Abstract
Gene fusions can act as oncogenic drivers and offer targets for cancer therapy. Since fusions are rare in colorectal cancer (CRC), their universal screening seems impractical. Our aim was to investigate gene fusions in 62 CRC cases with deficient MLH1 (dMLH1) and BRAFV600E wild-type (wt) status from a consecutive real-life series of 2079 CRCs. First, gene fusions were analysed using a novel FusionPlex Lung v2 RNA–based next-generation sequencing (NGS) panel, and these results were compared to a novel Idylla GeneFusion assay and pan-TRK immunohistochemistry (IHC). NGS detected seven (7/62, 11%) NTRK1 fusions (TPM3::NTRK1, PLEKHA6::NTRK1 and LMNA::NTRK1, each in two cases, and IRF2BP2::NTRK1 in one case). In addition, two ALK, four RET and seven BRAF fusions were identified. Idylla detected seven NTRK1 expression imbalances, in line with the NGS results (overall agreement 100%). Furthermore, Idylla detected the two NGS–identified ALK rearrangements as one specific ALK fusion and one ALK expression imbalance, whilst only two of the four RET fusions were discovered. However, Idylla detected several expression imbalances of ALK (n = 7) and RET (n = 1) that were found to be fusion negative with the NGS. Pan-TRK IHC showed clearly detectable, fusion partner-dependent staining patterns in the seven NTRK1 fusion cases. Overall agreement for pan-TRK antibody clone EPR17341 was 98% and for A7H6R 100% when compared to the NGS. Of the 62 CRCs, 43 were MLH1 promoter hypermethylated (MLH1ph) and 39 were RASwt. All fusion cases were both MLH1ph and RASwt. Our results show that kinase fusions (20/30, 67%) and most importantly targetable NTRK1 fusions (7/30, 23%) are frequent in CRCs with dMLH1/BRAFV600Ewt/MLH1ph/RASwt. NGS was the most comprehensive method in finding the fusions, of which a subset can be screened by Idylla or IHC, provided that the result is confirmed by NGS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Universal screening for mismatch repair deficiency (dMMR) amongst colorectal cancer (CRC) patients has been recommended to facilitate identification of Lynch syndrome (LS) and to direct optimal oncological treatment of those cases presenting a sporadic microsatellite unstable tumour [1]. For the treatment of adult patients with unresectable or metastatic CRC with dMMR or microsatellite instability (MSI), European Medicines Agency (EMA) has approved first-line monotherapy treatment of immuno-oncological drug pembrolizumab in 2020. Besides DNA-repair deficiency phenotype in CRC, gene fusions that act as oncogenic drivers offer targets for cancer therapy. To this end, larotrectinib became the first and entrectinib the second tumour agnostic, i.e. ‘histology-independent’, cancer treatment approved by EMA (2019 and 2020, respectively) in patients whose solid tumours display a neurotrophic tyrosine receptor kinase (NTRK) gene fusion and are advanced, have spread to other parts of the body or are not amenable to surgery, and who have no satisfactory alternative treatments [2]. The family of NTRK genes consists of NTRK1-3 encoding TRKA, TRKB and TRKC proteins that play a role in development and functioning of the nervous system, and act as drivers of oncogenesis in various cancers [3]. Only 0.2–0.3% of CRCs harbour NTRK gene fusions, which makes universal screening of CRC patients for this gene rearrangement impractical. However, recent studies have recognised an enrichment of NTRK fusions in a subset of CRCs presenting dMMR due to loss of MLH1 gene expression, BRAFV600E wild-type (wt), MLH1 promoter hypermethylation (MLH1ph) and RASwt [4,5,6,7].
Gene fusions can be studied using DNA–, RNA– or combined DNA/RNA–based next-generation sequencing (NGS), as well as with fluorescence in situ hybridization (FISH), reverse transcription-polymerase chain reaction (RT-PCR) and immunohistochemistry (IHC). ESMO recommendations propose that IHC can be used as a screening method (if no smooth muscle or neuronal differentiation is present) to enrich patients with NTRK fusions in an unselected population [2, 8]. FISH and RT-PCR are recommended to be used in tumour types that harbour high frequency of a specific NTRK fusion, such as ETV6::NTRK3, which is relatively infrequently found in CRC. DNA– and/or RNA–based NGS panels can be used either upfront or to confirm the presence of NTRK fusion in TRK immunopositive tumours or those devoid of other driver mutations, such as those in BRAF and RAS genes. Of the NGS platforms, RNA–based panels are favoured due to their ability to detect both known and novel fusions and higher sensitivity when compared to the DNA–based ones. Recently, a novel fully automated quantitative RT-PCR option has been introduced, the research use only (RUO) Idylla gene fusion assay, which can detect several oncogenic gene fusions.
The aim of this study was to investigate enrichment of NTRK and other oncogenic gene fusions in a cohort of dMLH1 and BRAFV600Ewt CRC cases (n = 62), which originated from universal dMMR screen of over two thousand consecutive CRC patients in a real-life diagnostic setting. First, gene fusions were analysed using a novel RNA–based FusionPlex Lung v2 NGS panel, and these results were then compared to a novel RNA–based Idylla GeneFusion assay and pan-TRK immunohistochemistry (IHC). In addition to MMR and BRAF mutation status, all 62 cases were analysed for MSI, MLH1ph and RAS mutation status.
Materials and methods
Sample selection
Universal dMMR screening for all newly diagnosed CRC cases has been routine in Helsinki University Hospital (HUH) since January 2018, using IHC for MMR proteins MLH1, MSH2, MSH6 and PMS2. In case of deficient MLH1 IHC, the dMMR screening algorithm leads to BRAFV600E mutation–targeted IHC testing to identify potential LS patients, i.e. immunonegative and thus BRAFV600Ewt, to be further tested for MLH1ph (methylation-positive cases interpreted as sporadic dMLH1). Our patient material consisted of consecutive CRC patients undergoing primary surgery at HUH between January 2018 and April 2020 (n = 2079), out of which 66 showed immunohistochemically confirmed loss of expression of MLH1 (and concomitant loss of PMS2) and BRAFV600Ewt status. Two of the 66 samples were not representative for our study and two were not available leading to a study cohort of 62 samples. The study was approved by the Ethics Committee of the HUH.
