMPRSS2), the cell-membrane-based protease needed for conformational changes in the SARS-CoV-2 spike protein leading to fusion with the host cell plasma membrane [152]. These proteins were found in most human cell types and most tissues, including epithelial, endothelial, mesenchymal, immune, muscular, and neural cells [60, 67, 160, 175]. Notably, there are tissue and cell-specific differences regarding ACE2 and TMPRSS2 expression that further vary upon underlying diseases. Cardiomyocytes, for example, express significant levels of the ACE2 receptor [173] but lack TMPRSS2 [166], thus, questioning direct cardiomyocyte infection. Endothelial cells, on the other hand, do express both receptors and thus might facilitate cardiac involvement in COVID-19. However, ACE2 expression in a normal liver is minimal and mainly restricted to bile ducts, but is significantly upregulated in hepatocytes in liver fibrosis, hinting at a higher prevalence of liver involvement in cirrhotic patients [119]. In addition, virus factors also play a role in organ tropism and disease involvement. The currently dominant Omicron variant has a higher affinity for ACE2 compared to Delta and inefficiently utilizes the cellular protease TMPRSS2 that promotes cell entry via plasma membrane fusion; thus, it is dependent on cell entry via the so-called endocytic pathway. These alterations might ultimately change the tropism from ACE2/TMPRSS2 co-expressing cells to ACE2-only expressing cells resulting in a different clinical presentation and organ involvement [109]. These findings underline the crucial role of ongoing autoptic investigations in the ongoing pandemic (Figs. 2, 3, and 4).
Radiographic images of pulmonary changes in COVID-19 over time: A Typical radiographic presentation of acute respiratory distress syndrome with bipulmonary infiltrates, so called “white lung”. B CT-image of a 39-year-old previously healthy man after 30 days of extracorporeal membrane oxygenation (ECMO) treatment displaying bipulmonary ground glass opacities and basal infiltrates in line with (prolonged) ARDS. C CT-image of a 62-year-old man showing interstitial fibrosis and cystic remodeling including subpleural areas after 3 weeks of ECMO treatment. D CT-image of 34-year-old woman after 70 days of ECMO treatment with advanced pulmonary remodeling
Histologic findings in acute (panel A and B) and post-acute (panel C and D) COVID-19. A Acute COVID-19 pneumonia with diffuse alveolar damage (DAD) characterized by hyaline membranes (arrows), alveolar septae necrosis and lymphocytic inflammatory infiltrate. HE staining, Magnification 50×, scale bar 200 μm. B Acute COVID-19 pneumonia with a capillary hyaline microthrombus (arrow). HE staining, Magnification 600×, scale bar 10 μm. C, D Post-acute COVID-19 fibrotic remodeling with thickened alveolar septae and prominent type-II-pneumocyte hyperplasia. HE staining, panel C: magnification 20×, scale bar 500 μm, panel D: magnification 100×, scale bar 100 μm
Respiratory tract morphology of COVID-19 patients with Hematoxylin & Eosin staining and SARS-CoV-2 RNA detection by fluorescence in situ hybridization, corresponding areas from consecutive slides. A Respiratory epithelial cells lining the tracheal mucosa with SARS-CoV-2 RNA (arrowhead, green signal). B Pulmonary alveolar capillary endothelial cells with detection of SARS-CoV-2 RNA (arrowheads, green signal). C Pulmonary intraalveolar detached pneumocytes (arrowheads, green signal) and intraalveolar macrophages (arrow, green signal) with SARS-CoV-2 RNA. Scale bars: left column, scale bars = 100 μm, center column, scale bars = 50 μm, right column, scale bars = 10 μm
Organ specific involvement in COVID-19
Lung and respiratory system
COVID-19 is associated with a broad spectrum of clinical respiratory syndromes ranging from mild upper airway symptoms, to progressive viral pneumonia leading to labored breathing and progressive hypoxemia. Intensive care treatment with invasive mechanical ventilatory support is required in approximately 10% of hospitalized cases [39, 76]. Pulmonary manifestation is the leading cause of death in COVID-19 patients, usually presenting with peripheral lung ground-glass opacities on computed tomography (CT) [131, 145], similarly detectable with post-mortem imaging in autopsies [86]. Histologically, this corresponds to the morphological hallmark of acute respiratory distress syndrome (ARDS), i.e., diffuse alveolar damage (DAD) with edema, hemorrhage, hyaline membranes, and pneumocyte damage [77].
The diffuse alveolar damage in SARS-CoV-2 [18, 135], and similarly in SARS-CoV [15, 101, 115], is associated with a prominent formation of fibrin thrombi. SARS-CoV-2-infection leads to an angiocentric inflammation in COVID-19-induced respiratory failure with a greater number of ACE2-positive endothelial cells [8]. Increasing clinical evidence shows that endothelial dysfunction is a common denominator after SARS-COV-2 infection [71]. Histologically, this is characterized by acute vascular inflammation and perivascular T cell recruitment leading to swelling and disruption of the endothelial cell barriers and an anomalous microvascular architecture [8, 71]. The resulting vascular injury, especially of endothelial cells [147] leads to (micro-) thrombosis [94], vasoconstriction, and intussusceptive angiogenesis. Intussusceptive, i.e., non-sprouting, angiogenesis is a morphogenetic process of intravascular septation that produces two lumina from a single vessel within minutes, and has so far been identified in cancer [5], inflammatory and autoimmune diseases and tissue regeneration [3]. Intussusceptive angiogenesis was increased threefold in COVID-19 patients compared to influenza A virus subtype H1N1 (A/H1N1, “swine flu”) associated ARDS and manifested early (within days) but persisted over 20 days after initial infection.
Pulmonary thrombosis in large vessels was only seen in some of the cases, whereas most showed obstructed microvasculature by fibrin strands, activated platelets, deformed neutrophils, and neutrophil extracellular traps [95, 125]. Although virus-associated thrombotic microangiopathies have been described in several cardio-respiratory diseases, e.g., influenza [142] or Parvo-B19-viral myocarditis [9], in COVID-19 lungs these pathologies were reported up to nine times more often compared to influenza A/H1N1 lungs [8]. The extensive microangiopathy observed in COVID-19 patients leads to hypoxia, intrapulmonary shunting via upregulation of the bronchial vasa privata [42], and an overall increase of pulmonary vascular resistance [125]. These autopsy findings were a turning point in the understanding of COVID-19 as an angiocentric disease providing potential explanation for the drastic and rapid clinical deterioration of COVID-19 patients. The massive impact on the microvasculature also provided an explanation for the difficulties observed in the management of mechanical ventilation and the relatively high need for extracorporeal membrane oxygenation therapy [14]. These histology-based autopsy studies were the propelling factor for the adaptation of anti-thrombotic treatment in COVID-19. The above-mentioned structural adaptation of the microvascular network, transmigration of lymphocytes, a shift toward M2 macrophages [170], and the “cytokine storm” observed in COVID-19 patients is considered as a response to SARS-CoV-2-induced cellular damage. This is supported by multimodular detection of viral components in endothelial cells [4, 8], lymphatic cells [104], but also, in type 1 and 2 pneumocytes [19, 104]. Mucus plugging was noted at autopsy in COVID-19 patients with and without history of asthma [88, 110].
Prominent pulmonary microvascular alterations observed in COVID-19 resemble those of distinct fibrotic lung diseases [6, 41, 91]. These findings are in line with histologic changes reported through the course of disease, from acute DAD to organizing changes with interstitial myofibroblastic proliferations, septal collagen deposition, and development of loose alveolar plugs of fibroblastic tissue [17]. Although the pathologic mechanisms underlying fibrotic remodeling in pulmonary thrombotic or thromboembolic occlusions are still not fully understood, thrombo-fibrosis appears to be promoted by hypoxia-induced activation of endothelial cells, incorporation of mesenchymal progenitor and immune cells with subsequent scarring of the pulmonary parenchyma [2, 6, 7, 164].
As the nasopharyngeal and oropharyngeal tissues are the main entry and (early) replication sites of SARS-CoV-2 [159], it was shown that SARS-CoV-2 infects respiratory, olfactory, and paranasal sinus epithelia with tropism to ciliated mucosa [112], and the adjacent tissues, and can explain the reason why anosmia and dysgeusia are prevalent symptoms in infected patients [85]. A similar pathogenesis has been proposed in the salivary gland, in which SARS-CoV-2-infected glands show inflammation, injury of the glandular parenchyma, and abnormalities in the saliva [100, 140]. Other hypotheses proposed the vagus nerve as a virus pathway from the respiratory to the central nervous system [163].
Cardiovascular system
Acute cardiac involvement, i.e., elevated cardiac injury markers such as troponin, arrhythmia, lowered ejection fraction, ventricular dyskinesia, etc., has been variably reported from 16% [154] to 36% [78] of COVID-19 patients on clinical admission and up to 78% in one magnetic resonance imaging (MRI) cohort-study [130]. These frequencies are in line with those reported for severe lung disease caused by the closely related betacoronavirus SARS-CoV [167].
At the beginning of the COVID-19 pandemic—especially in children—a vasculitis-like symptom complex with occasional cardiac involvement, partly resembling Kawasaki disease, was reported and termed (pediatric) inflammatory multisystem syndrome (P)IMS [31, 53], however, so far, the concept of (P)IMS is still debated.
Current hypotheses on cardiac involvement include direct injury to the myocardium by a viral infection, given that cardiomyocytes express significant levels of ACE-2 [173]. However, cardiomyocytes lack TMPRSS2 [166], questioning relevant direct cardiomyocyte infection. Another explanation for the cardiac involvement is the systemic release of pro-inflammatory mediators, e.g., interleukins (IL-1, IL-6), tumor necrosis factor alpha (TNF-α), interferon-γ (IFN-γ), and macrophage inflammatory protein (MIP), often called the cytokine storm [27, 36]. The mechanisms of indirect myocardial injury include increased vascular wall permeability and myocardial edema, which are in line with observed wall thickening in ultrasound or MRI, and a patchy hypoxia-induced inflammatory response and cellular damage [168]. Evidence for a direct SARS-CoV-2-specific cardiac involvement is based on a series of case reports describing clinical (peri-) myocarditis [65, 70, 73, 82, 168]. In the majority of these studies, except two reports on lymphocytic myocarditis [37, 51], the histopathological assessment did not show typical signs of viral myocarditis according to the Dallas criteria, i.e., lymphocyte infiltration and myocyte necrosis. Instead, increased numbers of macrophages were observed, often termed as “borderline myocarditis” [11, 59, 144, 155]. The presence of SARS-CoV-2 RNA in cardiac tissue has been reported, but the extent and compartment specific involvement are still debated [11, 51, 144, 157]. In analogy to overt vascular involvement in the lungs, cardiac endothelialitis was proposed either via direct virus infection [49, 50, 144] or indirect mechanisms [16, 58, 122]. An autopsy study of 95 COVID-19 cases failed to demonstrate myocarditis, but showed relevant SARS-CoV-2 virus loads in 43% of all cases, where the virus was primarily located in interstitial cells. Cardiac virus replication was found in 15% of these cases [96]. On the other hand, SARS-CoV-2, particularly the Omicron variant, seems to be able to enter cells independently of TMPRSS2 by the so-called endocytic pathway [109]. There are single case reports of virus-shaped particles [11] in cardiomyocytes; thus, direct infection might be possible in a subset of cases. However, the clinicopathological evidence remains scarce [24]. Another finding in COVID-19 autopsies was cardiac amyloidosis. Cardiac amyloidosis is not uncommon in autopsies, particularly in elderly patients, and is often not associated with clinical symptoms [124]. A direct pathophysiological link between COVID-19 and cardiac amyloidosis is debated, but appears to be rather unlikely. However, cardiac amyloidosis might predispose COVID-19 patients to heart failure regarding the higher incidence of amyloidosis seen in this cohort [62].
Altogether, it is not yet clear whether cardiac involvement in COVID-19 follows the classical inflammatory pathogenesis observed in myocarditis caused by other viruses, such as influenza virus, SARS-CoV-1, or coxsackie virus or whether microvascular/angiogenic or inflammatory pathogenesis might rather be the driving forces in COVID-19 cardiac involvement.
Kidneys
Kidney involvement is among the most frequent and severe organ complications in severe COVID-19, mostly manifesting as acute kidney damage and being associated with unfavorable outcome [20]. Next to diabetes, cardiovascular disease, and chronic respiratory disease, chronic kidney disease is the most important underlying disease associated with risk of severe COVID-19 [30]. Besides, patients with chronic kidney disease, dialysis, or kidney transplant are particularly vulnerable to COVID-19 with high morbidity and mortality [74]. Renal SARS-CoV-2 tropism was first demonstrated in autopsies [129] and is associated with increased disease severity and acute kidney injury [20]. Affected kidneys in most cases show acute tubular damage, followed by collapsing glomerulopathy or, likely unspecific or secondary, focal segmental glomerulosclerosis [48]. Microthrombi or thrombotic microangiopathy were observed but are difficult to attribute directly to the SARS-CoV-2 infection [146]. Specific proximal tubular dysfunction has also been described [156]. So far, it is unclear to what extent the renal involvement is mediated via direct viral effects on kidney cells, secondary effects of a severe course of disease with cytokine storm and hypoxia, or a combination thereof.
