Abstract
One hurdle in the development of zebrafish models of human disease is the presence of multiple zebrafish orthologs resulting from whole genome duplication in teleosts. Mutations in inositol polyphosphate 5-phosphatase K (INPP5K) lead to a syndrome characterized by variable presentation of intellectual disability, brain abnormalities, cataracts, muscle disease, and short stature. INPP5K is a phosphatase acting at position 5 of phosphoinositides to control their homeostasis and is involved in insulin signaling, cytoskeletal regulation, and protein trafficking. Previously, our group and others have replicated the human phenotypes in zebrafish knockdown models by targeting both INPP5K orthologs inpp5ka and inpp5kb. Here, we show that inpp5ka is the more closely related orthologue to human INPP5K. While both inpp5ka and inpp5kb mRNA expression levels follow a similar trend in the developing head, eyes, and tail, inpp5ka is much more abundantly expressed in these tissues than inpp5kb. In situ hybridization revealed a similar trend, also showing unique localization of inpp5kb in the pineal gland and retina indicating different transcriptional regulation. We also found that inpp5kb has lost its catalytic activity against its preferred substrate, PtdIns(4,5)P2. Since most human mutations are missense changes disrupting phosphatase activity, we propose that loss of inpp5ka alone can be targeted to recapitulate the human presentation. In addition, we show that the function of inpp5kb has diverged from inpp5ka and may play a novel role in the zebrafish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inositol polyphosphate 5-phosphatase K (INPP5K [MIM:607875]) is a highly conserved phosphatase that participates in the regulation of phosphoinositide (PI) signaling. Also referred to as skeletal muscle and kidney-enriched inositol phosphatase (SKIP), INPP5K is highly expressed in the brain, eyes, and muscles during development and adulthood (Ijuin et al. 2000; Gurung et al. 2003). In humans, homozygous or compound heterozygous mutations in INPP5K have been causally linked to a form of muscular dystrophy with cataracts and intellectual disability (MIM: 617404) also associated with short stature, and microcephaly with considerable variability in the age of onset and clinical presentation (Osborn et al. 2017; Wiessner et al. 2017; Yousaf et al. 2017; D’Amico et al. 2020; Hathazi et al. 2021). Similarities have been noted with Marinesco-Sjögren syndrome (MIM: 248800), a form of myopathy also associated with congenital cataracts, short stature, and cerebellar ataxia (Senderek et al. 2005; Krieger et al. 2013).
PIs are a category of lipid molecules that play crucial roles in signal transduction, ion channel regulation, cellular migration, membrane trafficking, vesicle transport, and many other processes (Di Paolo and De Camilli 2006; Balla 2013; Raghu et al. 2019). The seven unique members of this group are distinguished by their patterns of phosphorylation of the inositol head (PtdIns), which can occur at one or more of three positions (-3, -4, or -5). Production of PIs is regulated by an array of kinases and phosphatases (Balla 2013). INPP5K hydrolyzes the D-5 position of the inositol ring in both PtdIns(4,5)P2 and PtdIns(3,4,5)P3, with highest activity for PtdIns(4,5)P2 (Ijuin et al. 2000; Vandeput et al. 2006; Davies et al. 2015). INPP5K is largely localized to the endoplasmic reticulum (ER) (Gurung et al. 2003; Dong et al. 2018) but can translocate to membrane ruffles as part of a complex with the glucose-regulated protein GRP78/BiP to negatively regulate insulin receptor signaling via phosphatidylinositol-3-kinase (PI3K) (Ijuin and Takenawa 2003; Ijuin et al. 2015, 2016a, b).
Multiple zebrafish (Danio rerio) models of INPP5K loss of function have been generated using morpholino oligonucleotides (MOs) targeting both paralogs, inpp5ka and inpp5kb (Osborn et al. 2017; Wiessner et al. 2017; Hathazi et al. 2021). However, when the genes were targeted independently, knockdown of inpp5ka was sufficient to yield phenotypes typical of neurological and muscular disorders, such as microphthalmia, microcephaly, shortened body, reduced touch-evoked motility, and myopathy. In contrast, inpp5kb MOs produced a mild phenotype in a small subset of morphants (Osborn et al. 2017). In addition, we found inpp5ka expression to be significantly higher than inpp5kb in zebrafish embryos at 2 days post fertilization (dpf) (Osborn et al. 2017). These findings suggested that inpp5ka may be the most conserved human paralog and inpp5kb function may have diverged.
Due to a genome duplication event in teleost fish, about 30% of zebrafish genes have a paralog (Howe et al. 2013), but duplicated genes often acquire differential expression and function (Postlethwait et al. 1998; Ravi and Venkatesh 2018). In this study, we sought to better characterize expression patterns and function of inpp5ka and inpp5kb to understand whether they diverged and support the development of better models of INPP5K mutations in humans. We show that both inpp5ka and inpp5kb have a dynamic developmental expression in the eyes, head, and tail, but found that inpp5kb is expressed at lower levels and specifically enriched in the pineal gland and the inner nuclear layer of the retina. In addition, Inpp5kb lost the majority of its phosphatase activity for PtdIns(4,5)P2 which is the preferred substrate for INPP5K (Ijuin et al. 2000). Together, these data indicate that inpp5ka is the closest ortholog to INPP5K and suggest a unique role for inpp5kb within the zebrafish neural tissues.