MLH1 promoter hypermethylation analysis
MLH1 methylation status of the 62 CRC cases was determined with methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA). Here, SALSA MS-MLPA Probemix ME011-D1 Mismatch Repair Genes kit (MRC Holland, Amsterdam, the Netherlands) was used to determine the methylation status of the promoter regions of MMR genes (MLH1, MSH2, PMS2 and MSH6) using probes that contain a digestion site for the methylation-sensitive restriction endonuclease HhaI. At the same time, the BRAFV600E point mutation was detected with a probe specific for this mutation. The kit includes six probe pairs for MLH1 (covering A to D sites in MLH1 promoter and MLH1 intron 1 93 nt after exon 1). All reactions and analysis of the results were accomplished according to the manufacturer’s instructions and as described in Gylling et al. [9]. DNA amount of 250 ng extracted from formalin-fixed paraffin-embedded (FFPE) tissue samples was used for each MLPA reaction. As a threshold, ratio of > 0.15 (corresponding to > 15% of methylated DNA) was used to indicate promoter methylation. In addition, 45/62 CRC cases were analysed using bisulphite pyrosequencing of the MLH1 promoter region to detect methylated cytosines in tumour DNA at positions c.-269, c.-262, c.-252 and c.-250 of the MLH1 gene, showing 100% concordant results.
Idylla KRAS and NRAS-BRAF mutation tests, and MSI test
KRAS and NRAS mutations were identified, and the BRAFV600wt status confirmed, from the 62 CRC FFPE samples using real-time PCR–based CE-IVD–validated Idylla KRAS and NRAS-BRAF mutation assays (Biocartis NV, Mechelen, Belgium). Furthermore, the MSI status of the dMMR samples was confirmed with Idylla MSI test that was performed as described previously [10]. The Idylla KRAS Mutation Test detects 21 KRAS mutations in exons 2, 3 and 4, whereas the Idylla NRAS-BRAF Mutation Test detects 18 NRAS mutations in exons 2, 3 and 4, and five BRAF mutations in codon 600. For the analyses, 10-µm thick tissue slices were cut from the CRC tissue blocks. The tissue sample handling and analysis were performed according to the manufacturer’s protocol and 20 to 80% of tumour cell content was used.
Next generation sequencing (NGS) analysis
The cohort of 62 dMLH1/BRAFV600Ewt CRC FFPE samples underwent RNA–based targeted NGS analysis using novel RUO FusionPlex® Lung v2 (Invitae Corporation, San Francisco, CA) which identifies fusion transcripts of ALK, BRAF, EGFR, ERBB2, FGFR1, FGFR2, FGFR3, KRAS, MET, NRG1, NTRK1, NTRK2, NTRK3, NUTM1, PIK3CA, RET and ROS1 genes. Total nucleic acid was extracted from 10-µm FFPE tissue sections using QIASymphony RNA Kit (QIAGEN, Valencia, CA), and the quality was assessed by Qubit RNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA). RNA was then reversely transcribed, and the quality was checked by ABI StepOne Plus (Applied Biosystems/Thermo Fisher Scientific). Libraries were quantified with KAPA Library Quantification Kit Illumina® Platforms (ABI StepOne Plus). Libraries were paired-end sequenced at 2 × 150 cycles on Illumina Novaseq 6000 instrument using SP flow cell with Novaseq Xp workflow (Individual Lane loading). Data were analysed using the Archer Analysis v6.2.7 software (Archer/Invitae) for the presence of gene fusion using GRCh37 as the reference genome (Supplementary Table S1).
NTRK1 fusions were further validated with two independent RNA–based NGS platforms, FusionPlex Comprehensive Thyroid and Lung panel (CTL AK0070 v1.1, Invitae) and TruSight RNA Pan-Cancer panel (Illumina, San Diego, CA). FusionPlex CTL Kit detects fusion transcripts of ALK, AXL, BRAF, CCND1, FGFR1, FGFR2, FGFR3, MET, NRG1, NTRK1, NTRK2, NTRK3, PPARG, RAF1, RET, ROS1 and THADA genes. In short, total nucleic acid was extracted from 10-µm FFPE tissue sections using QIASymphony RNA Kit (QIAGEN), and the quality was assessed by Qubit RNA HS Assay Kit (Thermo Fisher Scientific). RNA was then reversely transcribed, and the quality was checked by ABI StepOne Plus (Applied Biosystems/Thermo Fisher Scientific). Libraries were quantified with Ion Library TaqMan Quantitation Kit (ABI StepOne Plus) and sequenced on either Ion S5 or Ion Proton systems (IonTorrent/Thermo Fisher Scientific). Data were analysed using the Archer analysis software (Suite_Analysis_v6.0.4; Invitae) for the presence of gene fusion using GRCh37 as the reference genome (Supplementary Table S1).
For Illumina TruSight RNA Pan-Cancer panel, total RNA was extracted from 10-µm FFPE tissue sections using RNeasy FFPE kit (QIAGEN) according to the manufacturer’s protocol. The RNA quality (DV200) was assessed using the Agilent Bioanalyzer 2100 instrument (Agilent, Santa Clara, CA). Targeted RNA libraries were prepared according to the Illumina TruSight RNA Pan-Cancer panel reference guide (Illumina), and the amount of input RNA was evaluated based on DV200 value. The final libraries were paired-end sequenced at 2 × 75 cycles on Illumina MiniSeq instrument using its High Output kit. Illumina DRAGEN software (v3.8.4) was used for the fusion calling and GRCh38 was used as the reference genome (Supplementary Table S1).
NGS analysis of oncogenic mutations in a metastasized case (#52) with synchronous CRC was done using an in-house cancer panel containing seven target genes (PIK3CA, EGFR, KIT, KRAS, MET, NRAS and PDGFRA) and exons 11–15 of BRAF and was performed as previously described [11].