Immune, lymphatic, and hematopoietic system
The inflammatory reaction in COVID-19 shows some differences to other viral lung diseases, such as influenza. The initial inflammatory response occurs earlier in patients infected by influenza while the intensity of inflammation is generally comparable. In contrast to influenza, one hallmark of COVID-19 is the missing or delayed limitation of the immune response due to prolonged viral persistence or impaired endothelial function and microcirculation resulting in increased risk of secondary complications and fibrotic remodeling [23]. The impaired immune response in severe COVID-19 seems to be the result of immature, dysfunctional neutrophils, and monocytes [136]. The high proportion of asymptomatic or mild disease courses in COVID-19 and the rapid immune response and production of antibodies following vaccination may be explained by cross-reactive immunity with other coronavirus strains [21, 99].
A common phenomenon in lymphatic tissues following viral infections is the expansion of the paracortical areas with extrafollicular activation of B cells caused by antigen-specific activated T cells. This extrafollicular B cell activation is the morphological correlate of rapid B cell responses and is accompanied by rather small, inactive germinal centers [22, 63]. Upon recognition of an antigen, B cells differentiate into short-lived antibody-forming cells, are active as frontline protectors, do not participate in class-switch, and are not able to differentiate to memory B cells [22, 26]. Lymph nodes of COVID-19 patients exhibit these features and are characterized by extrafollicular accumulation of plasmablasts [158, 171] and a decrease of memory B cells [61, 114].
Peripheral lymphopenia with loss of memory B cells is a typical feature of severe COVID-19, partly due to a redistribution of lymphocytes from lymphatic organs to the lung [33, 54, 102]. Lymphatic depletion may also be seen in the lymph nodes and the spleen due to a direct cytopathic effect of the virus [1]. In these patients, the absence of germinal center reaction is accompanied by a striking reduction in B cell lymphoma 6 protein (BCL6) positive germinal center B cells and also an early specific block in BCL6-positive T helper cell differentiation, together with an increase in cytokine-producing TH1 cells and aberrant extrafollicular TNF-α accumulation [75]. Also, a dominant population of CD8-positive T cells and a diffuse increase in M2 polarized macrophages have been described [61, 118].
Patients with a severe COVID-19 course exhibit a systemic inflammatory response with an altered pattern of inflammatory chemokines, cytokines, high ferritin levels, and IL-1/IL-6 pathway dysregulation. Although these features are not specific for COVID-19 [47, 98, 126], cytokine dysregulation plays an important role in the progression of severe COVID-19. In the context of the cytokine storm, hemophagocytosis in lymph nodes, spleen, and bone marrow have been put forward as possible histomorphologic features, although it is not observed universally [97, 174].
Secondary hemophagocytic lymphohistiocytosis, cytokine release syndrome, and macrophage activation syndrome are overlapping syndromes characterized by an activation of lymphocytes and macrophages with a subsequent excessive immune response, which leads to multiorgan damage. In addition, most reports describe an increase of bone marrow cellularity with left shift deviation. The increase in the total numbers of macrophages might be due to the prominent systemic inflammation, while the increase in CD8+ T cells is considered a direct effect of viral infection [38, 57, 107, 127]. However, histiocytic hyperplasia with secondary hemophagocytic lymphohistiocytosis was primarily seen in autopsy studies indicating terminal dysfunction, but not in those patients surviving the infection showing decreasing IL-6 levels [118, 126].
The concept of neutrophil extracellular trap formation (NETosis), followed by activation of the coagulation cascade, a process termed “immunothrombosis,” has been recognized as a pathogen eradication strategy that could play a central role inducing or exacerbating autoimmune or vascular pathologies in COVID-19 and other inflammatory conditions [116, 177].
Liver, pancreaticobiliary, and gastrointestinal system
Morphological alterations of the liver are quite common in living and deceased COVID-19 patients; however, they are mostly unspecific. Steatosis, mostly macrovesicular, is frequent and with a prevalence of 30–70% likely represents a preexistent finding and/or a possible result of hypoxia and drug-related adverse effects rather than a virus-induced pathology [46, 113]. Other changes, such as mild lobular hepatitis, centrilobular congestion and necrosis, platelet-fibrin microthrombi (15–70% of the cases), and cholestasis are likely related to the cytokine storm, the general hypoxia in COVID-19 patients, and septic complications [44, 46, 172]. Although in some studies viral RNA could be detected in a substantial number of liver samples [92], this result could not be confirmed by others [105].
Pancreatic enzyme elevation and acute pancreatitis have been described in SARS-CoV-2-infected patients, especially in severe and critical manifestations of COVID-19 [176]. While a potential causal relationship between viral infection and pancreaticobiliary organ damage is still not fully understood [32], it was shown that SARS-CoV-2 can infect and replicate in ex vivo in cultured human pancreatic islets, which was associated with morphological, transcriptional and functional changes [111, 161]. In autopsy studies, severe injury or remodeling of the pancreas during COVID-19 has not been reported so far [44]. Detailed examination, especially of the pancreas, is often hampered by its characteristic rapid post-mortem autolysis. A recently described severe manifestation of cholangiopathy, so severe that it might result in liver transplantation, most likely represents a variant of secondary sclerosing cholangitis in critically ill patients [45, 84].
Gastrointestinal symptoms such as anorexia, diarrhea, nausea, and vomiting have been reported in about 15% of patients with COVID-19 and some cases presenting with isolated gastrointestinal manifestation have been observed [56]. Viral RNA can be detected in fecal samples from patients with COVID-19, and viral RNA positivity can persist even after sputum samples become PCR negative [153]. Although fecal transmission was possible in a laboratory system as shown by infection of colon carcinoma cell lines (CACo) and gut organoids [93]. The true risk and relevance of SARS-CoV-2 transmission via a fecal-oral route remains unclear and seems to be of rather limited importance. Immunohistochemistry and single-cell transcriptome data revealed expression of ACE2 and TMPRSS2 in enterocytes of the small intestine and colon, most abundant in the ileum [60, 169]. Data of endoscopic samples or surgical specimens is sparse and so far, consists only of small case series or case reports. In COVID-19 patients with gastrointestinal symptoms, structural damage has been variable and ranges from limited and focal inflammation with interstitial edema accompanied by plasmacellular and lymphocytic infiltrates, up to substantial ulceration and necrosis of the mucosa. Notably, different studies confirmed viral RNA and antigens in intestinal epithelial cells as well as in macrophages and lymphocytes suggesting active SARS-CoV-2 replication in the intestine.
Nervous system and skeletal muscle
SARS-CoV-2 invasion in the central nervous system (CNS) occurs via the blood or nerves. A hematogenous route is supported by the fact that COVID-19 leads to viremia and SARS-CoV-2 targets brain endothelial cells [106, 108, 139]. Also, SARS-CoV-2 is not only transported through brain endothelial cells but also replicates in these [90]. The olfactory route and transport along vagal nerves have been suggested as a CNS entry port. Dysfunction of the olfactory system is a key symptom of COVID-19 and SARS-CoV-2 colonizes the nasal cavity. If SARS-CoV-2 additionally transits to CNS via olfactory and sensory nerve endings in the olfactory mucosa is debated with data in favor and opposing this concept [80, 108]. In COVID-19, there is vagal nerve dysfunction and SARS-CoV-2 viral proteins can be found in COVID-19 patients, thus a vagal route of SARS-CoV-2 CNS entry has to be considered [25, 52].
When considering the larger neuropathological studies [34, 106, 108, 132, 139], focal cerebral infarctions are seen in approximately 13% of autopsies. However, cerebral hypoxia in COVID-19 is not consistently defined so far. Global hypoxic-ischemic states, possibly resulting from respiratory failure in COVID-19, have to be distinguished from focal cerebral thromboembolic events and patients with systemic hypercoagulation syndrome. The cytokine storm results in a deterioration of the blood-brain barrier. In contrast to the microglial activation reported in many studies, only a few cases with encephalitis or meningitis have been described [29]. Microglia and astrocyte subpopulations associated with COVID-19 share features with those found in neurodegenerative diseases [137, 165], and synaptic signaling of upper-layer excitatory neurons, linked to cognitive function, are preferentially affected in COVID-19 [165]. Since patients with COVID-19 frequently succumb to bacterial superinfection, sepsis, ventilation, and polypharmacotherapy as well as long-term intensive care unit (ICU) stays with an isolating environment/deafferentation, comparative studies with similar clinical pictures will be required to identify COVID-19 specific encephalopathic changes. Up to now, there is no evidence of a CNS reservoir for viable SARS-CoV-2 virus [90], even though viral RNA or protein may be found at the CNS barriers. A future challenge is the identification of how the CNS contributes to symptoms of the post-COVID syndrome, such as fatigue, headaches, anosmia, muscle weakness, and cognitive dysfunction; it is still very early days. Although there are already large studies on post-COVID, with thousands of study participants [143], the time span is not sufficient to be able to say how long-term consequences will develop. The patients presenting with neurological signs of post-COVID are probably a heterogenous group with some having a dysregulated microbiome, others alteration of the vascular system or dysfunctional brainstem signaling, as well as others with ongoing low-level inflammation or autoimmunity triggered in susceptible hosts [128].
COVID-19 can affect the peripheral neural system meeting diagnostic criteria for acute polyradiculoneuropathy. Early in the course of the pandemic, it was suggested that similar to other viruses, SARS-CoV-2 might directly infect peripheral neurons or trigger Guillain-Barré syndrome (GBS) [55, 141]. However, later epidemiological studies found no link of GBS and SARS-CoV-2 [79]. Similarly, direct infection of skeletal muscle fibers or autoimmune myopathy/myositis triggered by SARS-CoV-2 have been suggested to cause myalgia, muscle weakness, and elevated creatine kinase (CK) levels that are frequently observed in COVID-19 patients [55] and were more pronounced in critically ill patients, compared to mildly affected individuals [103]. A post-mortem case control study could not detect signs of infection of skeletal muscle, but identified myositis with different levels of severity in COVID-19. Inflammation of the muscle was correlating with disease duration, supporting a postinfectious, immune-mediated pathology [12].
Endocrine organs
Thyroid dysfunction in COVID-19 occurs frequently, ranging from thyrotoxicosis, observed in 15 to 20% of patients, to hypothyroidism and nonthyroidal illness syndrome (NTIS) [28, 81]. The causes of thyroid impairment remain elusive and a multitude of putative explanations has been considered. Direct infection of thyroid follicular epithelium appears as a putative pathogenic process, given the high ACE2 expression compared, e.g., to the lungs [134]. However, studies analyzing SARS-COV-2 RNA or protein yielded contradictory results, with some studies reporting the presence of viral components [160], while others do not [28, 134]. Histologic analyses of tissues obtained from autopsies yielded conflicting results and the thyroid, as well as other endocrine organs, have not yet been sufficiently investigated in most autopsy studies [121, 134]. The presence of interstitial lymphoid infiltrates in the thyroid, commonly reported in COVID-19, is difficult to differentiate from pre-existing thyroiditis and is thus of unclear significance [134]. Regardless of the underlying pathogenesis, the degree of thyroid dysfunction directly correlates with the patients’ prognosis. The current knowledge concerning the effect of SARS-CoV-2 on other endocrine organs, such as the pituitary gland, parathyroid, adrenals, gonads, endocrine pancreas, and the diffuse neuroendocrine system, remains rudimentary [121].
Superinfections
SARS-CoV-2 induces epithelial damage, leading to impaired epithelial barrier function and consecutive invasion by secondary pathogens [123]. In a clinical context, invasive ventilation and other complications (e.g., sepsis and multi organ failure) can further aggravate those alterations. Overall, however, superinfections are not increased in the early phase of COVID-19 disease, occur with similar frequency as in other systemic viral infections (e.g., influenza), and are usually acquired via the respiratory system [72]. The combination of downregulation of proteins playing a key role for the innate immune system, e.g., the toll-like receptors, and corticosteroid treatment in severe COVID-19, might contribute to the increased susceptibility for life-threatening mycosis and bacterial superinfections with sepsis, especially in patients with long-term intensive care treatment for SARS-CoV-2 infection [10, 44, 61, 87, 120].