Methods
Animal care
Maintenance and husbandry of zebrafish (Danio rerio) breeders and larvae were performed following protocols approved by the Institutional Animal Care and Use Committee of George Washington University and Rutgers University. All animals were from AB or EK backgrounds.
Protein alignments
Clustal Omega and Jalview were used to align the sequences for all transcripts and define conservation (Sievers et al. 2011). Percent identities between the human INPP5K (NP_057616.2), zebrafish Inpp5ka (NP_001082962.2), and Inpp5kb (XP_021335021.1) were calculated using Jalview (Waterhouse et al. 2009).
Quantitative PCR analysis
Samples were collected at 1, 2, 3, 4, 5, and 30 dpf and at 1-year-old for adults. Whole zebrafish embryos and larvae or micro-dissected tissue from eyes, head, and tails were pooled and RNA was extracted using the ReliaPrep RNA Miniprep System kit (Promega, Madison, WI). RNA was treated with DNase I (New England Biolabs) and complementary DNA (cDNA) was synthesized using the iScript cDNA Synthesis kit (Bio-Rad). 600 ng of cDNA per sample were analyzed via qPCR using the SsoFast EvaGreen Supermix (Bio-Rad) or Power UP SYBR Green Master Mix (Thermo Fisher). All reactions were run with 3 technical replicates and repeated on at least 3 biological replicates for 40 cycles on a QuantStudio 3 RealTime PCR system and recorded with QuantStudio Design and Analysis software. Custom primers were designed for inpp5ka (ex9_F: 5′-TGGGACTGGATTGGGTTAT-3′; ex10_R:5′-GCTCCTCATTGAAAGACACC-3′) and inpp5kb (ex2_F: 5′-CGACCACTGACCTCTATGTG-3′; ex3_R: 5′-ATGAGGAGGTGACTCCATGT-3′) and housekeeping controls elongation factor 1 alpha eef1a (eef1a_F: 5′GGGCAAGGGCTCCTTCAA-3′; eef1a_R: 5′-CGCTCGGCCTTCAGTTTG-3′) and riboprotein L18 rpl18 (rpl18_F: 5′-GGCTAAGGTGATGTTTTCGTG-3′; rpl18_R: 5′-GCACATTGCCAATGTTCAGC-3′).
Whole-mount in situ hybridization
Full-length inpp5ka (NM_001089493.1) and inpp5kb (XM_021479346.1) cDNAs were cloned into the pCS2 + plasmid (Addgene). Digoxygenin-labeled sense and antisense probes were synthesized from the linearized plasmids using the DIG RNA Labeling Kit (SP6/T7) (Roche/MilliporeSigma). Whole-mount ISH was performed as previously described (Yan et al. 2009). Expression patterns were confirmed in at least 5 independent embryos per probe and representative images are shown.
RNA scope analysis
Zebrafish larvae were euthanized at 5 dpf by tricaine methanesulfonate and fixed in fresh 4% PFA for 24 h at 4 °C. Larvae were rinsed with 1 × PBS three times and cryoprotected with a 15% and 30% sucrose gradient with 0.025% sodium azide. The fish were embedded in tissue freeze medium (TFM) (General Data Healthcare) and frozen in 2-methyl butane with dry ice. Cryosections were performed using a Leica CM1850 UV Cryostat (Leica Microsystems) at 12 μm and mounted on SuperFrost Plus Slides (Fisher Scientific). The RNAscope assay was performed using the Multiplex Fluorescent Reagent Kit (Advanced Cell Diagnostics) according to the protocol provided for fixed-frozen tissue samples (UM 323,100). The following modifications were made: post fixation in 4% PFA at 4 °C was increased to 20 min, antigen retrieval was performed for 5 min using a microwave to help the target retrieval buffer reach boiling point, and RNAscope Protease Plus was used instead of RNAscope Protease III. The samples were hybridized using custom-designed RNAscope target probes for inpp5ka (Dr-inpp5ka-C1, Cat# 1,224,361-C1) and inpp5kb (Dr-inpp5kb-C2, Cat# 1,224,371-C2). In addition, two positive control genes were used for each target gene, polr2a (Dr-polr2a-C1) and ppib (Dr-ppib-C2). The negative control gene used in this experiment was DapB (DapB-C1,C2) expressed in Bacillus subtilis. We used the TSA Vivid Fluorophore 520 (Cat# 323,271) to develop the HRP-C1 signal and TS Vivid Fluorophore 650 (Cat# 323,273) to develop the HRP-C2 signal; both at a concentration of 1:750. Slides were coverslipped using ProLong™ Gold Antifade Mountant (Thermo Fisher). Images were taken using a Zeiss LSM800 confocal microscope and Zeiss Zen imaging software. Expression patterns were confirmed in at least 5 independent larvae per probe and representative images are shown.