Idylla gene fusion test
All 62 cases were further tested with the RUO real-time RT-PCR–based Idylla™ GeneFusion Assay (Biocartis NV) to detect ALK, ROS1, RET and MET exon 14 skipping and NTRK1/2/3 fusions in single cartridge and approximately in 3 h. For NTRK1/2/3, ALK, ROS and RET fusions, the detection was performed with expression imbalances indicating putative gene fusion irrespective of fusion partner based on the 3’ kinase overexpression caused by the partner gene. For ALK, ROS and RET fusions and MET ex14 skipping, the detection was additionally performed by assessing specific gene fusion variations of the most common variants. For the analysis, 10-µm sections were cut from the FFPE CRC tissue blocks. The tissue sample handling and analysis were performed according to the manufacturer’s protocol and 20 to 80% of tumour cell content was used.
Immunohistochemistry
For immunohistochemical analyses, 4-µm sections cut from the FFPE CRC tissue blocks (n = 62) were used. The MMR IHC was performed as described previously [10] and BRAFV600E mutation was detected by using the specific monoclonal ready-to-use antibody (clone VE1, 760–5095, Roche, Tucson, AZ), utilising detection with OptiView DAB kit (760–700, Roche) and additional Amplification Kit (760–099, Roche) [12]. Pan-TRK IHC was performed using ready-to-use monoclonal antibody (clone EPR17341, 790–7026, Roche) and detection by OptiView DAB kit (760–700, Roche). ALK IHC was performed using monoclonal antibody (clone 5A4, Novocastra™, Leica Biosystems, Newcastle Upon Tyne, UK) at dilution 1:50 and Roche’s OptiView DAB and Amplification kits. All above-described immunostainings were performed with the Ventana Benchmark ULTRA immunostainer (Roche). The other pan-TRK antibody used in this study (clone A7H6R, #92,991, Cell Signaling Technology Inc., Danvers, MA) was diluted 1:50 and stained with Autostainer (Agilent) using Envision Flex High pH detection kit (K8000, Agilent). Each slide had a non-neoplastic colorectal specimen as a positive external control (neural structures), and since we used (freshly cut) whole tissue sections of the tumour samples, most of them also contained positive internal control (neural structures in the muscularis propria). IHC stainings were analysed in a blinded manner by IU and AR. Positive pan-TRK immunoreactivity, indicating putative NTRK fusion, was determined when ≥ 1% of the cancer cells displayed cytoplasmic, membranous, nuclear and/or perinuclear immunopositivity.
Statistical analysis
The RNA–based NGS panel (FusionPlex Lung v2) was considered as the gold standard test against which the overall agreement, sensitivity and specificity, and the positive and negative predictive values (PPV and NPV, respectively) were calculated. Fisher’s exact test (GraphPad QuickCalcs: https://www.graphpad.com/quickcalcs/contingency2/) was used for comparison of the agreement between NGS and the Idylla test, and between NGS and the IHC methods. Clinicopathological characteristics between fusion positive and negative cases were compared using Mann–Whitney U-test (numerical variables) or Fisher’s exact test (categorical variables). All statistical tests were two-tailed and numeric variables are reported by median and range. P-value less than 0.05 was considered as statistically significant.
Results
Clinicopathological characteristics and MLH1 promoter hypermethylation and RAS mutation status of the CRC cases
Our study consisted of 62 CRC samples with dMLH1 and BRAFV600Ewt status as detected by IHC. Of these patients, 59.7% were females and the tumours localised mainly to the right colon (85.5%), were low-grade (69.4%), pT3 (59.7%), pN0 (64.5%) and M0 (80.6%) (Table 1). Of the cases, 42/60 were MLH1ph as detected by MS-MLPA. One case was not available for the analysis. One case was not analysable despite three attempts, but was successfully analysed using bisulphite pyrosequencing and found to be MLH1ph. Overall, 43/61 (70.5%) of the CRC cases were MLH1ph (Supplementary Table S2). KRAS mutations were detected in 20 cases by Idylla KRAS mutation test, and three NRAS mutations were detected by Idylla NRAS-BRAF mutation test (Supplementary Table S2). Overall, RAS mutations were found in 23 (37.1%) of the dMLH1/BRAFV600Ewt CRC cases. KRAS mutations were the most common in codon 12 (10 cases), followed by codon 61 (five), codon 146 (four) and codon 13 (one). NRAS mutations were found in codons 61 (two cases) and 12 (one case). MSI status was verified using Idylla MSI test, which resulted MSI in 62/62 of the cases (one sample, case 16, was initially MSS but after re-analysis using a more representative tissue block resulted MSI). Finally, the NRAS-BRAF test (n = 62) and the MS-MLPA (n = 60) analyses confirmed the BRAFV600Ewt status of all CRC cases.
RNA–based NGS fusion panel analysis
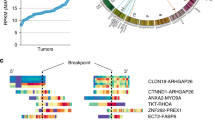
Expanded RNA–based NGS panel FusionPlex Lung v2 was used as the gold standard for detection of gene fusions in the 62 dMLH1/BRAFV600Ewt CRC samples. Seven in-frame NTRK1 fusions (Table 2) were identified (TPM3::NTRK1, PLEKHA6::NTRK1 and LMNA::NTRK1, each in two cases, and IRF2BP2::NTRK1 in one case), of which IRF2BP2::NTRK1 is a novel fusion in CRC (Fig. 1). FusionPlex CTL and/or TruSight RNA Pan-Cancer NGS panels confirmed the seven NTRK1 fusions (Supplementary Table S3). An NTRK1 fusion was thus identified in 7/62 (11.3%) of the dMLH1/BRAFV600Ewt, in 7/43 (16.3%) of the dMLH1/BRAFV600Ewt/MLH1ph and in 7/30 (23.3%) of the dMLH1/BRAFV600Ewt/MLH1ph/RASwt CRC samples. In addition to NTRK1 fusions, Lung v2 NGS detected two in-frame ALK fusions (EML4::ALK in both), four in-frame RET fusions (CCDC6::RET in three cases and NCOA4::RET in one case) and seven in-frame BRAF fusions (AGAP3::BRAF and TRIM24::BRAF both in two cases, and STARD3NL::BRAF, MKRN1::BRAF and LMTK2::BRAF each in one case) (Table 2). Analysis by the Lung v2 NGS failed in four cases (#7, 9, 11 and 26). These four samples were reanalysed using FusionPlex CTL NGS kit, and three were found to be fusion negative, whereas one failed with this panel as well (case 7). A kinase fusion was thus identified in 20/62 (32.3%) of the dMLH1/BRAFV600Ewt, in 20/43 (46.5%) of the dMLH1/BRAFV600Ewt/MLH1ph and in 20/30 (66.7%) of the dMLH1/BRAFV600Ewt/MLH1ph/RASwt CRC samples.