Open questions, limitations, and conclusions
Since the start of pandemics, our understanding of COVID-19 has greatly increased. Besides tremendous efforts in clinical and virological research, data derived from clinical and forensic autopsies represent an important pillar of the rapid increase in knowledge. Most autopsy-based findings provide information on the severe and later stages of the disease and help to delineate discrepancies between clinical presentation and actual organ involvement, acting as key drivers for the development of novel diagnostic and therapeutic tools, as well as feedback on the individual patient.
The value and importance of autopsies increased tremendously during the COVID-19 pandemic. This rejuvenated the field of clinical autopsies, which was slowly becoming an ever-diminishing branch of pathology. It also led to a substantial increase in recognition of the broad applicability of most novel technologies of biomedical research, even in post-mortem and autopsy tissues, resulting in data going far beyond the classical histopathological analyses. The use of these methods predicts a very clear-cut future for the further development of autopsies beyond COVID-19. However, it also became apparent that for many biomaterials and data, we are currently lacking comparable or matched control or reference autopsy cohorts. An increment of clinical autopsies is a mandatory stepping-stone as preparation for the next pandemic—and for the interim period—to gather reference values and biomaterials and enable optimal training of surgical and forensic pathologists.
The current immense knowledge on COVID-19 was only possible via interdisciplinary, systematic, and holistic analysis of findings from clinical observations, imaging, laboratory diagnostics, and autopsies. However, there is a considerable risk of over-interpretation of (unspecific) findings. So far, no other disease has been studied as extensively in such a short period of time and with such intensity by dozens of specialists. It is not surprising that many of the current studies, not only on autopsies, have limitations that have to be kept in mind. These include single-center designs, often with limited numbers of cases, and, importantly, lacking control populations. Bringing together national and international collaborative initiatives seems inevitable to effectively tackle the COVID-19 pandemic and serve as a preparedness infrastructure for the future. The German initiatives, the Network for Autopsies in Pandemics (DEFEAT PANDEMIcs) and the COVID-19 Autopsy Registry (DeRegCOVID), are national examples, demonstrating that joint approaches are feasible, effective, and can serve as a blueprint for other countries. These examples should serve as an initiator for international initiatives, which are being conducted in clinical medicine, such as the Solidarity study of the WHO (World Health Organization), but not yet in pathology. In addition, the network operates in an integrative, participatory, and interdisciplinary manner including virologists, biologists, and clinicians and is open to all new collaborations and complementary expertise.
Important open questions for future research include the (long-term) consequences of COVID-19, especially post-COVID conditions (“long COVID”). Autopsies and molecular methods will likely help to unravel the mechanisms of these conditions in specific organ systems. This might include viral persistence in tissues, aberrant immune phenotypes of resident cells, or even auto-immune processes. Additional open questions also include the (immune) pathological role of virus variants. The DEFEAT PANDEMIcs consortium recently started a detailed post-mortem virus sequencing in fatal cases. First autopsy reports indicate no differences in virus loads and organotropism between the wild type and the first relevant virus variant, B.1.1.7 [64].
In conclusion, the direct pulmonary effects of SARS-CoV-2 infection are well established and are the leading pathology in COVID-19, with data indicating disease specific mechanisms such as angiocentric inflammation with systemic (micro-) thrombosis and neoangiogenesis via intussusception. In comparison, it remains unclear to what extent virus invasion or replication might contribute to extrapulmonary organ injury, e.g., in the kidney. Such pathologies, including the observed hypercoagulability, might represent indirect effects of severe disease course and multi-organ failure, or the combination of both direct and indirect effects. Future joint studies integrating the knowledge and material acquired by autopsies, together with clinical studies, preclinical models, and humanized organoid systems, are needed to dissect these emerging questions.
References
Abdullaev A, Odilov A, Ershler M, Volkov A, Lipina T, Gasanova T et al (2021) Viral load and patterns of SARS-CoV-2 dissemination to the lungs, mediastinal lymph nodes, and spleen of patients with COVID-19 associated lymphopenia. Viruses 13(7):1410. https://doi.org/10.3390/v13071410
Ackermann M, Gaumann A, Mentzer SJ, Hinrichs JB, Warnecke G, Hoeper MM, Braubach P, Kuehnel M, Maegel L, Jonigk D (2017) Plexiform vasculopathy in chronic thromboembolic pulmonary hypertension. Am J Respir Crit Care Med 196:e48–e51. https://doi.org/10.1164/rccm.201703-0591IM
Ackermann M, Houdek JP, Gibney BC, Ysasi A, Wagner W, Belle J, Schittny JC, Enzmann F, Tsuda A, Mentzer SJ, Konerding MA (2014) Sprouting and intussusceptive angiogenesis in postpneumonectomy lung growth: mechanisms of alveolar neovascularization. Angiogenesis 17:541–551. https://doi.org/10.1007/s10456-013-9399-9
Ackermann M, Mentzer SJ, Jonigk D (2020) Pulmonary vascular pathology in Covid-19. Reply N Engl J Med 383:888–889. https://doi.org/10.1056/NEJMc2022068
Ackermann M, Morse BA, Delventhal V, Carvajal IM, Konerding MA (2012) Anti-VEGFR2 and anti-IGF-1R-Adnectins inhibit Ewing’s sarcoma A673-xenograft growth and normalize tumor vascular architecture. Angiogenesis 15:685–695. https://doi.org/10.1007/s10456-012-9294-9
Ackermann M, Stark H, Neubert L, Schubert S, Borchert P, Linz F, Wagner WL, Stiller W, Wielputz M, Hoefer A, Haverich A, Mentzer SJ, Shah HR, Welte T, Kuehnel M, Jonigk D (2020) Morphomolecular motifs of pulmonary neoangiogenesis in interstitial lung diseases Eur Respir J 55. https://doi.org/10.1183/13993003.00933-2019
Ackermann M, Tsuda A, Secomb TW, Mentzer SJ, Konerding MA (2013) Intussusceptive remodeling of vascular branch angles in chemically-induced murine colitis. Microvasc Res 87:75–82. https://doi.org/10.1016/j.mvr.2013.02.002
Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H, Tzankov A, Li WW, Li VW, Mentzer SJ, Jonigk D (2020) Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19 N Engl J Med 383:120-128. https://doi.org/10.1056/NEJMoa2015432
Ackermann M, Wagner WL, Rellecke P, Akhyari P, Boeken U, Reinecke P (2020) Parvovirus B19-induced angiogenesis in fulminant myocarditis. Eur Heart J 41:1309. https://doi.org/10.1093/eurheartj/ehaa092
Alanio A, Delliere S, Fodil S, Bretagne S, Megarbane B (2020) Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respir Med 8:e48–e49. https://doi.org/10.1016/S2213-2600(20)30237-X
Albert CL, Carmona-Rubio AE, Weiss AJ, Procop GG, Starling RC, Rodriguez ER (2020) The enemy within: sudden-onset reversible cardiogenic shock with biopsy-proven cardiac myocyte infection by severe acute respiratory syndrome Coronavirus 2. Circulation 142(19):1865–1870. https://doi.org/10.1161/CIRCULATIONAHA.120.050097
Aschman T, Schneider J, Greuel S, Meinhardt J, Streit S, Goebel HH, Buttnerova I, Elezkurtaj S, Scheibe F, Radke J, Meisel C, Drosten C, Radbruch H, Heppner FL, Corman VM, Stenzel W (2021) Association between SARS-CoV-2 infection and immune-mediated myopathy in patients who have died. JAMA Neurol 78:948–960. https://doi.org/10.1001/jamaneurol.2021.2004
Bacon ER, Ihle K, Lee PP, Waisman JR (2020) Building a rapid autopsy program—a step-by-step logistics guide. Translational Medicine Communications 5. https://doi.org/10.1186/s41231-020-00074-x
Barbaro RP, MacLaren G, Boonstra PS, Iwashyna TJ, Slutsky AS, Fan E, Bartlett RH, Tonna JE, Hyslop R, Fanning JJ, Rycus PT, Hyer SJ, Anders MM, Agerstrand CL, Hryniewicz K, Diaz R, Lorusso R, Combes A, Brodie D, Extracorporeal Life Support O (2020) Extracorporeal membrane oxygenation support in COVID-19: an international cohort study of the Extracorporeal Life Support Organization registry. Lancet 396:1071–1078. https://doi.org/10.1016/S0140-6736(20)32008-0
Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S (2020) COVID-19 Autopsies, Oklahoma, USA. Am J Clin Pathol 153:725–733. https://doi.org/10.1093/ajcp/aqaa062
Bernard I, Limonta D, Mahal LK, Hobman TC (2020) Endothelium infection and dysregulation by SARS-CoV-2: evidence and caveats in COVID-19. Viruses 13(1):29. https://doi.org/10.3390/v13010029
Bosmuller H, Matter M, Fend F, Tzankov A (2021) The pulmonary pathology of COVID-19. Virchows Arch 478:137–150. https://doi.org/10.1007/s00428-021-03053-1
Bosmuller H, Traxler S, Bitzer M, Haberle H, Raiser W, Nann D, Frauenfeld L, Vogelsberg A, Klingel K, Fend F (2020) The evolution of pulmonary pathology in fatal COVID-19 disease: an autopsy study with clinical correlation. Virchows Arch 477:349–357. https://doi.org/10.1007/s00428-020-02881-x
Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Deutsch G, Lacy JM, Williams T, Yarid N, Marshall DA (2020) Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet 396:320–332. https://doi.org/10.1016/S0140-6736(20)31305-2
Braun F, Lutgehetmann M, Pfefferle S, Wong MN, Carsten A, Lindenmeyer MT, Norz D, Heinrich F, Meissner K, Wichmann D, Kluge S, Gross O, Pueschel K, Schroder AS, Edler C, Aepfelbacher M, Puelles VG, Huber TB (2020) SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet 396:597–598. https://doi.org/10.1016/S0140-6736(20)31759-1
Braun J, Loyal L, Frentsch M, Wendisch D, Georg P, Kurth F, Hippenstiel S, Dingeldey M, Kruse B, Fauchere F, Baysal E, Mangold M, Henze L, Lauster R, Mall MA, Beyer K, Rohmel J, Voigt S, Schmitz J, Miltenyi S, Demuth I, Muller MA, Hocke A, Witzenrath M, Suttorp N, Kern F, Reimer U, Wenschuh H, Drosten C, Corman VM, Giesecke-Thiel C, Sander LE, Thiel A (2020) SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature 587:270–274. https://doi.org/10.1038/s41586-020-2598-9
Brighenti A, Andrulis M, Geissinger E, Roth S, Muller-Hermelink HK, Rudiger T (2005) Extrafollicular proliferation of B cells in the absence of follicular hyperplasia: a distinct reaction pattern in lymph nodes correlated with primary or recall type responses. Histopathology 47:90–100. https://doi.org/10.1111/j.1365-2559.2005.02173.x