Phosphatase assay
Full-length inpp5ka (NM_001089493.1) and inpp5kb (XM_021479346.1) cDNAs were generated by gene synthesis and cloned into the pGEX-1 to generate GST-fusion proteins (Genewiz/Azenta Life Sciences). GST-human INPP5K and GST were used as positive and negative controls respectively (Weissner et al. 2017). Constructs were transformed into BL21 DE3 pLysS, induced with 100uM IPTG overnight, and harvested by centrifugation. Cells were lysed in assay buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 10 mM MgCl2) plus 1% Triton X-100, EDTA-free protease inhibitors (Roche Diagnostics) and turbonuclease (Sigma). GST fusion proteins were affinity purified over gluthione sepharose 4B (GE Healthcare). After extensive washing, aliquots of beads were run on Coomassie gels to determine the abundance of full-length fusion proteins. Beads bearing equal amounts of fusion proteins were incubated in assay buffer containing 135 μM PtdIns(4,5)P2diC8 or PtdIns(3,4,5)P3diC8, including control wells with no enzyme or no substrate lipid, and incubated for 1 h at 37C. Free phosphate was measured using the Malachite Green assay kit (Echelon Biosciences). Results of three independent experiments were presented as mean ± standard deviation. To minimize variability between purifications, all constructs were freshly prepared and purified in parallel for each experiment.
Results
Zebrafish and human INPP5K protein alignments
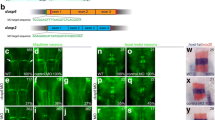
To determine whether inpp5ka and inpp5kb lead to functionally divergent proteins, we first analyzed their protein sequence. Protein sequence alignment of INPP5K (NP_057616.2), Inpp5ka (NP_001082962.2), and Inpp5kb (XP_021335021.1) revealed 45.8% and 43.9% identity between the human orthologue and Inpp5ka and Inpp5kb respectively, while the zebrafish proteins showed 62.2% identity with each other (Fig. 1). Higher conservation was present in the phosphatase domain with 49.7% amino acid sequence identity between the human protein and either zebrafish protein. It is important to note that while the primary isoforms listed above are the most similar to the human gene, both inpp5ka and inpp5kb have additional predicted transcripts that are not found in humans. One transcript has an alternative exon 1 (inpp5ka: XM_009291785.3 and inpp5kb: XM_021479345.1) adding an N-terminal sequence of 63 amino acids for Inpp5ka and 48 amino acids for Inpp5kb. A small 27 base pair alternative exon 9 was also identified in predicted transcripts both inpp5ka (XM_00557623.4) and inpp5kb (XM_005155275.4). All expression and functional analyses performed in this study were based on the sequence of the primary isoforms most similar to the human gene, but the design for the primer and probes did not exclude other predicted isoforms.
Protein alignment of INPP5K, Inpp5ka, and Inpp5kb highlighting conserved amino acids required for phosphatase activity. The start and end of the catalytic domain in the human protein are marked with large red arrowheads. Amino acids required for phosphatase activity have been denoted with asterisks (*). Small arrowheads indicate residues that are altered by missense variants in humans. Hs, Homo sapiens; Dr, Danio rerio
Divergent expression and localization of INPP5K orthologs in zebrafish larva
Analysis of inpp5ka and inpp5kb mRNA obtained from whole zebrafish embryos had shown higher expression of inpp5ka (Osborn et al. 2017). We used qPCR to quantify expression patterns throughout the first five days of development. We found that inpp5ka (NM_001089493.1) was consistently expressed much more abundantly than inpp5kb (XM_021479346.1) (Fig. 2A). The developmental expression trend was similar for inpp5ka and inpp5kb. For comparison across the two genes fold changes for the whole embryo were quantified compared to inpp5kb expression at 1 dpf since it showed the lowest levels. Both genes showed relatively low levels at 1 and 2 dpf, but expression increased after 3 dpf and we saw 5.8, 3.2, and 3.6 higher expression of inpp5ka at 3, 4, and 5 dpf, respectively (Fig. 2A, fold change calculated to 1 dpf inpp5kb. inpp5ka: 1 dpf 8.5 ± 0.8, 2 dpf 11.1 ± 1.5, 3 dpf 51.9 ± 0.4, 4 dpf 112.1 ± 3.6, 5 dpf 84.9 ± 5.7; inpp5kb: 2 dpf 2.1 ± 0.1, 3 dpf 9.0 ± 2.3, 4 dpf 35.5 ± 3.9, 5 dpf 23.6 ± 2.5. p > 0.0001 at 3, 4, and 5 dpf).