Fusion-positive cases (n = 20) had significantly higher preponderance of MLH1ph (P = 0.0002) and RASwt genotype (P < 0.0001) compared to fusion negative cases (n = 42), but no statistically significant differences in clinicopathological features including age, sex, pTNM stage, grade or tumour site were observed between the negative and positive cases (Supplementary Table S4). One NTRK1, one ALK and one RET fusion case presented distant metastases (cases 12, 35 and 43) and were all RASwt. Furthermore, one NTRK1-rearranged case with distant metastases (case 52) had synchronous sigma tumour with proficient (p)MMR and KRASG12V mutation, as was also shown to be the case for the peritoneal metastasis, whilst the NTRK1-rearranged tumour in colon ascendens was dMMR and RASwt.
Idylla gene fusion assay
All 62 CRC samples with dMLH1 and BRAFV600Ewt status were next analysed using Idylla gene fusion assay. NTRK1 expression imbalance was identified in seven cases, which coincided with the Lung v2 NGS fusion cases (Table 2 and Supplementary Table S2). The concordance between Idylla gene fusion test NTRK1 expression imbalances and NTRK1 fusions detected by Lung v2 NGS was 100% (59/59 valid Idylla tests, P < 0.0001; 3/62 Idylla analyses were invalid). Using the NGS as a reference, the sensitivity, specificity, PPV and NPV of Idylla gene fusion test to identify expression imbalance in NTRK1 fusion cases were 100% for each. Idylla detected the Lung v2 NGS identified EML4(e6)::ALK(e20) fusion as a specific ALK fusion. The second ALK fusion identified by the Lung v2 NGS (EML4(e21)::ALK(e20)) is not included in the set of specific fusions detected by Idylla, and it was thus detected as an ALK expression imbalance. However, Idylla reported seven additional ALK expression imbalances in cases, which were not ALK fusion–positive by Lung v2 NGS. Thus, concordance between Idylla ALK specific fusion or expression imbalance and the Lung v2 NGS was 88.1% (52/59 valid Idylla tests, P = 0.021), and the sensitivity, specificity, PPV and NPV were 100%, 87.7%, 22.2% and 100%, respectively. In addition, two combined RET–specific fusion and RET expression imbalance results were detected in line with the Lung v2 NGS results, whilst two RET fusions were not detected by the Idylla analysis (specific fusion detection not included and no RET expression imbalance reported). Further, one false positive RET expression imbalance was reported. Thus, concordance between Idylla RET–specific fusion and/or expression imbalance and the Lung v2 NGS was 94.9% (56/59 valid Idylla tests, P = 0.010), and the sensitivity, specificity, PPV and NPV were 50%, 98.2%, 66.7% and 96.4%, respectively. Initially, seven Idylla results were invalid due to improper/inappropriate RNA amplification, of which four cases were successfully re-analysed (Table 2 and Supplementary Table S2). It should be pointed out that none of the initially invalid samples in the Idylla analyses was the same as the four samples that failed with the Lung v2 NGS. In addition, some individual targets remained invalid in the Idylla analyses despite re-runs, which was especially common in the expression imbalance of NTRK3 (12/59, 20.3%; Supplementary Table S2).
Pan-TRK and ALK IHC
Pan-TRK IHC was used to screen for NTRK gene fusions in the 62 CRC samples using two antibody clones and scored in blinded manner by two observers independently. With clone EPR17341 eight and with clone A7H6R seven, CRC samples were scored as immunopositive by both observers. However, staining intensity was higher and subcellular localization clearer using the clone EPR17341 protocol and scoring results using this clone are shown in Table 2. All samples showed cytoplasmic immunopositivity with variable intensity that was relatively diffuse (50–100% of the tumour cells being positive), and four samples also expressed membranous and two perinuclear staining (Table 2, Fig. 2 and Supplementary Fig. S1). No nuclear staining was observed. The overall agreement between pan-TRK IHC (EPR17341) and NGS was 98.4% (61/62, P < 0.0001). Using the NGS as a reference, the sensitivity of pan-TRK (EPR17341) IHC was 100% and the specificity 98.2% with PPV of 87.5% and NPV of 100%. For the clone A7H6R, these parameters were 100%. Importantly, one EPR17341 pan-TRK IHC positive sample, expressing the lowest cytoplasmic staining intensity and being negative with clone A7H6R, did not contain an NTRK fusion but an ALK fusion (case 12) (Table 2 and Supplementary Figs. S1 and S2). This case, and the other ALK fusion case, showed strong cytoplasmic ALK immunopositivity, whilst the rest of the samples were completely negative (Table 2 and Supplementary Fig. S2).
Pan-TRK (clone EPR17341) immunostaining patterns in CRC samples. A Strong cytoplasmic staining with moderate membranous staining (Case 19; TPM3::NTRK1). B Moderate cytoplasmic staining with perinuclear staining (Case 29; LMNA::NTRK1). C Weak cytoplasmic staining with moderate membranous staining (Case 50; PLEKHA6::NTRK1). D Moderate cytoplasmic staining (Case 59; IRF2BP2::NTRK1). Original magnification 400 ×
Discussion
We screened 2079 CRC resection specimens for dMLH1 and BRAFV600Ewt using IHC and identified 64 such cases of which 62 were available for this study. The BRAFV600E IHC result was confirmed in this study by MS-MLPA and Idylla NRAS-BRAF test, which both were 100% concordant. The MSI status was confirmed with Idylla MSI test. Since NTRK gene fusions have been shown to be enriched in this subgroup of CRC [4, 7] and solid tumours with NTRK fusion can be treated with larotrectinib or entrectinib [2], we were especially interested to investigate this targetable gene rearrangement using several techniques. First, RNA–based Lung v2 NGS panel was used as the gold standard, and seven (7/62, 11.3%) NTRK1 fusions were found in our CRC cohort with dMLH1/BRAFV600Ewt. Westphalen et al. recently showed the prevalence of NTRK fusions in CRC to be 0.22% in a large real-world cohort of adult CRC cases (n = 34 590) using NGS–based database approach [7]. This figure is comparable to previous screening studies for NTRK fusions in CRC that have found the prevalence to be 0.14–0.35% using upfront NGS or IHC with NGS confirmation [4,5,6, 13,14,15,16]. Thus, our NTRK fusion frequency of 0.34% (7/2079) is at the higher end.