Budinger GRS, Misharin AV, Ridge KM, Singer BD, Wunderink RG (2021) Distinctive features of severe SARS-CoV-2 pneumonia J Clin Invest 131. https://doi.org/10.1172/JCI149412
Buja LM, Stone JR (2021) A novel coronavirus meets the cardiovascular system: Society for Cardiovascular Pathology Symposium 2021. Cardiovasc Pathol 53:107336. https://doi.org/10.1016/j.carpath.2021.107336
Bulfamante G, Bocci T, Falleni M, Campiglio L, Coppola S, Tosi D, Chiumello D, Priori A (2021) Brainstem neuropathology in two cases of COVID-19: SARS-CoV-2 trafficking between brain and lung. J Neurol 268:4486–4491. https://doi.org/10.1007/s00415-021-10604-8
Chappell CP, Draves KE, Giltiay NV, Clark EA (2012) Extrafollicular B cell activation by marginal zone dendritic cells drives T cell-dependent antibody responses. J Exp Med 209:1825–1840. https://doi.org/10.1084/jem.20120774
Chen C, Zhou Y, Wang DW (2020) SARS-CoV-2: a potential novel etiology of fulminant myocarditis. Herz 45:230–232. https://doi.org/10.1007/s00059-020-04909-z
Chen W, Tian Y, Li Z, Zhu J, Wei T, Lei J (2021) Potential interaction between SARS-CoV-2 and thyroid: a review Endocrinology 162 https://doi.org/10.1210/endocr/bqab004
Chou SH, Beghi E, Helbok R, Moro E, Sampson J, Altamirano V, Mainali S, Bassetti C, Suarez JI, McNett M, Consortium GC-N, Consortium E (2021) Global incidence of neurological manifestations among patients hospitalized with COVID-19-A report for the GCS-NeuroCOVID consortium and the ENERGY Consortium. JAMA Netw Open 4:e2112131 https://doi.org/10.1001/jamanetworkopen.2021.12131
Clark A, Jit M, Warren-Gash C, Guthrie B, Wang HHX, Mercer SW, Sanderson C, McKee M, Troeger C, Ong KL, Checchi F, Perel P, Joseph S, Gibbs HP, Banerjee A, Eggo RM, COVI CMMID, (2020) Global, regional, and national estimates of the population at increased risk of severe COVID-19 due to underlying health conditions in 2020: a modelling study. Lancet Glob Health 8:1003–1017. https://doi.org/10.1016/S2214-109x(20)30264-3
Davies P, Evans C, Kanthimathinathan HK, Lillie J, Brierley J, Waters G, Johnson M, Griffiths B, du Pre P, Mohammad Z, Deep A, Playfor S, Singh D, Inwald D, Jardine M, Ross O, Shetty N, Worrall M, Sinha R, Koul A, Whittaker E, Vyas H, Scholefield BR, Ramnarayan P (2020) Intensive care admissions of children with paediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 (PIMS-TS) in the UK: a multicentre observational study. Lancet Child Adolesc Health 4:669–677. https://doi.org/10.1016/S2352-4642(20)30215-7
de Madaria E, Capurso G (2021) COVID-19 and acute pancreatitis: examining the causality. Nat Rev Gastroenterol Hepatol 18:3–4. https://doi.org/10.1038/s41575-020-00389-y
De Biasi S, Lo Tartaro D, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, Mattioli M, Paolini A, Gozzi L, Jaacoub D, Faltoni M, Volpi S, Milic J, Sita M, Sarti M, Pucillo C, Girardis M, Guaraldi G, Mussini C, Cossarizza A (2020) Expansion of plasmablasts and loss of memory B cells in peripheral blood from COVID-19 patients with pneumonia. Eur J Immunol 50:1283–1294. https://doi.org/10.1002/eji.202048838
Deigendesch N, Sironi L, Kutza M, Wischnewski S, Fuchs V, Hench J, Frank A, Nienhold R, Mertz KD, Cathomas G, Matter MS, Siegemund M, Tolnay M, Schirmer L, Probstel AK, Tzankov A, Frank S (2020) Correlates of critical illness-related encephalopathy predominate postmortem COVID-19 neuropathology. Acta Neuropathol 140:583–586. https://doi.org/10.1007/s00401-020-02213-y
Delorey TM, Ziegler CGK, Heimberg G, Normand R, Yang Y, Segerstolpe A, Abbondanza D, Fleming SJ, Subramanian A, Montoro DT, Jagadeesh KA, Dey KK, Sen P, Slyper M, Pita-Juarez YH, Phillips D, Biermann J, Bloom-Ackermann Z, Barkas N, Ganna A, Gomez J, Melms JC, Katsyv I, Normandin E, Naderi P, Popov YV, Raju SS, Niezen S, Tsai LT, Siddle KJ, Sud M, Tran VM, Vellarikkal SK, Wang Y, Amir-Zilberstein L, Atri DS, Beechem J, Brook OR, Chen J, Divakar P, Dorceus P, Engreitz JM, Essene A, Fitzgerald DM, Fropf R, Gazal S, Gould J, Grzyb J, Harvey T, Hecht J, Hether T, Jane-Valbuena J, Leney-Greene M, Ma H, McCabe C, McLoughlin DE, Miller EM, Muus C, Niemi M, Padera R, Pan L, Pant D, Pe’er C, Pfiffner-Borges J, Pinto CJ, Plaisted J, Reeves J, Ross M, Rudy M, Rueckert EH, Siciliano M, Sturm A, Todres E, Waghray A, Warren S, Zhang S, Zollinger DR, Cosimi L, Gupta RM, Hacohen N, Hibshoosh H, Hide W, Price AL, Rajagopal J, Tata PR, Riedel S, Szabo G, Tickle TL, Ellinor PT, Hung D, Sabeti PC, Novak R, Rogers R, Ingber DE, Jiang ZG, Juric D, Babadi M, Farhi SL, Izar B, Stone JR, Vlachos IS, Solomon IH, Ashenberg O, Porter CBM, Li B, Shalek AK, Villani AC, Rozenblatt-Rosen O, Regev A (2021) COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595:107–113. https://doi.org/10.1038/s41586-021-03570-8
Deng Q, Hu B, Zhang Y, Wang H, Zhou X, Hu W, Cheng Y, Yan J, Ping H, Zhou Q (2020) Suspected myocardial injury in patients with COVID-19: evidence from front-line clinical observation in Wuhan, China. Int J Cardiol 311:116–121. https://doi.org/10.1016/j.ijcard.2020.03.087
Dettmeyer R, Lasczkowski G, Weber A, Wolter T, Kernbach-Wighton G (2020) [Histopathological findings following SARS-CoV-2 infection with and without treatment-Report of three autopsies] Rechtsmedizin (Berl):1–8 https://doi.org/10.1007/s00194-020-00408-x
Dewaele K, Claeys R (2020) Hemophagocytic lymphohistiocytosis in SARS-CoV-2 infection. Blood 135:2323. https://doi.org/10.1182/blood.2020006505
Docherty AB, Harrison EM, Green CA, Hardwick HE, Pius R, Norman L, Holden KA, Read JM, Dondelinger F, Carson G, Merson L, Lee J, Plotkin D, Sigfrid L, Halpin S, Jackson C, Gamble C, Horby PW, Nguyen-Van-Tam JS, Ho A, Russell CD, Dunning J, Openshaw PJ, Baillie JK, Semple MG, investigators IC, (2020) Features of 20 133 UK patients in hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: prospective observational cohort study. BMJ 369:m1985. https://doi.org/10.1136/bmj.m1985
Duarte-Neto AN, Monteiro RAA, da Silva LFF, Malheiros D, de Oliveira EP, Theodoro-Filho J, Pinho JRR, Gomes-Gouvea MS, Salles APM, de Oliveira IRS, Mauad T, Saldiva PHN, Dolhnikoff M (2020) Pulmonary and systemic involvement in COVID-19 patients assessed with ultrasound-guided minimally invasive autopsy. Histopathology 77:186–197. https://doi.org/10.1111/his.14160
Ebina M, Shimizukawa M, Shibata N, Kimura Y, Suzuki T, Endo M, Sasano H, Kondo T, Nukiwa T (2004) Heterogeneous increase in CD34-positive alveolar capillaries in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 169:1203–1208. https://doi.org/10.1164/rccm.200308-1111OC
Eckermann M, Frohn J, Reichardt M, Osterhoff M, Sprung M, Westermeier F, Tzankov A, Werlein C, Kuhnel M, Jonigk D, Salditt T (2020) 3D virtual pathohistology of lung tissue from Covid-19 patients based on phase contrast X-ray tomography Elife 9. https://doi.org/10.7554/eLife.60408
Edler C, Schroder AS, Aepfelbacher M, Fitzek A, Heinemann A, Heinrich F, Klein A, Langenwalder F, Lutgehetmann M, Meissner K, Puschel K, Schadler J, Steurer S, Mushumba H, Sperhake JP (2020) Dying with SARS-CoV-2 infection-an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int J Legal Med 134:1275–1284. https://doi.org/10.1007/s00414-020-02317-w
Evert K, Dienemann T, Brochhausen C, Lunz D, Lubnow M, Ritzka M, Keil F, Trummer M, Scheiter A, Salzberger B, Reischl U, Boor P, Gessner A, Jantsch J, Calvisi DF, Evert M, Schmidt B, Simon M (2021) Autopsy findings after long-term treatment of COVID-19 patients with microbiological correlation. Virchows Arch 479:97–108. https://doi.org/10.1007/s00428-020-03014-0
Faruqui S, Okoli FC, Olsen SK, Feldman DM, Kalia HS, Park JS, Stanca CM, Figueroa Diaz V, Yuan S, Dagher NN, Sarkar SA, Theise ND, Kim S, Shanbhogue K, Jacobson IM (2021) Cholangiopathy after severe COVID-19: clinical features and prognostic implications. Am J Gastroenterol 116:1414–1425. https://doi.org/10.14309/ajg.0000000000001264
Fassan M, Mescoli C, Sbaraglia M, Guzzardo V, Russo FP, Fabris R, Trevenzoli M, Pelizzaro F, Cattelan AM, Basso C, Navalesi P, Farinati F, Vettor R, Dei Tos AP (2021) Liver histopathology in COVID-19 patients: a mono-Institutional series of liver biopsies and autopsy specimens. Pathol Res Pract 221:153451. https://doi.org/10.1016/j.prp.2021.153451
Feld J, Tremblay D, Thibaud S, Kessler A, Naymagon L (2020) Ferritin levels in patients with COVID-19: A poor predictor of mortality and hemophagocytic lymphohistiocytosis. Int J Lab Hematol 42:773–779. https://doi.org/10.1111/ijlh.13309
Ferlicot S, Jamme M, Gaillard F, Oniszczuk J, Couturier A, May O, Grunenwald A, Sannier A, Moktefi A, Le Monnier O, Petit-Hoang C, Maroun N, Brodin-Sartorius A, Michon A, Dobosziewicz H, Andreelli F, Guillet M, Izzedine H, Richard C, Dekeyser M, Arrestier R, Sthele T, Lefevre E, Mathian A, Legendre C, Mussini C, Verpont MC, Pallet N, Amoura Z, Essig M, Snanoudj R, Brocheriou-Spelle I, Francois H, Belenfant X, Geri G, Daugas E, Audard V, Buob D, Massy ZA, Zaidan M, collaboration AP-HUIC-r (2021) The spectrum of kidney biopsies in hospitalized patients with COVID-19, acute kidney injury, and/or proteinuria. Nephrol Dial Transplant https://doi.org/10.1093/ndt/gfab042
Fox SE, Lameira FS, Rinker EB, Vander Heide RS (2020) Cardiac endotheliitis and multisystem inflammatory syndrome after COVID-19. Ann Intern Med 173:1025–1027. https://doi.org/10.7326/L20-0882
Fox SE, Li G, Akmatbekov A, Harbert JL, Lameira FS, Brown JQ, Vander Heide RS (2020) Unexpected features of cardiac pathology in COVID-19 infection circulation 142:1123–1125 https://doi.org/10.1161/CIRCULATIONAHA.120.049465
Gauchotte G, Venard V, Segondy M, Cadoz C, Esposito-Fava A, Barraud D, Louis G (2021) SARS-Cov-2 fulminant myocarditis: an autopsy and histopathological case study. Int J Legal Med 135:577–581. https://doi.org/10.1007/s00414-020-02500-z
Glatzel M, Hagel C, Matschke J, Sperhake J, Deigendesch N, Tzankov A, Frank S (2021) Neuropathology associated with SARS-CoV-2 infection. Lancet 397:276. https://doi.org/10.1016/S0140-6736(21)00098-2
Grimaud M, Starck J, Levy M, Marais C, Chareyre J, Khraiche D, Leruez-Ville M, Quartier P, Leger PL, Geslain G, Semaan N, Moulin F, Bendavid M, Jean S, Poncelet G, Renolleau S, Oualha M (2020) Acute myocarditis and multisystem inflammatory emerging disease following SARS-CoV-2 infection in critically ill children. Ann Intensive Care 10:69. https://doi.org/10.1186/s13613-020-00690-8
Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for C (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720 https://doi.org/10.1056/NEJMoa2002032