inpp5ka and inpp5kb mRNAs differ in expression levels in zebrafish larvae. A Gene expression determined by qPCR. inpp5ka is more highly expressed in whole body lysates through 5 dpf. B Larval tissues were excised from the eye, head, and tail for localized gene expression analysis excluding the area of the trunk around the yolk. C, D Expression for both inpp5ka (C) and inpp5kb (D) is low in the tail and increases in the eyes and brain. By 5 dpf, both are most highly expressed in the eyes. Results from the whole body from (A) are shown as reference. E, F inpp5ka (E) maintains higher expression levels than inpp5kb (F) in the tail/muscle and head at 30 dpf and in adult fish. Both genes are expressed at very high levels in the eyes. Values are averages ± SEM. 2-way ANOVA results for tissue and developmental timepoint (time) are also listed. *p < 0.05, **p < 0.01, ***p < 0.001
Loss of INPP5K in humans affects the muscle, brain, and eyes and knockdown of inpp5ka in zebrafish larvae resulted in morphological abnormalities in the eyes and skeletal muscle (Osborn et al. 2017; Wiessner et al. 2017; Hathazi et al. 2021). We dissected the heads, eyes, and tails of developing larvae for tissue-specific expression analysis (Fig. 2B). Tissue-specific analysis revealed that, while inpp5ka was consistently expressed at higher levels than inpp5kb, both paralogs exhibit the greatest expression in the eyes, intermediate expression in the head, and low levels in the tail. For comparison, we calculated fold changes compared to expression levels of inpp5kb at 1 dpf. inpp5ka expression increased dramatically in the eyes by 5 dpf (Fig. 2C). Both genes showed similar increases in the head by 4 dpf with tail levels remaining consistently low (Fig. 2C, D, fold change relative to 1 dpf inpp5kb in the tail. inpp5ka: 3 dpf head 145.2 ± 19.3, eyes 80.4 ± 31.6; 4 dpf head 271.2 ± 41.5, eyes 528.1 ± 129.8; 5 dpf 218.5 ± 2.7, eyes 653.1 ± 99.6. inpp5kb: 3 dpf head 30.4 ± 3.4, eyes 28.6 ± 4.4; 4 dpf head 217.3 ± 128.8, eyes 157.7 ± 8.4; 5 dpf head 85.8 ± 7.9, eyes 276.9 ± 40.6). Since expression levels in the developmental time course seemed to be dropping after 4 dpf, we asked whether tissue-specific expression differences were still present in juvenile (30 dpf) and adult (1 year). Both inpp5ka and inpp5kb showed very high expression levels in the eyes which were comparable at both ages (Fig. 2E, F). mRNA expression in the head was higher for inpp5ka and increased in adulthood. While expression in the tail at 30 dpf and in dissected muscle at 1 year was low for inpp5ka, but still 7.6 to 11 times higher than inpp5kb (Fig. 2E, F, fold change relative to 30 dpf inpp5kb tail. inpp5ka: 30 dpf tail 7.6 ± 1.4, head 33.9 ± 6.9, eyes 828.9 ± 320.6; 360 dpf tail 11.3 ± 2.1, head 114.5 ± 21.0, eyes 965.9 ± 83.8. inpp5kb: 30 dpf head 16.3 ± 7.1, eyes 1967.1 ± 484.7; 360 dpf tail 0.99 ± 0.22, head 19.6 ± 5.1, eyes 1597.9 ± 615.8). Overall, inpp5ka showed consistently higher levels of expression than inpp5kb throughout the body apart from the eyes where expression of both genes was the highest at juvenile and adult timepoints.
To confirm the expression patterns, we first conducted in situ hybridization on whole-mount larvae at 3 dpf when expression begins to diverge. inpp5ka antisense probes reflected the results of qPCR expression assays. inpp5ka mRNA was most abundant in the head and eyes, with lower expression in the tail (Fig. 3A–C). As expected, inpp5kb antisense targeting revealed lower expression throughout the head and eyes (Fig. 3D, E). However, in contrast with inpp5ka, inpp5kb was abundantly expressed in the pineal gland (Fig. 3F), a neuroendocrine organ which responds to light and plays a role in circadian rhythm (Cahill 1996; Vatine et al. 2011; Livne et al. 2016). These findings suggested that in addition to lower expression, inpp5kb may have also diverged in its expression pattern. To better define the cellular distribution of the two mRNAs we then performed fluorescent RNA scope in situ on tissue cryosections at 5 dpf when expression levels are significantly increased via qPCR. This analysis revealed additional differences. inpp5ka was evenly distributed throughout the brain, in all layers of the retina, and was also present around the nuclei of lens cells (Fig. 4A, B). Expression in the muscle at the same exposure was much lower reflecting qPCR results (Fig. 4C). inpp5kb showed striking differences in distribution with very high expression in the pineal gland (Fig. 4D, Suppl.Fig. 1A) and in the inner nuclear layer of the retina (Fig. 4E, Suppl. Fig. 1B), though expression was still noted in the brain, other retinal layers, and the lens. As seen for inpp5ka, inpp5kb expression in the muscle was very low (Fig. 4F) though nuclei are sparse in this tissue and staining is less concentrated than in the brain and eyes (Suppl.Fig. 1C). Thus, the mRNA expression levels of inpp5kb in the brain and eye appear to be primarily driven by specific expression patterns while inpp5ka has a more even distribution pattern.