NTRK1 fusions with TPM3, LMNA and TPR partners have been shown to be the most common ones in CRC [6, 7] and PLEKHA6::NTRK1 fusions have also previously been described in this disease [17, 18]. Of these, we found NTRK1 to partner with TPM3, LMNA and PLEKHA6. IRF2BP2::NTRK1 fusion was found in a single case of our series, and to our knowledge, this is the first time it has been reported in CRC. Interestingly, the IRF2BP2::NTRK1 containing tumour was the only one located to the left colon (sigma) amongst our NTRK1 fusion–positive cases. We were, however, unable to find any NTRK3 fusions, although Lung v2 NGS can detect a wide range of NTRK3 fusions. This may relate to the fact that the estimated proportion of NTRK3 fusions of all NTRK fusions is only 11% in CRC [7]. Strengths of our study included the use of real-life diagnostic tissue material originating from a cohort of over two thousand consecutive surgically treated primary CRC patients and the use of multiple techniques, including detection of the NTRK1 fusions using three independent RNA–based NGS platforms. Weakness of the study was that we investigated gene fusions only in a subgroup of CRC. To this end, we may have missed a few NTRK fusion–positive CRCs, since a small proportion (11–19%) of NTRK fusions has been found in microsatellite-stable CRCs [4, 6]. Four samples failed with Lung v2 NGS, three of which were found to be fusion-negative by the FusionPlex CTL NGS analysis, whereas the fourth one failed with CTL panel as well.
Westphalen et al. found CRC to be the only cancer type in which NTRK fusions are associated with sporadic MSI [7]. Interestingly, Kim et al. have recently described that NTRK fusions in CRC develop along the serrated pathway, in which sporadic dMLH1 is a major molecular event, and these fusions can already be present in premalignant sessile serrated lesions [19]. Additionally, several other NGS–based studies have suggested 2.6–7.3% occurrence of NTRK fusions in dMMR/MSI CRCs [15, 16, 18, 20, 21]. In the dMLH1/BRAFV600Ewt subgroup, frequency of NTRK fusions has been reported to be 5–28% [4, 16, 17] and in the subgroup of dMLH1/MLH1ph 14–19% [18, 19], which are in line with our frequencies of 11% (7/62) and 16% (7/43), respectively. Yet other studies have reported NTRK fusions to occur in dMLH1/BRAFV600Ewt/MLH1ph/RASwt subgroup of CRC with as high frequency as 17–44% [15,16,17,18, 22] being comparable to our prevalence of 23.3% (7/30).
We also evaluated the performance of the novel fully automated Idylla gene fusion assay and pan-TRK IHC. Idylla gene fusion test was 100% specific and sensitive to detect NTRK1 expression imbalance in the seven NTRK1 fusion CRC cases. However, Idylla reported initially invalid result in seven cases (7/62, 11%), of which three remained invalid after re-analysis, and additional 12 isolated invalid results for NTRK3 expression imbalance. Interestingly, the high frequency of NTRK3 invalids might originate from promoter methylation causing loss of NTRK3 expression, which has been reported in over 11% of CRC cases [23]. Furthermore, our study shows that specificity and sensitivity of pan-TRK IHC are optimal in CRC samples. Although specificity of this method varies between different tumour types, it has previously been reported to be 100% for pan-TRK in CRC [4, 13, 17, 24]. Pan-TRK IHC positive CRCs are characterised by cytoplasmic staining with additional positivity in other subcellular compartments in a fusion partner-dependent manner. Membranous staining has been linked to TPM3, TPR and PLEKHA6; perinuclear staining to LMNA and MUC2 and nuclear staining to ETV6 [6, 24,25,26]. We also detected moderate membranous staining along with variable intensity of cytoplasmic staining with TPM3 and PLEKHA6 fusion partners and perinuclear staining with LMNA. IRF2BP2::NTRK1 fusion represents only 2% of all NTRK fusions [7], but it has been reported in lung, thyroid and prostate cancers [3]. In lung cancer, it shows cytoplasmic immunostaining [25], which was the case with our CRC sample as well.
In addition to NTRK1 fusions, Lung v2 NGS detected two ALK fusions (2/62, 3.2%) in our CRC cohort, which both partnered with the most common ALK fusion partner EML4 in CRC [27]. ALK IHC showed strong cytoplasmic staining in both cases, whilst the rest of the samples were completely negative. Our results thus suggest a prevalence of 0.10% (2/2079) for ALK fusions in CRC, which is comparable to previously published ALK fusion prevalence of 0.05–0.6% in CRC [27,28,29,30]. The Idylla fusion assay detected the EML4(e6)::ALK(e20) fusion as a specific fusion and the EML4(e21)::ALK(e20) fusion as an ALK expression imbalance. The EML4(e21)::ALK(e20) fusion with non-canonical breakpoint of EML4 gene at exon 21 has been reported to constitute only about 2% of the ALK-rearrangements in non-small cell lung cancer, where EML4(e6)::ALK(e20) is the most common EML4::ALK variant [31]. The less frequent EML4(e21)::ALK(e20) variant is not covered by the Idylla’s fusion-specific detection, which is designed to catch the most relevant gene fusions in lung cancer. However, EML4(e21)::ALK(e20) fusion variant seems to be more frequent molecular event in CRC [22, 27, 29, 30]. In addition to the NGS–identified ALK fusions, Idylla detected seven false positive ALK expression imbalances. Based on our study, detection of specific ALK fusion seems to be a valid result, whereas all expression imbalance results need to be validated by a more specific method.