Guidon AC, Amato AA (2020) COVID-19 and neuromuscular disorders. Neurology 94:959–969. https://doi.org/10.1212/WNL.0000000000009566
Guo M, Tao W, Flavell RA, Zhu S (2021) Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol 18:269–283. https://doi.org/10.1038/s41575-021-00416-6
Gupta A, Weitzman S, Abdelhaleem M (2008) The role of hemophagocytosis in bone marrow aspirates in the diagnosis of hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 50:192–194. https://doi.org/10.1002/pbc.21441
Gustine JN, Jones D (2021) Immunopathology of hyperinflammation in COVID-19. Am J Pathol 191:4–17. https://doi.org/10.1016/j.ajpath.2020.08.009
Halushka MK, Vander Heide RS (2021) Myocarditis is rare in COVID-19 autopsies: cardiovascular findings across 277 postmortem examinations. Cardiovasc Pathol 50:107300. https://doi.org/10.1016/j.carpath.2020.107300
Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203:631–637. https://doi.org/10.1002/path.1570
Haslbauer JD, Matter MS, Stalder AK, Tzankov A (2021) Histomorphological patterns of regional lymph nodes in COVID-19 lungs. Pathologe https://doi.org/10.1007/s00292-021-00945-6
Haslbauer JD, Tzankov A, Mertz KD, Schwab N, Nienhold R, Twerenbold R, Leibundgut G, Stalder AK, Matter M, Glatz K (2021) Characterisation of cardiac pathology in 23 autopsies of lethal COVID-19. J Pathol Clin Res 7:326–337. https://doi.org/10.1002/cjp2.212
Haslbauer JD, Zinner C, Stalder AK, Schneeberger J, Menter T, Bassetti S, Mertz KD, Went P, Matter MS, Tzankov A (2021) Vascular damage, Thromboinflammation, Plasmablast activation, T-cell dysregulation and pathological histiocytic response in pulmonary draining lymph nodes of COVID-19. Front Immunol 12:763098. https://doi.org/10.3389/fimmu.2021.763098
Heinrich F, Meissner K, Langenwalder F, Puschel K, Norz D, Hoffmann A, Lutgehetmann M, Aepfelbacher M, Bibiza-Freiwald E, Pfefferle S, Heinemann A (2021) Postmortem stability of SARS-CoV-2 in nasopharyngeal mucosa Emerg Infect Dis 27. https://doi.org/10.3201/eid2701.203112
Henwood M, Lake D, Allen F, Sange M (2021) Myocarditis in SARS-CoV-2 negative patients with suspected preceding infection BMJ Case Rep 14https://doi.org/10.1136/bcr-2020-239513
Hirschbuhl K, Schaller T, Kling E, Markl B, Claus R (2020) Autopsy of patients with COVID-19: a balance of fear and curiosity. Pathol Res Pract 216:153039. https://doi.org/10.1016/j.prp.2020.153039
Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271–280 e8. https://doi.org/10.1016/j.cell.2020.02.052
Hopfer H, Herzig MC, Gosert R, Menter T, Hench J, Tzankov A, Hirsch HH, Miller SE (2021) Hunting coronavirus by transmission electron microscopy—a guide to SARS-CoV-2-associated ultrastructural pathology in COVID-19 tissues. Histopathology 78:358–370. https://doi.org/10.1111/his.14264
Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH 3rd et al (2020) SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 82(2):429–446 e14. https://doi.org/10.1016/j.cell.2020.05.042
Hu H, Ma F, Wei X, Fang Y (2021) Coronavirus fulminant myocarditis treated with glucocorticoid and human immunoglobulin. Eur Heart J 42:206. https://doi.org/10.1093/eurheartj/ehaa190
Huertas A, Montani D, Savale L, Pichon J, Tu L, Parent F, Guignabert C, Humbert M (2020) Endothelial cell dysfunction: a major player in SARS-CoV-2 infection (COVID-19)? Eur Respir J 56. https://doi.org/10.1183/13993003.01634-2020
Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP (2020) Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect 26:1395–1399. https://doi.org/10.1016/j.cmi.2020.06.025
Inciardi RM, Lupi L, Zaccone G, Italia L, Raffo M, Tomasoni D, Cani DS, Cerini M, Farina D, Gavazzi E, Maroldi R, Adamo M, Ammirati E, Sinagra G, Lombardi CM, Metra M (2020) Cardiac involvement in a patient with coronavirus disease 2019 (COVID-19). JAMA Cardiol 5:819–824. https://doi.org/10.1001/jamacardio.2020.1096
Jager KJ, Kramer A, Chesnaye NC, Couchoud C, Sanchez-Alvarez JE, Garneata L, Collart F, Hemmelder MH, Ambuhl P, Kerschbaum J, Legeai C, Del Pino YPMD, Mircescu G, Mazzoleni L, Hoekstra T, Winzeler R, Mayer G, Stel VS, Wanner C, Zoccali C, Massy ZA (2020) Results from the ERA-EDTA Registry indicate a high mortality due to COVID-19 in dialysis patients and kidney transplant recipients across Europe. Kidney Int 98:1540–1548. https://doi.org/10.1016/j.kint.2020.09.006
Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, Piechocka-Trocha A, Lefteri K, Osborn M, Bals J, Bartsch YC, Bonheur N, Caradonna TM, Chevalier J, Chowdhury F, Diefenbach TJ, Einkauf K, Fallon J, Feldman J, Finn KK, Garcia-Broncano P, Hartana CA, Hauser BM, Jiang C, Kaplonek P, Karpell M, Koscher EC, Lian X, Liu H, Liu J, Ly NL, Michell AR, Rassadkina Y, Seiger K, Sessa L, Shin S, Singh N, Sun W, Sun X, Ticheli HJ, Waring MT, Zhu AL, Alter G, Li JZ, Lingwood D, Schmidt AG, Lichterfeld M, Walker BD, Yu XG, Padera RF, Jr., Pillai S, Massachusetts Consortium on Pathogen Readiness Specimen Working G (2020) Loss of Bcl-6-expressing T follicular helper cells and Germinal centers in COVID-19. Cell 183:143–157 e113 https://doi.org/10.1016/j.cell.2020.08.025
Karagiannidis C, Mostert C, Hentschker C, Voshaar T, Malzahn J, Schillinger G, Klauber J, Janssens U, Marx G, Weber-Carstens S, Kluge S, Pfeifer M, Grabenhenrich L, Welte T, Busse R (2020) Case characteristics, resource use, and outcomes of 10 021 patients with COVID-19 admitted to 920 German hospitals: an observational study. Lancet Resp Med 8:853–862. https://doi.org/10.1016/S2213-2600(20)30316-7
Katzenstein AL, Bloor CM, Leibow AA (1976) Diffuse alveolar damage—the role of oxygen, shock, and related factors. A review. Am J Pathol 85:209–228
Kawakami R, Sakamoto A, Kawai K, Gianatti A, Pellegrini D, Nasr A, Kutys B, Guo L, Cornelissen A, Mori M, Sato Y, Pescetelli I, Brivio M, Romero M, Guagliumi G, Virmani R, Finn AV (2021) Pathological evidence for SARS-CoV-2 as a cause of myocarditis: JACC Review Topic of the Week. J Am Coll Cardiol 77:314–325. https://doi.org/10.1016/j.jacc.2020.11.031
Keddie S, Pakpoor J, Mousele C, Pipis M, Machado PM, Foster M, Record CJ, Keh RYS, Fehmi J, Paterson RW, Bharambe V, Clayton LM, Allen C, Price O, Wall J, Kiss-Csenki A, Rathnasabapathi DP, Geraldes R, Yermakova T, King-Robson J, Zosmer M, Rajakulendran S, Sumaria S, Farmer SF, Nortley R, Marshall CR, Newman EJ, Nirmalananthan N, Kumar G, Pinto AA, Holt J, Lavin TM, Brennan KM, Zandi MS, Jayaseelan DL, Pritchard J, Hadden RDM, Manji H, Willison HJ, Rinaldi S, Carr AS, Lunn MP (2021) Epidemiological and cohort study finds no association between COVID-19 and Guillain-Barre syndrome. Brain 144:682–693. https://doi.org/10.1093/brain/awaa433
Khan M, Yoo SJ, Clijsters M, Backaert W, Vanstapel A, Speleman K, Lietaer C, Choi S, Hether TD, Marcelis L, Nam A, Pan L, Reeves JW, Van Bulck P, Zhou H, Bourgeois M, Debaveye Y, De Munter P, Gunst J, Jorissen M, Lagrou K, Lorent N, Neyrinck A, Peetermans M, Thal DR, Vandenbriele C, Wauters J, Mombaerts P, Van Gerven L (2021) Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 184(5932–5949):e5915. https://doi.org/10.1016/j.cell.2021.10.027
Khoo B, Tan T, Clarke SA, Mills EG, Patel B, Modi M, Phylactou M, Eng PC, Thurston L, Alexander EC, Meeran K, Comninos AN, Abbara A, Dhillo WS (2021) Thyroid function before, during, and after COVID-19. J Clin Endocrinol Metab 106:e803–e811. https://doi.org/10.1210/clinem/dgaa830
Kim IC, Kim JY, Kim HA, Han S (2020) COVID-19-related myocarditis in a 21-year-old female patient. Eur Heart J 41:1859. https://doi.org/10.1093/eurheartj/ehaa288
Klein A, Edler C, Fitzek A, Frob D, Heinemann A, Meissner K, Mushumba H, Puschel K, Schroder AS, Sperhake JP, Ishorst-Witte F, Aepfelbacher M, Heinrich F (2020) [The first COVID-19 hotspot in a retirement home in Hamburg]. Rechtsmedizin (Berl):1–7 https://doi.org/10.1007/s00194-020-00404-1
Klindt C, Jensen BE, Brandenburger T, Feldt T, Killer A, Schimmoller L, Antoch G, Senff T, Hauka S, Timm J, Bahners BH, Seidl M, Esposito I, Luedde T, Bode JG, Keitel V (2021) Secondary sclerosing cholangitis as a complication of severe COVID-19: a case report and review of the literature. Clin Case Rep 9:e04068. https://doi.org/10.1002/ccr3.4068
Klingenstein M, Klingenstein S, Neckel PH, Mack AF, Wagner AP, Kleger A et al (2020) Evidence of SARS-CoV2 entry protein ACE2 in the human nose and olfactory bulb. Cells Tissues Organs 209(4–6):155–164. https://doi.org/10.1159/000513040
Kniep I, Heinemann A, Edler C, Sperhake JP, Puschel K, Ondruschka B, Schroder AS (2021) COVID-19 lungs in post-mortem computed tomography. Rechtsmedizin (Berl):1–3 https://doi.org/10.1007/s00194-021-00462-z
Koehler P, Bassetti M, Chakrabarti A, Chen SCA, Colombo AL, Hoenigl M, Klimko N, Lass-Florl C, Oladele RO, Vinh DC, Zhu LP, Boll B, Bruggemann R, Gangneux JP, Perfect JR, Patterson TF, Persigehl T, Meis JF, Ostrosky-Zeichner L, White PL, Verweij PE, Cornely OA, European Confederation of Medical Mycology, International Society for Human Animal Mycology, Asia Fungal Working Group, INFOCUS LATAM/ISHAM Working Group, ISHAM Pan Africa Mycology Working Group, European Society for Clinical Microbiology, Infectious Diseases Fungal Infection Study Group, ESCMID Study Group for Infections in Critically Ill Patients, Interregional Association of Clinical Microbiology and Antimicrobial Chemotherapy, Medical Mycology Society of Nigeria, Medical Mycology Society of China Medicine Education Association, Infectious Diseases Working Party of the German Society for Haematology and Medical Oncology, Association of Medical Microbiology, Infectious Disease Canada (2021) Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect Dis 21:e149–e162. https://doi.org/10.1016/S1473-3099(20)30847-1
Konopka KE, Wilson A (2019) Myers JL (2020) Postmortem lung findings in a patient with asthma and coronavirus disease. Chest 158:e99–e101. https://doi.org/10.1016/j.chest.2020.04.032
Krasemann S, Dittmayer C, v. Stillfried S, Meinhardt J, Heinrich F, Hartmann K, Pfefferle S, Thies E, v. Manitius R, Aschman T, Radke J, Osterloh A, Schmid S, Buhl EM, Ihlow J, Elezkurtaj S, Horst D, Hocke AC, Timm S, Bachmann S, Corman V, Goebel HH, Matschke J, Stanelle-Bertram S, Gabriel G, Seilhean D, Adle-Biassette H, Ondruschka B, Ochs M, Stenzel W, Heppner FL, Boor P, Radbruch H, Laue M, Glatzel M (2022) Assessing and improving the validity of COVID-19 autopsy studies—a multicenter approach to establish essential standards for immunohistochemical and ultrastructural analyses. MedRxiv https://doi.org/10.1101/2022.01.13.22269205