inpp5ka and inpp5kb mRNAs differ in localization at 3 dpf. A–C In situ hybridization in 3 dpf larvae shows that inpp5ka mRNA is highly expressed throughout the head and eyes. Scale bars: 500µm in (A), 100µm in (C). D–F inpp5kb expression is concentrated to the pineal gland. The pineal gland is indicated by the black arrow
Cellular distribution of inpp5ka and inpp5kb at 5 dpf by RNA scope in situ. A–C inpp5ka shows even distribution in the brain and eye (A, B), also labeling lens cells (B). Muscle expression is sparse and at lower levels (C). D–F inpp5kb brain expression is highest in the pineal gland (arrow in D). Increased expression is also noted in the inner nuclear layer of the retina with lower expression in photoreceptors, retinal ganglion cells, and lens cells (E, also see Suppl. Figure 1B). Sparse expression is present in muscle cells (F, also see Suppl Fig. 1C). G–I Merged images including both probes (inpp5ka in green and inpp5kb in red) and DAPI to counterstain the nuclei. Scale bars: 100 µm for (A), (D), and (G), 50 µm for all other panels
Divergence in phosphatase activity of human and zebrafish orthologs of INPP5K
To evaluate the preservation of the PI phosphatase activity in the zebrafish isoforms, we conducted a malachite phosphatase assay to examine the activity of INPP5K and the two zebrafish Inpp5k isoforms against PIP3 and the preferred substrate PtdIns(4,5)P2 (Fig. 5A). We found that zebrafish Inpp5ka and human INPP5K were both highly active against PtsIns(4,5)P2 as expected. This activity was specific, as illustrated by the lack of phosphatase activity against PIP3. However, compared to Inpp5ka, Inpp5kb was nearly inactive against PtdIns(4,5)P2. Inpp5ka yielded 409 pmol of free phosphate vs 20 pmol for Inpp5kb, indicating that Inpp5ka had a 20-fold higher activity compared to Inpp5kb (Fig. 5B).
Inpp5ka and Inpp5kb exhibit different phosphatase activity. A Phosphatase activity of human INPP5K, Inpp5ka, and Inpp5kb in the malachite assay. Human INPP5K and Inpp5ka demonstrate high activity for the PI(4,5)P2 substrate. PIP3 did not elicit activity from any isoform. B Inpp5ka is more significantly active against diC8PI(4,5)P2 compared to Inpp5kb. Values are averages ± SEM. ***p < 0.001 following a t-test
The INPP5K protein is primarily composed of a 5-phosphatase domain between amino acids 16–318 and a SKITCH domain between amino acids 321–448. Most mutations identified in humans are missense and have been shown to reduce or ablate phosphatase activity (Osborn et al. 2017; Wiessner et al. 2017). We wondered whether the loss in activity in Inpp5kb could be caused by changes in amino acids identified to be critical for the catalytic activity of INPP5K. Basing this analysis on the available crystal structures of other Type II inositol phosphate 5-phosphatases, INPP5B and SYNJ1 (Trésaugues et al. 2014; Paesmans et al. 2020), we found that all sites were conserved in Inpp5ka and Inpp5kb and there were no major changes that could explain differences in activity (asterisks in Fig. 1). We also assessed whether residues known to be affected by pathogenic variants in patients were conserved in Inpp5kb, and these amino acids were all maintained (small arrowheads in Fig. 1) (D’Amico et al. 2020; Osborn et al. 2017; Hathazi et al. 2021; Wiessner et al. 2017; Yousaf et al. 2017). Thus, possible changes in known residues do not explain the difference in function between Inpp5ka and Inpp5kb.
Discussion
INPP5K mutations in humans cause a distinct neurodevelopmental syndrome with variable presentation of intellectual disability, cataracts, short stature, and muscle disease (Osborn et al. 2017; Wiessner et al. 2017; Yousaf et al. 2017; D’Amico et al. 2020). Multiple zebrafish models have been developed to study inpp5ka/b function using morpholino oligonucleotides either blocking translation or knocking down mRNA expression (Osborn et al. 2017; Wiessner et al. 2017; Hathazi et al. 2021). However, the presence of duplicated inpp5k genes, inpp5ka and inpp5kb, in zebrafish complicates the development of both candidate loss-of-function or point mutations zebrafish models since both orthologs may need to be targeted. Initial functional data from our previous studies had shown that inpp5ka knockdown alone was sufficient to replicate the findings in the double gene knockdown (Osborn et al. 2017). In this study, we show that inpp5ka and inpp5kb have diverged in expression levels, patterns, and function following teleost whole genome duplication (WGD). inpp5ka, rather than inpp5kb, maintains a higher sequence identity and similar expression pattern to human INPP5K, suggesting that genetic removal of this gene may be sufficient to recapitulate the human mutation.