Besides NTRK1 and ALK fusions, Lung v2 NGS detected four RET fusions (4/62, 6.5%) and seven BRAF fusions (7/62, 11.3%) in our CRC cohort. We found RET to partner with CCDC6 in three cases and NCOA4 in one case, both of which seem to be quite common RET fusion partners in CRC [32, 33]. BRAF fusions with partners AGAP3, TRIM24 and MKRN1 found in our study have also been previously reported to occur in CRC [15, 16, 34]. STARD3NL::BRAF fusion has earlier been described at least in one paediatric sarcoma [35], whereas to our best knowledge, LMTK2::BRAF has not been reported before in any tumour type. Two of the four Lung v2 NGS–detected RET fusions were detected by the Idylla assay as both RET–specific fusion and expression imbalance. The Idylla gene fusion test does not include BRAF fusions. Taken together, the Idylla platform identified two specific RET fusions that were in line with the NGS results, but did not report expression imbalance of two NGS–detected RET fusions (detection of these specific fusions is not included in the Idylla assay).
Upfront RNA–based NGS analysis is the most comprehensive, sensitive and specific method to identify gene fusions, but it is also time-consuming and requires more labour, expertise and financial resources when compared to other methods. The Idylla platform offers the fastest turnaround time with moderate cost, whilst IHC is the most affordable option. It is however clear that both Idylla expression imbalance and pan-TRK IHC results need to be validated using an alternative method, preferably an RNA–based NGS [2, 8]. To our knowledge, this is the first publication where the RUO FusionPlex Lung v2 NGS and the Idylla GeneFusion assay have been used in detecting gene fusions. As there is a tumour agnostic oncological treatment for cancer patients with an NTRK fusion, we would like to propose that one should screen for this gene rearrangement in CRC patients with dMLH1/BRAFV600Ewt/MLH1ph using Idylla gene fusion test or pan-TRK IHC, followed by an RNA–based NGS confirmation of the positive cases, or alternatively using upfront RNA–based NGS depending on local resources.
Availability of data and materials
The data obtained during the current study are available from the corresponding author AR on reasonable request.
Code availability
Not applicable.
Abbreviations
- ALK:
-
Anaplastic lymphoma kinase
- BRAF:
-
V-raf murine sarcoma viral oncogene homolog B1
- CRC:
-
Colorectal cancer
- CTL:
-
Comprehensive Thyroid and Lung panel
- dMMR:
-
Deficient MMR
- EMA:
-
European Medicines Agency
- FFPE:
-
Formalin-fixed paraffin-embedded
- HUH:
-
Helsinki University Hospital
- IHC:
-
Immunohistochemistry
- KRAS:
-
Kirsten rat sarcoma viral oncogene homolog
- LS:
-
Lynch syndrome
- MLH1:
-
MutL homolog 1
- MMR:
-
Mismatch repair
- MSI:
-
Microsatellite instability
- MS-MLPA:
-
Methylation-specific multiplex ligation-dependent probe amplification
- NGS:
-
Next-generation sequencing
- NPV:
-
Negative predictive value
- NRAS:
-
Neuroblastoma ras viral oncogene homolog
- NTRK:
-
Neurotrophic tyrosine receptor kinase
- pMMR:
-
Proficient MMR
- PPV:
-
Positive predictive value
- RAS:
-
Rat sarcoma virus
- RET:
-
rearranged during transfection
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- RUO:
-
Research use only
- TRK:
-
Tropomyosin receptor kinase
- wt:
-
Wild-type
References
Luchini C, Bibeau F, Ligtenberg MJL, Singh N, Nottegar A, Bosse T, Miller R, Riaz N, Douillard JY, Andre F, Scarpa A (2019) ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol 30:1232–1243
Yoshino T, Pentheroudakis G, Mishima S, Overman MJ, Yeh KH, Baba E, Naito Y, Calvo F, Saxena A, Chen LT, Takeda M, Cervantes A, Taniguchi H, Yoshida K, Kodera Y, Kitagawa Y, Tabernero J, Burris H, Douillard JY (2020) JSCO-ESMO-ASCO-JSMO-TOS: international expert consensus recommendations for tumour-agnostic treatments in patients with solid tumours with microsatellite instability or NTRK fusions. Ann Oncol 31:861–872
Amatu A, Sartore-Bianchi A, Bencardino K, Pizzutilo EG, Tosi F, Siena S (2019) Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann Oncol 30:viii5-viii15
Chou A, Fraser T, Ahadi M, Fuchs T, Sioson L, Clarkson A, Sheen A, Singh N, Corless CL, Gill AJ (2019) NTRK gene rearrangements are highly enriched in MLH1/PMS2 deficient, BRAF wild-type colorectal carcinomas—a study of 4569 cases. Mod Pathol 33:924–932
Forsythe A, Zhang W, Phillip Strauss U, Fellous M, Korei M, Keating K (2020) A systematic review and meta-analysis of neurotrophic tyrosine receptor kinase gene fusion frequencies in solid tumors. Ther Adv Med Oncol 12:1758835920975613
Lasota J, Chłopek M, Lamoureux J, Christiansen J, Kowalik A, Wasąg B, Felisiak-Gołąbek A, Agaimy A, Biernat W, Canzonieri V, Centonze G, Chmielik E, Daum O, Dubová M, Dziuba I, Goertz S, Góźdź S, Guttmejer-Nasierowska A, Haglund C, Hałoń A, Hartmann A, Inaguma S, Iżycka-Świeszewska E, Kaczorowski M, Kita P, Kołos M, Kopczyński J, Michal M, Milione M, Okoń K, Pęksa R, Pyzlak M, Ristimäki A, Ryś J, Szostak B, Szpor J, Szumiło J, Teresiński L, Waloszczyk P, Wejman J, Wesołowski W, Miettinen M (2020) Colonic adenocarcinomas harboring NTRK fusion genes: a clinicopathologic and molecular genetic study of 16 cases and review of the literature. Am J Surg Pathol 44:162–173