Krasemann S, Haferkamp U, Pfefferle S, Woo MS, Heinrich F, Schweizer M, Appelt-Menzel A, Cubukova A, Barenberg J, Leu J, Hartmann K, Thies E, Littau JL, Sepulveda-Falla D, Zhang L, Ton K, Liang Y, Matschke J, Ricklefs F, Sauvigny T, Sperhake J, Fitzek A, Gerhartl A, Brachner A, Geiger N, Konig EM, Bodem J, Franzenburg S, Franke A, Moese S, Muller FJ, Geisslinger G, Claussen C, Kannt A, Zaliani A, Gribbon P, Ondruschka B, Neuhaus W, Friese MA, Glatzel M, Pless O (2022) The blood-brain barrier is dysregulated in COVID-19 and serves as a CNS entry route for SARS-CoV-2 Stem. Cell Rep 17:307–320. https://doi.org/10.1016/j.stemcr.2021.12.011
Kropski JA, Richmond BW, Gaskill CF, Foronjy RF, Majka SM (2018) Deregulated angiogenesis in chronic lung diseases: a possible role for lung mesenchymal progenitor cells (2017 Grover Conference Series). Pulm Circ 8:2045893217739807. https://doi.org/10.1177/2045893217739807
Lagana SM, Kudose S, Iuga AC, Lee MJ, Fazlollahi L, Remotti HE, Del Portillo A, De Michele S, de Gonzalez AK, Saqi A, Khairallah P, Chong AM, Park H, Uhlemann AC, Lefkowitch JH, Verna EC (2020) Hepatic pathology in patients dying of COVID-19: a series of 40 cases including clinical, histologic, and virologic data. Mod Pathol 33:2147–2155. https://doi.org/10.1038/s41379-020-00649-x
Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, van Donselaar E, Riesebosch S, Kuijpers HJH, Schipper D, van de Wetering WJ, de Graaf M, Koopmans M, Cuppen E, Peters PJ, Haagmans BL, Clevers H (2020) SARS-CoV-2 productively infects human gut enterocytes. Science 369:50–54. https://doi.org/10.1126/science.abc1669
Lax SF, Skok K, Zechner P, Kessler HH, Kaufmann N, Koelblinger C, Vander K, Bargfrieder U, Trauner M (2020) Pulmonary arterial thrombosis in COVID-19 with fatal outcome : results from a prospective, single-center, clinicopathologic case series. Ann Intern Med 173:350–361. https://doi.org/10.7326/M20-2566
Leppkes M, Knopf J, Naschberger E, Lindemann A, Singh J, Herrmann I, Sturzl M, Staats L, Mahajan A, Schauer C, Kremer AN, Volkl S, Amann K, Evert K, Falkeis C, Wehrfritz A, Rieker RJ, Hartmann A, Kremer AE, Neurath MF, Munoz LE, Schett G, Herrmann M (2020) Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine 58:102925. https://doi.org/10.1016/j.ebiom.2020.102925
Lindner D, Fitzek A, Brauninger H, Aleshcheva G, Edler C, Meissner K, Scherschel K, Kirchhof P, Escher F, Schultheiss HP, Blankenberg S, Puschel K, Westermann D (2020) Association of cardiac infection with SARS-CoV-2 in confirmed COVID-19 autopsy cases. JAMA Cardiol 5:1281–1285. https://doi.org/10.1001/jamacardio.2020.3551
Liu B, Li M, Zhou Z, Guan X, Xiang Y (2020) Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J Autoimmun 111:102452. https://doi.org/10.1016/j.jaut.2020.102452
Loscocco GG, Malandrino D, Barchiesi S, Berni A, Poggesi L, Guglielmelli P, Vannucchi AM (2020) The HScore for secondary hemophagocytic lymphohistiocytosis, calculated without a marrow biopsy, is consistently low in patients with COVID-19. Int J Lab Hematol 42:e270–e273. https://doi.org/10.1111/ijlh.13310
Loyal L, Braun J, Henze L, Kruse B, Dingeldey M, Reimer U, Kern F, Schwarz T, Mangold M, Unger C, Dorfler F, Kadler S, Rosowski J, Gurcan K, Uyar-Aydin Z, Frentsch M, Kurth F, Schnatbaum K, Eckey M, Hippenstiel S, Hocke A, Muller MA, Sawitzki B, Miltenyi S, Paul F, Mall MA, Wenschuh H, Voigt S, Drosten C, Lauster R, Lachman N, Sander LE, Corman VM, Rohmel J, Meyer-Arndt L, Thiel A, Giesecke-Thiel C (2021) Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science 374:eabh1823 https://doi.org/10.1126/science.abh1823
Lozada-Nur F, Chainani-Wu N, Fortuna G, Sroussi H (2020) Dysgeusia in COVID-19: possible mechanisms and implications. Oral Surg Oral Med Oral Pathol Oral Radiol 130:344–346. https://doi.org/10.1016/j.oooo.2020.06.016
Magro C, Mulvey JJ, Berlin D, Nuovo G, Salvatore S, Harp J, Baxter-Stoltzfus A, Laurence J (2020) Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res 220:1–13. https://doi.org/10.1016/j.trsl.2020.04.007
Malik P, Patel U, Mehta D, Patel N, Kelkar R, Akrmah M, Gabrilove JL, Sacks H (2021) Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ Evid Based Med 26:107–108. https://doi.org/10.1136/bmjebm-2020-111536
Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, Miao X, Li Y, Hu B (2020) Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol 77:683–690. https://doi.org/10.1001/jamaneurol.2020.1127
Martines RB, Ritter JM, Matkovic E, Gary J, Bollweg BC, Bullock H, Goldsmith CS, Silva-Flannery L, Seixas JN, Reagan-Steiner S, Uyeki T, Denison A, Bhatnagar J, Shieh WJ, Zaki SR, Group C-PW (2020) Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect Dis 26:2005–2015. https://doi.org/10.3201/eid2609.202095
Massoth LR, Desai N, Szabolcs A, Harris CK, Neyaz A, Crotty R, Chebib I, Rivera MN, Sholl LM, Stone JR, Ting DT, Deshpande V (2021) Comparison of RNA in situ hybridization and immunohistochemistry techniques for the detection and localization of SARS-CoV-2 in human tissues. Am J Surg Pathol 45:14–24. https://doi.org/10.1097/PAS.0000000000001563
Matschke J, Lutgehetmann M, Hagel C, Sperhake JP, Schroder AS, Edler C, Mushumba H, Fitzek A, Allweiss L, Dandri M, Dottermusch M, Heinemann A, Pfefferle S, Schwabenland M, Sumner Magruder D, Bonn S, Prinz M, Gerloff C, Puschel K, Krasemann S, Aepfelbacher M, Glatzel M (2020) Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol 19:919–929. https://doi.org/10.1016/S1474-4422(20)30308-2
Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, Across Speciality Collaboration UK (2020) COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395:1033–1034. https://doi.org/10.1016/S0140-6736(20)30628-0
Meinhardt J, Radke J, Dittmayer C, Franz J, Thomas C, Mothes R, Laue M, Schneider J, Brunink S, Greuel S, Lehmann M, Hassan O, Aschman T, Schumann E, Chua RL, Conrad C, Eils R, Stenzel W, Windgassen M, Rossler L, Goebel HH, Gelderblom HR, Martin H, Nitsche A, Schulz-Schaeffer WJ, Hakroush S, Winkler MS, Tampe B, Scheibe F, Kortvelyessy P, Reinhold D, Siegmund B, Kuhl AA, Elezkurtaj S, Horst D, Oesterhelweg L, Tsokos M, Ingold-Heppner B, Stadelmann C, Drosten C, Corman VM, Radbruch H, Heppner FL (2021) Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci 24:168–175. https://doi.org/10.1038/s41593-020-00758-5
Meng B, Abdullahi A, Ferreira I, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, Zepeda SK, Papa G, Kemp SA, Ikeda T, Toyoda M, Tan TS, Kuramochi J, Mitsunaga S, Ueno T, Shirakawa K, Takaori-Kondo A, Brevini T, Mallery DL, Charles OJ, Collaboration C-NBC-, Genotype to Phenotype Japan Consortium m, Ecuador CC, Bowen JE, Joshi A, Walls AC, Jackson L, Martin D, Smith KGC, Bradley J, Briggs JAG, Choi J, Madissoon E, Meyer K, Mlcochova P, Ceron-Gutierrez L, Doffinger R, Teichmann SA, Fisher AJ, Pizzuto MS, de Marco A, Corti D, Hosmillo M, Lee JH, James LC, Thukral L, Veesler D, Sigal A, Sampaziotis F, Goodfellow IG, Matheson NJ, Sato K, Gupta RK (2022) Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature https://doi.org/10.1038/s41586-022-04474-x
Menter T, Haslbauer JD, Nienhold R, Savic S, Hopfer H, Deigendesch N, Frank S, Turek D, Willi N, Pargger H, Bassetti S, Leuppi JD, Cathomas G, Tolnay M, Mertz KD, Tzankov A (2020) Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 77:198–209. https://doi.org/10.1111/his.14134
Muller JA, Gross R, Conzelmann C, Kruger J, Merle U, Steinhart J, Weil T, Koepke L, Bozzo CP, Read C, Fois G, Eiseler T, Gehrmann J, van Vuuren J, Wessbecher IM, Frick M, Costa IG, Breunig M, Gruner B, Peters L, Schuster M, Liebau S, Seufferlein T, Stenger S, Stenzinger A, MacDonald PE, Kirchhoff F, Sparrer KMJ, Walther P, Lickert H, Barth TFE, Wagner M, Munch J, Heller S, Kleger A (2021) SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nat Metab 3:149–165. https://doi.org/10.1038/s42255-021-00347-1
Nakayama T, Lee IT, Jiang S, Matter MS, Yan CH, Overdevest JB, Wu CT, Goltsev Y, Shih LC, Liao CK, Zhu B, Bai Y, Lidsky P, Xiao Y, Zarabanda D, Yang A, Easwaran M, Schurch CM, Chu P, Chen H, Stalder AK, McIlwain DR, Borchard NA, Gall PA, Dholakia SS, Le W, Xu L, Tai CJ, Yeh TH, Erickson-Direnzo E, Duran JM, Mertz KD, Hwang PH, Haslbauer JD, Jackson PK, Menter T, Andino R, Canoll PD, DeConde AS, Patel ZM, Tzankov A, Nolan GP, Nayak JV (2021) Determinants of SARS-CoV-2 entry and replication in airway mucosal tissue and susceptibility in smokers. Cell Rep Med 2:100421. https://doi.org/10.1016/j.xcrm.2021.100421
Nardo AD, Schneeweiss-Gleixner M, Bakail M, Dixon ED, Lax SF, Trauner M (2021) Pathophysiological mechanisms of liver injury in COVID-19. Liver Int 41:20–32. https://doi.org/10.1111/liv.14730
Newell KL, Clemmer DC, Cox JB, Kayode YI, Zoccoli-Rodriguez V, Taylor HE, Endy TP, Wilmore JR, Winslow GM (2021) Switched and unswitched memory B cells detected during SARS-CoV-2 convalescence correlate with limited symptom duration. PLoS One 16:e0244855. https://doi.org/10.1371/journal.pone.0244855
Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, Yan KW, Chan KH, Tsang NC, Guan Y, Yuen KY, Peiris JS (2003) Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773–1778. https://doi.org/10.1016/s0140-6736(03)13413-7
Nicolai L, Leunig A, Brambs S, Kaiser R, Joppich M, Hoffknecht ML, Gold C, Engel A, Polewka V, Muenchhoff M, Hellmuth JC, Ruhle A, Ledderose S, Weinberger T, Schulz H, Scherer C, Rudelius M, Zoller M, Keppler OT, Zwissler B, von Bergwelt-Baildon M, Kaab S, Zimmer R, Bulow RD, von Stillfried S, Boor P, Massberg S, Pekayvaz K, Stark K (2021) Vascular neutrophilic inflammation and immunothrombosis distinguish severe COVID-19 from influenza pneumonia. J Thromb Haemost 19:574–581. https://doi.org/10.1111/jth.15179
Nie X, Qian L, Sun R, Huang B, Dong X, Xiao Q, Zhang Q, Lu T, Yue L, Chen S, Li X, Sun Y, Li L, Xu L, Li Y, Yang M, Xue Z, Liang S, Ding X, Yuan C, Peng L, Liu W, Yi X, Lyu M, Xiao G, Xu X, Ge W, He J, Fan J, Wu J, Luo M, Chang X, Pan H, Cai X, Zhou J, Yu J, Gao H, Xie M, Wang S, Ruan G, Chen H, Su H, Mei H, Luo D, Zhao D, Xu F, Li Y, Zhu Y, Xia J, Hu Y, Guo T (2021) Multi-organ proteomic landscape of COVID-19 autopsies. Cell 184(775–791):e714. https://doi.org/10.1016/j.cell.2021.01.004
Nunez-Torron C, Ferrer-Gomez A, Moreno Moreno E, Perez-Mies B, Villarrubia J, Chamorro S, Lopez-Jimenez J, Palacios J, Piris-Villaespesa M, Garcia-Cosio M (2021) Secondary haemophagocytic lymphohistiocytosis in COVID-19: correlation of the autopsy findings of bone marrow haemophagocytosis with HScore. J Clin Pathol https://doi.org/10.1136/jclinpath-2020-207337
Paizis G, Tikellis C, Cooper ME, Schembri JM, Lew RA, Smith AI, Shaw T, Warner FJ, Zuilli A, Burrell LM, Angus PW (2005) Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut 54:1790–1796. https://doi.org/10.1136/gut.2004.062398