Polyploidization by WGD is a significant driver of evolution (Postlethwait et al. 1998; Sémon and Wolfe 2007). During the period of re-diploidization that follows a WGD event, most redundant genes are eliminated via genomic rearrangements and mutations causing one duplicated copy to become a pseudogene. However, a duplicated gene may be preserved and gradually diverge in expression patterns and/or function during evolution leading to gene adaptation through sub-functionalization or neo-functionalization (Sémon and Wolfe 2007; Kassahn et al. 2009). While inpp5ka is broadly and highly expressed throughout the zebrafish larvae, inpp5kb is significantly less expressed. Interestingly, both genes show the highest expression in the eye which is maintained to adulthood. inpp5ka is evenly distributed in retinal layers during development, matching the Inpp5k expression patterns identified in single-cell RNA sequencing (scRNAseq) datasets generated for the chick, and embryonic murine retina and the adult human retina (Lukowski et al. 2019; Balasubramanian et al. 2021; Yamagata et al. 2021). Considering that congenital cataracts are one of the most consistent phenotypes linked to INPP5K mutations in humans (Osborn et al. 2017; Wiessner et al. 2017; Yousaf et al. 2017) the high expression of inpp5ka in lens cells found in the RNA scope experiments is also of note.
In contrast, we found that inpp5kb is highly enriched in the inner nuclear layer closer where amacrine cells and bipolar cells, both interneurons modulating the transmission of visual signals, are located (Connaughton et al. 2004; Zhao et al. 2009). Additional differential expression was found in the brain revealing widespread and sustained expression of inpp5ka while inpp5kb is highly enriched in the pineal gland. The pineal gland is thought to be the master regulator for circadian rhythm in vertebrates. Melatonin is the key circadian hormone secreted by the pineal gland in zebrafish (Cahill 1996) and is thought to play a role in locomotor activity (Livne et al. 2016), as well in the timing of reproduction and feeding (Piccinetti et al. 2013). It will be interesting in the future to determine the role of Inpp5kb in pineal and retinal functions independently of its phosphatase activity. This divergence in expression patterns further supports sub-functionalization of inpp5kb.
Additionally, we found that Inpp5kb exhibits minimal phosphatase activity against the traditional substrate of INPP5K, PtdIns(4,5)P2 (Ijuin et al. 2000; Vandeput et al. 2006). In humans, much of the pathology resulting from mutations within INPP5K have been attributed to the dysregulation of phosphoinositide homeostasis (Osborn et al. 2017; Wiessner et al. 2017; McGrath et al. 2020; Hathazi et al. 2021). Most known mutations in INPP5K are missense variants occurring in the catalytic phosphatase domain reducing or ablating conversion of PtdIns(4,5)P2 to PtdIns(4)P (Osborn et al. 2017; Wiessner et al. 2017). In the muscle, INPP5K is involved in insulin signaling through the PI3K/Akt/mTOR pathway (Ijuin and Takenawa 2015; Ijuin et al. 2015), but studies in a muscle-specific Inpp5k mouse knock-out line also determined that abnormal accumulation of PtdIns(4,5)P2 led to a severe disruption in lysosome recycling (McGrath et al. 2020). Interestingly, lysosome enlargement and autophagy inhibition found in the Inpp5k-deficient muscle were not dependent of Akt/mTOR signaling, suggesting an independent additional role for PtdIns(4,5)P2 in muscle maintenance in the autophagic lysosome reformation pathway (McGrath et al. 2020). In addition, increased levels of D3-phosphoglycerate dehydrogenase (PHGDH) have been found in fibroblasts obtained from individuals with INPP5K phosphatase mutations, indicating further metabolic disruptions (Hathazi et al. 2021). It remains unclear whether INPP5K has additional functions independent of its phosphatase activity. It has been involved in endoplasmic reticulum (ER) organization by multiple groups despite the absence of its substrates in the ER (Dong et al. 2018; Ramos et al. 2019). It is recruited there via interactions with the ER transmembrane protein ARL6IP1 but it is not known whether it is acting on neighboring membranes or performing other functions as part of a complex (Dong et al. 2018). Studies on the different functions and interactions of wild-type and phosphatase-dead zebrafish Inpp5ka and of Inpp5kb and their different isoforms may help shed light on the diverse subcellular activities of these proteins.