Westphalen CB, Krebs MG, Le Tourneau C, Sokol ES, Maund SL, Wilson TR, Jin DX, Newberg JY, Fabrizio D, Veronese L, Thomas M, de Braud F (2021) Genomic context of NTRK1/2/3 fusion-positive tumours from a large real-world population. NPJ Precis Oncol 5:69
Marchiò C, Scaltriti M, Ladanyi M, Iafrate AJ, Bibeau F, Dietel M, Hechtman JF, Troiani T, López-Rios F, Douillard JY, Andrè F, Reis-Filho JS (2019) ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol 30:1417–1427
Gylling A, Ridanpää M, Vierimaa O, Aittomäki K, Avela K, Kääriäinen H, Laivuori H, Pöyhönen M, Sallinen SL, Wallgren- Pettersson C, Järvinen HJ, Mecklin JP, Peltomäki P (2009) Large genomic rearrangements and germline epimutations in Lynch syndrome. Int J Cancer 124:2333–2340
Ukkola I, Nummela P, Pasanen A, Kero M, Lepistö A, Kytölä S, Bützow R, Ristimäki A (2021) Detection of microsatellite instability with Idylla MSI assay in colorectal and endometrial cancer. Virchows Arch 479:471–479
Holm M, Andersson E, Osterlund E, Ovissi A, Soveri LM, Anttonen AK, Kytölä S, Aittomäki K, Osterlund P, Ristimäki A (2020) Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, Idylla, and next generation sequencing. PLoS One 15:e0239819
Thiel A, Heinonen M, Kantonen J, Gylling A, Lahtinen L, Korhonen M, Kytölä S, Mecklin JP, Orpana A, Peltomäki P, Ristimäki A (2013) BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Arch 463:613–621
Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, Jungbluth AA, Zehir A, Benayed R, Drilon A, Hyman DM, Ladanyi M, Sireci AN, Hechtman JF (2020) NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol 33:38–46
Rosen EY, Goldman DA, Hechtman JF, Benayed R, Schram AM, Cocco E, Shifman S, Gong Y, Kundra R, Solomon JP, Bardelli A, Scaltriti M, Drilon A, Iasonos A, Taylor BS, Hyman DM (2020) TRK fusions are enriched in cancers with uncommon histologies and the absence of canonical driver mutations. Clin Cancer Res 26:1624–1632
Cocco E, Benhamida J, Middha S, Zehir A, Mullaney K, Shia J, Yaeger R, Zhang L, Wong D, Villafania L, Nafa K, Scaltriti M, Drilon A, Saltz L, Schram AM, Stadler ZK, Hyman DM, Benayed R, Ladanyi M, Hechtman JF (2019) Colorectal carcinomas containing hypermethylated MLH1 promoter and wild-type BRAF/KRAS are enriched for targetable kinase fusions. Canc Res 79:1047–1053
Sato K, Kawazu M, Yamamoto Y, Ueno T, Kojima S, Nagae G, Abe H, Soda M, Oga T, Kohsaka S, Sai E, Yamashita Y, Iinuma H, Fukayama M, Aburatani H, Watanabe T, Mano H (2019) Fusion kinases identified by genomic analyses of sporadic microsatellite instability–high colorectal cancers. Clin Cancer Res 25:378–389
Bocciarelli C, Caumont C, Samaison L, Cariou M, Aline-Fardin A, Doucet L, Roudié J, Terris B, Merlio JP, Marcorelles P, Cappellen D, Uguen A (2021) MSI-High RAS-BRAF wild-type colorectal adenocarcinomas with MLH1 loss have a high frequency of targetable oncogenic gene fusions whose diagnoses are feasible using methods easy-to-implement in pathology laboratories. Hum Pathol 114:99–109
Wang J, Yi Y, Xiao Y, Dong L, Liang L, Teng L, Ying JM, Lu T, Liu Y, Guan Y, Pang J, Zhou L, Lu J, Zhang Z, Liu X, Liang X, Zeng X, Yi X, Zhou W, Xia X, Yang L, Zhang J, Kopetz S, Futreal PA, Wu H, Liang Z (2019) Prevalence of recurrent oncogenic fusion in mismatch repair-deficient colorectal carcinoma with hypermethylated MLH1 and wild-type BRAF and KRAS. Mod Pathol 32:1053–1064
Kim JH, Hong JH, Choi YL, Lee JA, Seo MK, Lee MS, An SB, Sung MJ, Cho NY, Kim SS, Shin YK, Kim S, Kang GH (2021) NTRK oncogenic fusions are exclusively associated with the serrated neoplasia pathway in the colorectum and begin to occur in sessile serrated lesions. J Pathol 255:399–411
Berrino E, Bragoni A, Annaratone L, Fenocchio E, Carnevale-Schianca F, Garetto L, Aglietta M, Sarotto I, Casorzo L, Venesio T, Sapino A, Marchiò C (2021) Pursuit of gene fusions in daily practice: evidence from real-world data in wild-type and microsatellite instable patients. Cancers (Basel) 13:3376
Guo Y, Guo XL, Wang S, Chen X, Shi J, Wang J, Wang K, Klempner SJ, Wang W, Xiao M (2020) Genomic alterations of NTRK, POLE, ERBB2, and microsatellite instability status in Chinese patients with colorectal cancer. Oncologist 25:e1671–e1680
Vaňková B, Vaněček T, Ptáková N, Hájková V, Dušek MM, Michal M, Švajdler P, Daum OO, Daumová M, Michal M, Mezencev R, Švajdler M (2020) Targeted next generation sequencing of MLH1 -deficient, MLH1 promoter hypermethylated, and BRAF/RAS -wild-type colorectal adenocarcinomas is effective in detecting tumors with actionable oncogenic gene fusions. Genes Chromosomes Cancer 59:562
Chen Z, Huang Z, Luo Y, Zou Q, Bai L, Tang G, Wang X, Cao G, Huang M, Xiang J, Yu H (2021) Genome-wide analysis identifies critical DNA methylations within NTRKs genes in colorectal cancer. J Transl Med 19:73
Solomon JP, Benayed R, Hechtman JF, Ladanyi M (2019) Identifying patients with NTRK fusion cancer. Ann Oncol 30:viii16-viii22
Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, Arcila ME, Dogan S, Klimstra DS, Ladanyi M, Jungbluth AA (2017) Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol 41:1547–1551