Park A, Iwasaki A (2020) Type I and type III interferons—induction, signaling, evasion, and application to combat COVID-19 cell host. Microbe 27:870–878. https://doi.org/10.1016/j.chom.2020.05.008
Parolin M, Parisotto M, Zanchetta F, Sartorato P, De Menis E (2021) Coronaviruses and endocrine system: a systematic review on evidence and shadows. Endocr Metab Immune Disord Drug Targets 21:1242–1251. https://doi.org/10.2174/1871530320666200905123332
Perico L, Benigni A, Casiraghi F, Ng LFP, Renia L, Remuzzi G (2021) Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat Rev Nephrol 17:46–64. https://doi.org/10.1038/s41581-020-00357-4
Polak SB, Van Gool IC, Cohen D, von der Thusen JH, van Paassen J (2020) A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol 33:2128–2138. https://doi.org/10.1038/s41379-020-0603-3
Porcari A, Bussani R, Merlo M, Varra GG, Pagura L, Rozze D, Sinagra G (2021) Incidence and characterization of concealed cardiac amyloidosis among unselected elderly patients undergoing post-mortem examination. Front Cardiovasc Med 8:749523. https://doi.org/10.3389/fcvm.2021.749523
Price LC, McCabe C, Garfield B, Wort SJ (2020) Thrombosis and COVID-19 pneumonia: the clot thickens! Eur Respir J 56 https://doi.org/10.1183/13993003.01608-2020
Prieto-Perez L, Fortes J, Soto C, Vidal-Gonzalez A, Alonso-Riano M, Lafarga M, Cortti MJ, Lazaro-Garcia A, Perez-Tanoira R, Trascasa A, Antonio A, Cordoba R, Rodriguez-Pinilla SM, Cedeno O, Peces-Barba G, Fernandez-Ormaechea I, Diez Medrano MJ, de Las Lopez, Heras M, Cabello A, Petkova E, Alvarez B, Carrillo I, Silva AM, Castellanos M, Calpena S, Valverde-Monge M, Fresneda D, Rubio-Martin R, Cornejo I, Blanco Astilleros, de Cordova L, de la Fuente S, Recuero S, Gorgolas M, Piris MA (2020) Histiocytic hyperplasia with hemophagocytosis and acute alveolar damage in COVID-19 infection. Mod Pathol 33:2139–2146. https://doi.org/10.1038/s41379-020-0613-1
Prilutskiy A, Kritselis M, Shevtsov A, Yambayev I, Vadlamudi C, Zhao Q, Kataria Y, Sarosiek SR, Lerner A, Sloan JM, Quillen K, Burks EJ (2020) SARS-CoV-2 infection-associated hemophagocytic lymphohistiocytosis. Am J Clin Pathol 154:466–474. https://doi.org/10.1093/ajcp/aqaa124
Proal AD, VanElzakker MB (2021) Long COVID or post-acute sequelae of COVID-19 (PASC): an overview of biological factors that may contribute to persistent symptoms. Front Microbiol 12:698169. https://doi.org/10.3389/fmicb.2021.698169
Puelles VG, Lutgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schroder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Pueschel K, Aepfelbacher M, Huber TB (2020) Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 383:590–592. https://doi.org/10.1056/NEJMc2011400
Puntmann VO, Carerj ML, Wieters I, Fahim M, Arendt C, Hoffmann J, Shchendrygina A, Escher F, Vasa-Nicotera M, Zeiher AM, Vehreschild M, Nagel E (2020) Outcomes of cardiovascular magnetic resonance imaging in patients recently recovered from coronavirus disease 2019 (COVID-19). JAMA Cardiol 5:1265–1273. https://doi.org/10.1001/jamacardio.2020.3557
Raptis CA, Hammer MM, Short RG, Shah A, Bhalla S, Bierhals AJ, Filev PD, Hope MD, Jeudy J, Kligerman SJ, Henry TS (2020) Chest CT and coronavirus disease (COVID-19): a critical review of the literature to date. AJR Am J Roentgenol 215:839–842. https://doi.org/10.2214/AJR.20.23202
Remmelink M, De Mendonca R, D’Haene N, De Clercq S, Verocq C, Lebrun L, Lavis P, Racu ML, Trepant AL, Maris C, Rorive S, Goffard JC, De Witte O, Peluso L, Vincent JL, Decaestecker C, Taccone FS, Salmon I (2020) Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Crit Care 24:495. https://doi.org/10.1186/s13054-020-03218-5
Sapino A, Facchetti F, Bonoldi E, Gianatti A, Barbareschi M, Societa Italiana di Anatomia Patologica e Citologia S (2020) The autopsy debate during the COVID-19 emergency: the Italian experience. Virchows Arch 476:821–823 https://doi.org/10.1007/s00428-020-02828-2
Scappaticcio L, Pitoia F, Esposito K, Piccardo A, Trimboli P (2020) Impact of COVID-19 on the thyroid gland: an update. Rev Endocr Metab Disord https://doi.org/10.1007/s11154-020-09615-z
Schaller T, Hirschbuhl K, Burkhardt K, Braun G, Trepel M, Markl B, Claus R (2020) Postmortem Examination of Patients With COVID-19. JAMA 323:2518–2520. https://doi.org/10.1001/jama.2020.8907
Schulte-Schrepping J, Reusch N, Paclik D, Bassler K, Schlickeiser S, Zhang B, Kramer B, Krammer T, Brumhard S, Bonaguro L, De Domenico E, Wendisch D, Grasshoff M, Kapellos TS, Beckstette M, Pecht T, Saglam A, Dietrich O, Mei HE, Schulz AR, Conrad C, Kunkel D, Vafadarnejad E, Xu CJ, Horne A, Herbert M, Drews A, Thibeault C, Pfeiffer M, Hippenstiel S, Hocke A, Muller-Redetzky H, Heim KM, Machleidt F, Uhrig A, Bosquillon de Jarcy L, Jurgens L, Stegemann M, Glosenkamp CR, Volk HD, Goffinet C, Landthaler M, Wyler E, Georg P, Schneider M, Dang-Heine C, Neuwinger N, Kappert K, Tauber R, Corman V, Raabe J, Kaiser KM, Vinh MT, Rieke G, Meisel C, Ulas T, Becker M, Geffers R, Witzenrath M, Drosten C, Suttorp N, von Kalle C, Kurth F, Handler K, Schultze JL, Aschenbrenner AC, Li Y, Nattermann J, Sawitzki B, Saliba AE, Sander LE, Deutsche C-OI (2020) Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182:1419–1440 e1423 https://doi.org/10.1016/j.cell.2020.08.001
Schwabenland M, Salie H, Tanevski J, Killmer S, Lago MS, Schlaak AE, Mayer L, Matschke J, Puschel K, Fitzek A, Ondruschka B, Mei HE, Boettler T, Neumann-Haefelin C, Hofmann M, Breithaupt A, Genc N, Stadelmann C, Saez-Rodriguez J, Bronsert P, Knobeloch KP, Blank T, Thimme R, Glatzel M, Prinz M, Bengsch B (2021) Deep spatial profiling of human COVID-19 brains reveals neuroinflammation with distinct microanatomical microglia-T-cell interactions. Immunity 54(1594–1610):e1511. https://doi.org/10.1016/j.immuni.2021.06.002
Sekulic M, Harper H, Nezami BG, Shen DL, Sekulic SP, Koeth AT, Harding CV, Gilmore H, Sadri N (2020) Molecular Detection of SARS-CoV-2 Infection in FFPE samples and histopathologic findings in fatal SARS-CoV-2 cases. Am J Clin Pathol 154:190–200. https://doi.org/10.1093/ajcp/aqaa091
Solomon IH, Normandin E, Bhattacharyya S, Mukerji SS, Keller K, Ali AS, Adams G, Hornick JL, Padera RF Jr, Sabeti P (2020) Neuropathological features of Covid-19. N Engl J Med 383:989–992. https://doi.org/10.1056/NEJMc2019373
Song J, Li Y, Huang X, Chen Z, Li Y, Liu C, Chen Z, Duan X (2020) Systematic analysis of ACE2 and TMPRSS2 expression in salivary glands reveals underlying transmission mechanism caused by SARS-CoV-2. J Med Virol 92:2556–2566. https://doi.org/10.1002/jmv.26045
Studart-Neto A, Guedes BF, Tuma RLE, Camelo Filho AE, Kubota GT, Iepsen BD, Moreira GP, Rodrigues JC, Ferrari MMH, Carra RB, Spera RR, Oku MHM, Terrim S, Lopes CCB, Passos Neto CEB, Fiorentino MD, JCC DES, Baima JPS, TFF DAS, Moreno CAM, Silva AMS, Heise CO, MendonCa RH, Fortini I, Smid J, Adoni T, GonCalves MRR, Pereira SLA, Pinto LF, Gomes HR, Zanoteli E, Brucki SMD, Conforto AB, Castro LHM, Nitrini R (2020) Neurological consultations and diagnoses in a large, dedicated COVID-19 university hospital. Arq Neuropsiquiatr 78:494–500 https://doi.org/10.1590/0004-282X20200089
Sugiyama MG, Gamage A, Zyla R, Armstrong SM, Advani S, Advani A, Wang C, Lee WL (2016) Influenza virus infection induces platelet-endothelial adhesion which contributes to lung injury. J Virol 90:1812–1823. https://doi.org/10.1128/JVI.02599-15
Taquet M, Geddes JR, Husain M, Luciano S, Harrison PJ (2021) 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry 8:416–427. https://doi.org/10.1016/S2215-0366(21)00084-5
Tavazzi G, Pellegrini C, Maurelli M, Belliato M, Sciutti F, Bottazzi A, Sepe PA, Resasco T, Camporotondo R, Bruno R, Baldanti F, Paolucci S, Pelenghi S, Iotti GA, Mojoli F, Arbustini E (2020) Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur J Heart Fail 22:911–915. https://doi.org/10.1002/ejhf.1828
Thompson BT, Chambers RC, Liu KD (2017) Acute respiratory distress syndrome. N Engl J Med 377:1904–1905. https://doi.org/10.1056/NEJMc1711824
Tiwari NR, Phatak S, Sharma VR, Agarwal SK (2021) COVID-19 and thrombotic microangiopathies. Thromb Res 202:191–198. https://doi.org/10.1016/j.thromres.2021.04.012
Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H (2020) Endothelial cell infection and endotheliitis in COVID-19. Lancet 395:1417–1418. https://doi.org/10.1016/S0140-6736(20)30937-5
von Stillfried S, Acker T, Aepfelbacher M, Baretton G, Bulow RD, Burrig KF, Holtherm HU, Jonigk D, Knuchel R, Majeed RW, Rohrig R, Wienstroer J, Boor P (2021) Cooperative approach of pathology and neuropathology in the COVID-19 pandemic : German registry for COVID-19 autopsies (DeRegCOVID) and German network for autopsies in pandemics (DEFEAT PANDEMIcs). Pathologe https://doi.org/10.1007/s00292-020-00897-3
von Stillfried S, Boor P (2021) Detection methods for SARS-CoV-2 in tissue. Pathologe https://doi.org/10.1007/s00292-021-00920-1
von Stillfried S, Bülow RD, Röhrig R, Boor P, Böcker J, Schmidt J, Tholen P, Majeed R, Wienströer J, Weis J, Bremer J, Knüchel R, Breitbach A, Cacchi C, Freeborn B, Wucherpfennig S, Spring O, Braun G, Römmele C, Märkl B, Claus R, Dhillon C, Schaller T, Sipos E, Hirschbühl K, Wittmann M, Kling E, Kröncke T, Heppner FL, Meinhardt J, Radbruch H, Streit S, Horst D, Elezkurtaj S, Quaas A, Göbel H, Hansen T, Titze U, Lorenzen J, Reuter T, Woloszyn J, Baretton G, Hilsenbeck J, Meinhardt M, Pablik J, Sommer L, Holotiuk O, Meinel M, Mahlke N, Esposito I, Crudele G, Seidl M, Amann KU, Coras R, Hartmann A, Eichhorn P, Haller F, Lange F, Schmid KW, Ingenwerth M, Rawitzer J, Theegarten D, Birngruber CG, Wild P, Gradhand E, Smith K, Werner M, Schilling O, Acker T, Gattenlöhner S, Stadelmann C, Metz I, Franz J, Stork L, Thomas C, Zechel S, Ströbel P, Wickenhauser C, Fathke C, Harder A, Ondruschka B, Dietz E, Edler C, Fitzek A, Fröb D, Heinemann A, Heinrich F, Klein A, Kniep I, Lohner L, Möbius D, Püschel K, Schädler J, Schröder A-S, Sperhake J-P, Aepfelbacher M, Fischer N, Lütgehetmann M, Pfefferle S, Glatzel M, Krasemann S, Matschke J, Jonigk D, Werlein C, Schirmacher P, Domke LM, Hartmann L, Klein IM, Schwab C, Röcken C, Friemann J, Langer D, Roth W, Strobl S, Rudelius M, Stock KF, Weichert W, Delbridge C, Kasajima A, Kuhn P-H, Slotta-Huspenina J, Weirich G, Barth P, Wardelmann E, Evert K, Büttner A, Manhart J, Nigbur S, Bittmann I, Fend F, Bösmüller H, Granai M, Klingel K, Warm V, Steinestel K, Umathum VG, Rosenwald A, Kurz F, Vogt N (2022) First report from the German COVID-19 autopsy registry. Lancet Reg Health - Europe https://doi.org/10.1016/j.lanepe.2022.100330