Overall, we propose that targeting the phosphatase domain in Inpp5ka would lead to a reliable model for INPP5K mutations in humans. Whether Inpp5kb evolved to perform a different function in specific subsets of cells and how it lost its phosphatase activity in the zebrafish remains to be studied.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Balasubramanian R, Min X, Quinn PMJ et al (2021) Phase transition specified by a binary code patterns the vertebrate eye cup. Sci Adv 7:eabj9846. https://doi.org/10.1126/sciadv.abj9846
Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93:1019–1137. https://doi.org/10.1152/physrev.00028.2012
Cahill GM (1996) Circadian regulation of melatonin production in cultured zebrafish pineal and retina. Brain Res 708:177–181. https://doi.org/10.1016/0006-8993(95)01365-2
Connaughton VP, Graham D, Nelson R (2004) Identification and morphological classification of horizontal, bipolar, and amacrine cells within the zebrafish retina. J Comp Neurol 477:371–385. https://doi.org/10.1002/cne.20261
D’Amico A, Fattori F, Nicita F et al (2020) A recurrent pathogenic variant of INPP5K underlies autosomal recessive congenital muscular dystrophy with cataracts and intellectual disability: evidence for a founder effect in Southern Italy. Front Genet 11:565868. https://doi.org/10.3389/fgene.2020.565868
Davies EM, Kong AM, Tan A et al (2015) Differential SKIP expression in PTEN-deficient glioblastoma regulates cellular proliferation and migration. Oncogene 34:3711–3727. https://doi.org/10.1038/onc.2014.303
Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443:651–657. https://doi.org/10.1038/nature05185
Dong R, Zhu T, Benedetti L et al (2018) The inositol 5-phosphatase INPP5K participates in the fine control of ER organization. J Cell Biol 217:3577–3592. https://doi.org/10.1083/jcb.201802125
Gurung R, Tan A, Ooms LM et al (2003) Identification of a novel domain in two mammalian inositol-polyphosphate 5-phosphatases that mediates membrane ruffle localization. The inositol 5-phosphatase skip localizes to the endoplasmic reticulum and translocates to membrane ruffles following epidermal growth factor stimulation. J Biol Chem 278:11376–11385. https://doi.org/10.1074/jbc.m209991200
Hathazi D, Cox D, D’Amico A, et al (2021) INPP5K and SIL1 associated pathologies with overlapping clinical phenotypes converge through dysregulation of PHGDH. Brain awab133. https://doi.org/10.1093/brain/awab133
Howe K, Clark MD, Torroja CF et al (2013) The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. https://doi.org/10.1038/nature12111
Ijuin T, Takenawa T (2003) SKIP negatively regulates insulin-induced GLUT4 translocation and membrane ruffle formation. Mol Cell Biol 23:1209–1220. https://doi.org/10.1128/mcb.23.4.1209-1220.2003
Ijuin T, Takenawa T (2015) Improvement of insulin signaling in myoblast cells by an addition of SKIP-binding peptide within Pak1 kinase domain. Biochem Biophys Res Commun 456:41–46. https://doi.org/10.1016/j.bbrc.2014.11.031
Ijuin T, Mochizuki Y, Fukami K et al (2000) Identification and characterization of a novel inositol polyphosphate 5-phosphatase. J Biol Chem 275:10870–10875
Ijuin T, Hatano N, Hosooka T, Takenawa T (2015) Regulation of insulin signaling in skeletal muscle by PIP3 phosphatase, SKIP, and endoplasmic reticulum molecular chaperone glucose-regulated protein 78. Biochem Biophys Acta 1853:3192–3201. https://doi.org/10.1016/j.bbamcr.2015.09.009
Ijuin T, Hatano N, Takenawa T (2016a) Glucose-regulated protein 78 (GRP78) binds directly to PIP3 phosphatase SKIP and determines its localization. Genes to Cells 21:457–465. https://doi.org/10.1111/gtc.12353
Ijuin T, Hosooka T, Takenawa T (2016b) Phosphatidylinositol 3,4,5-trisphosphate phosphatase SKIP links endoplasmic reticulum stress in skeletal muscle to insulin resistance. Mol Cell Biol 36:108–118. https://doi.org/10.1128/mcb.00921-15
Kassahn KS, Dang VT, Wilkins SJ et al (2009) Evolution of gene function and regulatory control after whole-genome duplication: Comparative analyses in vertebrates. Genome Res 19:1404–1418. https://doi.org/10.1101/gr.086827.108
Krieger M, Roos A, Stendel C et al (2013) SIL1 mutations and clinical spectrum in patients with Marinesco-Sjogren syndrome. Brain 136:3634–3644. https://doi.org/10.1093/brain/awt283
Livne ZB-M, Alon S, Vallone D et al (2016) Genetically blocking the zebrafish pineal clock affects circadian behavior. Plos Genet 12:e1006445. https://doi.org/10.1371/journal.pgen.1006445
Lukowski SW, Lo CY, Sharov AA et al (2019) A single-cell transcriptome atlas of the adult human retina. Embo J 38:100811. https://doi.org/10.15252/embj.2018100811
McGrath MJ, Eramo MJ, Gurung R, et al (2020) Defective lysosome reformation during autophagy causes skeletal muscle disease. J Clin Invest 131. https://doi.org/10.1172/jci135124