Conde E, Hernandez S, Sanchez E, Regojo RM, Camacho C, Alonso M, Martinez R, Lopez-Rios F (2021) Pan-TRK immunohistochemistry: an example-based practical approach to efficiently identify patients with NTRK fusion cancer. Arch Pathol Lab Med 145:1031–1040
Lasota J, Chłopek M, Wasąg B, Kowalik A, Christiansen J, Lamoureux J, Kuźniacka A, Felisiak-Gołąbek A, Liu Y, Reyes TAR, Saha R, Agaimy A, Behenska K, Biernat W, Cattaneo L, Centonze G, Daum O, Daumova M, Domagała P, Dziuba I, Geppert CE, Góźdź S, Nasierowska-Guttmejer A, Hałoń A, Hartmann A, Inaguma S, Iżycka-Świeszewska E, Kaczorowski M, Kołos M, Kopczyński J, Michal M, Milione M, Okoń K, Pęksa R, Pyzlak M, Ryś J, Waloszczyk P, Wejman J, Miettinen M (2020) Colorectal adenocarcinomas harboring ALK fusion genes: a clinicopathologic and molecular genetic study of 12 cases and review of the literature. Am J Surg Pathol 44:1224–1234
Houang M, Toon CW, Clarkson A, Sioson L, de Silva K, Watson N, Singh NR, Chou A, Gill AJ (2015) ALK and ROS1 overexpression is very rare in colorectal adenocarcinoma. Appl Immunohistochem Mol Morphol 23:134–138
Lee J, Kim HC, Hong JY, Wang K, Kim SY, Jang J, Kim ST, Park JO, Lim HY, Kang WK, Park YS, Lee J, Lee WY, Park YA, Huh JW, Yun SH, Do IG, Kim SH, Balasubramanian S, Stephens PJ, Ross JS, Li GG, Hornby Z, Ali SM, Miller VA, Kim KM, Ou SH (2015) Detection of novel and potentially actionable anaplastic lymphoma kinase (ALK) rearrangement in colorectal adenocarcinoma by immunohistochemistry screening. Oncotarget 6:24320–24332
Yakirevich E, Resnick MB, Mangray S, Wheeler M, Jackson CL, Lombardo KA, Lee J, Kim KM, Gill AJ, Wang K, Gowen K, Sun J, Miller VA, Stephens PJ, Ali SM, Ross JS, Safran H (2016) Oncogenic ALK fusion in rare and aggressive subtype of colorectal adenocarcinoma as a potential therapeutic target. Clin Cancer Res 22:3831–3840
Xia P, Zhang L, Li P, Liu E, Li W, Zhang J, Li H, Su X, Jiang G (2021) Molecular characteristics and clinical outcomes of complex ALK rearrangements identified by next-generation sequencing in non-small cell lung cancers. J Transl Med 19:30
Le Rolle AF, Klempner SJ, Garrett CR, Seery T, Sanford EM, Balasubramanian S, Ross JS, Stephens PJ, Miller VA, Ali SM, Chiu VK (2015) Identification and characterization of RET fusions in advanced colorectal cancer. Oncotarget 6:28929–28937
Shi M, Wang W, Zhang J, Li B, Lv D, Wang D, Wang S, Cheng D, Ma T (2021) Identification of RET fusions in a Chinese multicancer retrospective analysis by next-generation sequencing. Cancer Sci. https://doi.org/10.1111/cas.15181
Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J, Yelensky R, Lipson D, Ali SM, Elvin JA, Vergilio JA, Roels S, Miller VA, Nakamura BN, Gray A, Wong MK, Stephens PJ (2016) The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 138:881–890
Rankin A, Johnson A, Roos A, Kannan G, Knipstein J, Britt N, Rosenzweig M, Haberberger J, Pavlick D, Severson E, Vergilio JA, Squillace R, Erlich R, Sathyan P, Cramer S, Kram D, Ross J, Miller V, Reddy P, Alexander B, Ali SM, Ramkissoon S (2021) Targetable BRAF and RAF1 alterations in advanced pediatric cancers. Oncologist 26:e153–e163
Acknowledgements
We thank Merja Haukka, Pia Saarinen, Saila Saarinen and laboratory staff in the laboratory of Genetics for excellent technical assistance. This study was supported by Cancer Foundation Finland, Finska Läkaresällskapet, Helsinki University Hospital Research Funds, Medicinska Understödsföreningen Liv och Hälsa, Sigrid Jusélius Foundation, University of Helsinki, Academy of Finland (grant no. 330606) and Jane and Aatos Erkko Foundation. We thank Invitae Corporation for Archer Research Grant 2020 and Biocartis NV for providing the assays.
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital. This study was funded by Cancer Foundation Finland, Finska Läkaresällskapet, Helsinki University Hospital Research Funds, Medicinska Understödsföreningen Liv och Hälsa, Sigrid Jusélius Foundation, University of Helsinki, Academy of Finland (grant no. 330606) and Jane and Aatos Erkko Foundation. FusionPlex® Lung v2 panels were provided by Invitae Corporation (Archer Research Grant 2020). Biocartis NV provided Idylla platform and cartridges free of charge. The funding sources, Invitae or Biocartis, had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Iiris Ukkola, Pirjo Nummela, Soili Kytölä and Ari Ristimäki contributed to the study conception and design. Material preparation, data collection and raw data analysis were performed by Iiris Ukkola, Pirjo Nummela, Mia Kero, Hanna Tammio, Jenni Tuominen, Soili Kytölä and Ari Ristimäki and all authors had a role in detailed analysis of the data. The first draft of the manuscript was written by Iiris Ukkola, Pirjo Nummela and Ari Ristimäki and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Ethics Committee of the Helsinki University Central Hospital, and all methods were performed in accordance with the relevant guidelines and regulations. Data were anonymised prior to use for the study.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ukkola, I., Nummela, P., Kero, M. et al. Gene fusions and oncogenic mutations in MLH1 deficient and BRAFV600E wild-type colorectal cancers. Virchows Arch 480, 807–817 (2022). https://doi.org/10.1007/s00428-022-03302-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-022-03302-x