von Stillfried S, Bulow RD, Rohrig R, Knuchel-Clarke R, Boor P, DeRegCovid (2020) Autopsy registry can facilitate COVID-19 research. EMBO Mol Med 12:e12885. https://doi.org/10.15252/emmm.202012885
Wang MY, Zhao R, Gao LJ, Gao XF, Wang DP, Cao JM (2020) SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol 10:587269. https://doi.org/10.3389/fcimb.2020.587269
Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W (2020) Detection of SARS-CoV-2 in Different types of clinical specimens. JAMA 323:1843–1844. https://doi.org/10.1001/jama.2020.3786
Wei JF, Huang FY, Xiong TY, Liu Q, Chen H, Wang H, Huang H, Luo YC, Zhou X, Liu ZY, Peng Y, Xu YN, Wang B, Yang YY, Liang ZA, Lei XZ, Ge Y, Yang M, Zhang L, Zeng MQ, Yu H, Liu K, Jia YH, Prendergast BD, Li WM, Chen M (2020) Acute myocardial injury is common in patients with COVID-19 and impairs their prognosis. Heart 106:1154–1159. https://doi.org/10.1136/heartjnl-2020-317007
Wenzel P, Kopp S, Gobel S, Jansen T, Geyer M, Hahn F, Kreitner KF, Escher F, Schultheiss HP, Munzel T (2020) Evidence of SARS-CoV-2 mRNA in endomyocardial biopsies of patients with clinically suspected myocarditis tested negative for COVID-19 in nasopharyngeal swab. Cardiovasc Res 116:1661–1663. https://doi.org/10.1093/cvr/cvaa160
Werion A, Belkhir L, Perrot M, Schmit G, Aydin S, Chen Z, Penaloza A, De Greef J, Yildiz H, Pothen L, Yombi JC, Dewulf J, Scohy A, Gerard L, Wittebole X, Laterre PF, Miller SE, Devuyst O, Jadoul M, Morelle J, Cliniques universitaires Saint-Luc C-RG, (2020) SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int 98:1296–1307. https://doi.org/10.1016/j.kint.2020.07.019
Wichmann D, Sperhake JP, Lutgehetmann M, Steurer S, Edler C, Heinemann A, Heinrich F, Mushumba H, Kniep I, Schroder AS, Burdelski C, de Heer G, Nierhaus A, Frings D, Pfefferle S, Becker H, Bredereke-Wiedling H, de Weerth A, Paschen HR, Sheikhzadeh-Eggers S, Stang A, Schmiedel S, Bokemeyer C, Addo MM, Aepfelbacher M, Puschel K, Kluge S (2020) Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med 173:268–277. https://doi.org/10.7326/M20-2003
Wildner NH, Ahmadi P, Schulte S, Brauneck F, Kohsar M, Lutgehetmann M, Beisel C, Addo MM, Haag F, Schulze Zur Wiesch J (2021) B cell analysis in SARS-CoV-2 versus malaria: increased frequencies of plasmablasts and atypical memory B cells in COVID-19. J Leukoc Biol 109:77–90. https://doi.org/10.1002/JLB.5COVA0620-370RR
Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C (2020) Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. https://doi.org/10.1038/s41586-020-2196-x
Wong DWL, Klinkhammer BM, Djudjaj S, Villwock S, Timm MC, Buhl EM et al (2021) Multisystemic cellular tropism of SARS-CoV-2 in autopsies of COVID-19 patients. Cells 10(8):1900. https://doi.org/10.3390/cells10081900
Wu CT, Lidsky PV, Xiao Y, Lee IT, Cheng R, Nakayama T, Jiang S, Demeter J, Bevacqua RJ, Chang CA, Whitener RL, Stalder AK, Zhu B, Chen H, Goltsev Y, Tzankov A, Nayak JV, Nolan GP, Matter MS, Andino R, Jackson PK (2021) SARS-CoV-2 infects human pancreatic beta cells and elicits beta cell impairment. Cell Metab 33(1565–1576):e1565. https://doi.org/10.1016/j.cmet.2021.05.013
Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8:420–422. https://doi.org/10.1016/S2213-2600(20)30076-X
Yachou Y, El Idrissi A, Belapasov V, Ait Benali S (2020) Neuroinvasion, neurotropic, and neuroinflammatory events of SARS-CoV-2: understanding the neurological manifestations in COVID-19 patients. Neurol Sci 41:2657–2669. https://doi.org/10.1007/s10072-020-04575-3
Yanagihara T, Jones KD (2020) Demystifying morphomolecular alterations of vasculature in interstitial lung diseases. Eur Respir J 55 https://doi.org/10.1183/13993003.02446-2019
Yang AC, Kern F, Losada PM, Agam MR, Maat CA, Schmartz GP, Fehlmann T, Stein JA, Schaum N, Lee DP, Calcuttawala K, Vest RT, Berdnik D, Lu N, Hahn O, Gate D, McNerney MW, Channappa D, Cobos I, Ludwig N, Schulz-Schaeffer WJ, Keller A, Wyss-Coray T (2021) Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 595:565–571. https://doi.org/10.1038/s41586-021-03710-0
Yang J, Chen T, Zhou Y (2021) Mediators of SARS-CoV-2 entry are preferentially enriched in cardiomyocytes. Hereditas 158:4. https://doi.org/10.1186/s41065-020-00168-4
Yu CM, Wong RS, Wu EB, Kong SL, Wong J, Yip GW, Soo YO, Chiu ML, Chan YS, Hui D, Lee N, Wu A, Leung CB, Sung JJ (2006) Cardiovascular complications of severe acute respiratory syndrome. Postgrad Med J 82:140–144. https://doi.org/10.1136/pgmj.2005.037515
Zeng JH, Liu YX, Yuan J, Wang FX, Wu WB, Li JX, Wang LF, Gao H, Wang Y, Dong CF, Li YJ, Xie XJ, Feng C, Liu L (2020) First case of COVID-19 complicated with fulminant myocarditis: a case report and insights. Infection 48:773–777. https://doi.org/10.1007/s15010-020-01424-5
Zhang H, Kang ZJ, Gong HY, Xu D, Wang J, Li ZX, Li ZF, Cui XG, Xiao JR, Zhan J, Meng T, Zhou W, Liu JM, Xu HJ (2020) Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 69:1010–1018. https://doi.org/10.1136/gutjnl-2020-320953
Zhang L, Wang Y, Wu G, Xiong W, Gu W, Wang CY (2018) Macrophages: friend or foe in idiopathic pulmonary fibrosis? Respir Res 19:170. https://doi.org/10.1186/s12931-018-0864-2
Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, Rosain J, Bilguvar K, Ye J, Bolze A, Bigio B, Yang R, Arias AA, Zhou Q, Zhang Y, Onodi F, Korniotis S, Karpf L, Philippot Q, Chbihi M, Bonnet-Madin L, Dorgham K, Smith N, Schneider WM, Razooky BS, Hoffmann HH, Michailidis E, Moens L, Han JE, Lorenzo L, Bizien L, Meade P, Neehus AL, Ugurbil AC, Corneau A, Kerner G, Zhang P, Rapaport F, Seeleuthner Y, Manry J, Masson C, Schmitt Y, Schluter A, Le Voyer T, Khan T, Li J, Fellay J, Roussel L, Shahrooei M, Alosaimi MF, Mansouri D, Al-Saud H, Al-Mulla F, Almourfi F, Al-Muhsen SZ, Alsohime F, Al Turki S, Hasanato R, van de Beek D, Biondi A, Bettini LR, D’Angio M, Bonfanti P, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Oler AJ, Tompkins MF, Alba C, Vandernoot I, Goffard JC, Smits G, Migeotte I, Haerynck F, Soler-Palacin P, Martin-Nalda A, Colobran R, Morange PE, Keles S, Colkesen F, Ozcelik T, Yasar KK, Senoglu S, Karabela SN, Rodriguez-Gallego C, Novelli G, Hraiech S, Tandjaoui-Lambiotte Y, Duval X, Laouenan C, Clinicians C-S, Clinicians C, Imagine CG, French CCSG, Co VCC, Amsterdam UMCC-B, Effort CHG, Group N-UTCI, Snow AL, Dalgard CL, Milner JD, Vinh DC, Mogensen TH, Marr N, Spaan AN, Boisson B, Boisson-Dupuis S, Bustamante J, Puel A, Ciancanelli MJ, Meyts I, Maniatis T, Soumelis V, Amara A, Nussenzweig M, Garcia-Sastre A, Krammer F, Pujol A, Duffy D, Lifton RP, Zhang SY, Gorochov G, Beziat V, Jouanguy E, Sancho-Shimizu V, Rice CM, Abel L, Notarangelo LD, Cobat A, Su HC, Casanova JL (2020) Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370 https://doi.org/10.1126/science.abd4570
Zhao CL, Rapkiewicz A, Maghsoodi-Deerwester M, Gupta M, Cao W, Palaia T, Zhou J, Ram B, Vo D, Rafiee B, Hossein-Zadeh Z, Dabiri B, Hanna I (2021) Pathological findings in the postmortem liver of patients with coronavirus disease 2019 (COVID-19). Hum Pathol 109:59–68. https://doi.org/10.1016/j.humpath.2020.11.015
Zheng YY, Ma YT, Zhang JY, Xie X (2020) COVID-19 and the cardiovascular system. Nat Rev Cardiol 17:259–260. https://doi.org/10.1038/s41569-020-0360-5
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. https://doi.org/10.1016/S0140-6736(20)30566-3
Zhou L, Niu Z, Jiang X, Zhang Z, Zheng Y, Wang Z, Zhu Y, Gao L, Huang H, Wang X, Sun Q (2020) SARS-CoV-2 Targets by the pscRNA profiling of ACE2, TMPRSS2 and Furin proteases. iScience 23:101744 https://doi.org/10.1016/j.isci.2020.101744
Zippi M, Hong W, Traversa G, Maccioni F, De Biase D, Gallo C, Fiorino S (2020) Involvement of the exocrine pancreas during COVID-19 infection and possible pathogenetic hypothesis: a concise review. Infez Med 28:507–515
Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, Blair C, Weber A, Barnes BJ, Egeblad M, Woods RJ, Kanthi Y, Knight JS (2020) Neutrophil extracellular traps in COVID-19. JCI Insight 5 https://doi.org/10.1172/jci.insight.138999
Acknowledgements
The authors would like to thank Nikki Chapman for editing the manuscript. The authors thank Barbara Klinkhammer for taking pictures of SARS-CoV-2 RNA in situ hybridization.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was prepared as part of the project DEFEAT PANDEMICs, funded by the Federal Ministry of Education and Research (01KX2021) and the German Registry of COVID-19 Autopsies (www.DeRegCOVID.ukaachen.de), funded by the Federal Ministry of Health (ZMVI1-2520COR201).
Author information
Authors and Affiliations
Contributions
The idea for the article was provided by Peter Boor. Till Acker, Martin Aepfelbacher, Kerstin U. Amann, Gustavo Baretton, Peter Barth, Peter Boor, Irene Esposito, Matthias Evert, Falko Fend, Markus Glatzel, Torsten Hansen, Arndt Hartmann, Axel Heinemann, Frank L. Heppner, David Horst, Danny Jonigk, Jan C. Kamp, Bruno Märkl, Benjamin Ondruschka, Susanne Pfefferle, Andreas Rosenwald, Wilfried Roth, Martina Rudelius, Peter Schirmacher, Julia Slotta-Huspenina, Saskia von Stillfried, Ulf Titze, Joachim Weis, Tobias Welte, and Thorsten Wiech performed the literature search and drafted the work. Christopher Werlein, Rainer M. Bohle, Andreas Büttner, Reinhard Büttner, Reinhard Dettmeyer, Philip Eichhorn, Sefer Elezkurtaj, Katja Evert, Nikolaus Gaßler, Stefan Gattenlöhner, Heike Göbel, Elise Gradhand, Julia Hilsenbeck, Gita Mall, Jessica Pablik, Alexander Quaas, Helena Radbruch, Christoph Röcken, Kevin Smith, Linna Sommer, Saskia von Stillfried, Konrad Stock, Philipp Ströbel, Stephanie Strobl, Gregor Weirich, Martin Werner, Claudia Wickenhauser, and Peter Wild contributed data. All authors critically revised the work.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies involving animals performed by any of the authors. All autopsies were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All autopsies were conducted after consent for autopsy was obtained from the deceased or next of kin, or autopsies were requested by the health authorities or by the prosecutor’s office. Each participating center had local ethical approval.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Table 1
COVID-19 Autopsies in DEFEAT PANDEMIcs consortium (DOCX 16 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jonigk, D., Werlein, C., Acker, T. et al. Organ manifestations of COVID-19: what have we learned so far (not only) from autopsies?. Virchows Arch 481, 139–159 (2022). https://doi.org/10.1007/s00428-022-03319-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-022-03319-2