Osborn DPS, Pond HL, Mazaheri N et al (2017) Mutations in INPP5K cause a form of congenital muscular dystrophy overlapping Marinesco-Sjögren syndrome and dystroglycanopathy. Am J Hum Genet 100:537–545. https://doi.org/10.1016/j.ajhg.2017.01.019
Paesmans J, Martin E, Deckers B et al (2020) A structure of substrate-bound Synaptojanin1 provides new insights in its mechanism and the effect of disease mutations. Elife 9:e64922. https://doi.org/10.7554/elife.64922
Piccinetti CC, Migliarini B, Olivotto I et al (2013) Melatonin and peripheral circuitries: insights on appetite and metabolism in Danio rerio. Zebrafish 10:275–282. https://doi.org/10.1089/zeb.2012.0844
Postlethwait JH, Yan YL, Gates MA et al (1998) Vertebrate genome evolution and the zebrafish gene map. Nat Genet 18:345–349. https://doi.org/10.1038/ng0498-345
Raghu P, Joseph A, Krishnan H et al (2019) Phosphoinositides: regulators of nervous system function in health and disease. Front Mol Neurosci 12:208. https://doi.org/10.3389/fnmol.2019.00208
Ramos AR, Ghosh S, Suhel T et al (2019) Phosphoinositide 5-phosphatases SKIP and SHIP2 in ruffles, the endoplasmic reticulum and the nucleus: an update. Adv Biological Regul 75:100660. https://doi.org/10.1016/j.jbior.2019.100660
Ravi V, Venkatesh B (2018) The divergent genomes of teleosts. Annu Rev Anim Biosci 6:47–68. https://doi.org/10.1146/annurev-animal-030117-014821
Sémon M, Wolfe KH (2007) Consequences of genome duplication. Curr Opin Genet Dev 17:505–512. https://doi.org/10.1016/j.gde.2007.09.007
Senderek J, Krieger M, Stendel C et al (2005) Mutations in SIL1 cause Marinesco-Sjögren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet 37:1312–1314. https://doi.org/10.1038/ng1678
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539–539. https://doi.org/10.1038/msb.2011.75
Trésaugues L, Silvander C, Flodin S et al (2014) Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases. Structure 22:744–755. https://doi.org/10.1016/j.str.2014.01.013
Vandeput F, Backers K, Villeret V et al (2006) The influence of anionic lipids on SHIP2 phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase activity. Cell Signal 18:2193–2199. https://doi.org/10.1016/j.cellsig.2006.05.010
Vatine G, Vallone D, Gothilf Y, Foulkes NS (2011) It’s time to swim! Zebrafish and the circadian clock. Febs Lett 585:1485–1494. https://doi.org/10.1016/j.febslet.2011.04.007
Waterhouse AM, Procter JB, Martin DMA et al (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Wiessner M, Roos A, Munn CJ et al (2017) Mutations in INPP5K, encoding a phosphoinositide 5-phosphatase, cause congenital muscular dystrophy with cataracts and mild cognitive impairment. Am J Hum Genet 100:523–536. https://doi.org/10.1016/j.ajhg.2017.01.024
Yamagata M, Yan W, Sanes JR (2021) A cell atlas of the chick retina based on single-cell transcriptomics. Elife 10:e63907. https://doi.org/10.7554/elife.63907
Yan B, Neilson KM, Moody SA (2009) foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev Biol 329:80–95. https://doi.org/10.1016/j.ydbio.2009.02.019
Yousaf S, Sheikh SA, Riazuddin S, et al (2017) INPP5K variant causes autosomal recessive congenital cataract in a Pakistani family. Clinical Genetics. https://doi.org/10.1111/cge.13143
Zhao X-F, Ellingsen S, Fjose A (2009) Labelling and targeted ablation of specific bipolar cell types in the zebrafish retina. Bmc Neurosci 10:107. https://doi.org/10.1186/1471-2202-10-107
Acknowledgements
The authors would like to thank all members of the Manzini laboratory for helpful discussion, Kathleen Flaherty and Heather Pond for management of the zebrafish colonies at Rutgers University and George Washington University, respectively, and Himani Majumdar for assistance with the in situ hybridization protocols.
Funding
This work was funded by the National Institutes of Health R01NS109149 to M.C.M, Wellcome Trust (105616/Z/14/Z), and the Medical Research Council (MRC/N010035/1) to L.E.S. Additional support was provided to M.C.M. from the Robert Wood Johnson Foundation (grant #74260). B.F.K was supported by training grant T32NS115700 from the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
M.C.M. and E.S.C. conceived the study. D.S., B.M.G., E.S.C., L.E.R., S.M., N.B., B.F.K., L.C., and L.T. performed experiments and collected data. D.S., B.M.G., E.S.C., L.T., and M.C.M. analyzed the data. D.S., B.M.G., and M.C.M. wrote the manuscript with contributions from S.A.M. and L.E.S. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Communicated by Caroline Brennan.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shukla, D., Gural, B.M., Cauley, E.S. et al. Duplicated zebrafish (Danio rerio) inositol phosphatases inpp5ka and inpp5kb diverged in expression pattern and function. Dev Genes Evol 233, 25–34 (2023). https://doi.org/10.1007/s00427-023-00703-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-023-00703-z