Abstract
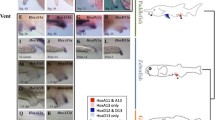
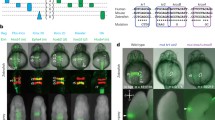
Hoxa2 genes provide critical patterning signals during development, and their regulation and function have been extensively studied. We report a previously uncharacterized significant sequence divergence of a highly conserved hindbrain hoxa2b enhancer element in the family syngnathidae (pipefishes, seahorses, pipehorses, seadragons). We compared the hox cis-regulatory element variation in the Gulf pipefish and two species of seahorse against eight other species of fish, as well as human and mouse. We annotated the hoxa2b enhancer element binding sites across three species of seahorse, four species of pipefish, and one species of ghost pipefish. Finally, we performed in situ hybridization analysis of hoxa2b expression in Gulf pipefish embryos. We found that all syngnathid fish examined share a modified rhombomere 4 hoxa2b enhancer element, despite the fact that this element has been found to be highly conserved across all vertebrates examined previously. Binding element sequence motifs and spacing between binding elements have been modified for the hoxa2b enhancer in several species of pipefish and seahorse, and that the loss of the Prep/Meis binding site and further space shortening happened after ghost pipefish split from the rest of the syngnathid clade. We showed that expression of this gene in rhombomere 4 is lower relative to the surrounding rhombomeres in developing Gulf pipefish embryos, reflecting previously published functional tests for this enhancer. Our findings highlight the benefits of studying highly derived, diverse taxa for understanding of gene regulatory evolution and support the hypothesis that natural mutations can occur in deeply conserved pathways in ways potentially related to phenotypic diversity.






Similar content being viewed by others
Data Availability
Upon publication, all raw sequencing data described in this study will be available via the NCBI Genbank. In situ images will be hosted by the Cresko Lab web server (http://creskolab.uoregon.edu) and distributed by the Cresko Laboratory GitHub account.
References
Alexander T, Nolte C, Krumlauf R (2009) Hox genes and segmentation of the hindbrain and axial skeleton. AnnRev Cell Dev 25:431–456. https://doi.org/10.1146/annurev.cellbio.042308.113423
Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A (1998) Zebrafish hox clusters and vertebrate genome evolution. Science 282:1711–1714. https://doi.org/10.1126/science.282.5394.1711
Benedetti I, Sassi D, Stefanelli A (1991) Mauthner neurons in syngnathid bony fishes. Acta Embryologiae et Morphologiae Experimentalis 12:75–76
Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F (1998) The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J 17:1434–1445. https://doi.org/10.1093/emboj/17.5.1434
Brown R (2010) Craniofacial development in pipefish: a morphological and molecular analysis. University of Oregon
Brudno M, Do CB, Cooper GM, Kim MF, Davydov E (2003a) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic. DNA Genome Res 13:721–731. https://doi.org/10.1101/gr.926603
Brudno M, Malde S, Poliakov A, Do CB, Couronne O, Dubchak I (2003b) Glocal alignment: finding rearrangements during alignment. Bioinformatics 19:i54–i62. https://doi.org/10.1093/bioinformatics/btg1005
Burglin TR, Affolter M (2016) Homeodomain proteins: an update. Chromosoma 125:497–521. https://doi.org/10.1007/s00412-015-0543-8
Carroll SB (1995) Homeotic genes and the evolution of arthropods and chordates. Nature 376:479–485. https://doi.org/10.1038/376479a0
Carroll SB (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. https://doi.org/10.1016/j.cell.2008.06.030
Carroll SB, Grenier JK, Weatherbee SD (2013) From DNA to diversity: molecular genetics and the evolution of animal design. Blackwell Pub, Malden, Massachusetts
Chan SK, Ryoo HD, Gould A, Krumlauf R, Mann RS (1997) Switching the in vivo specificity of a minimal Hox-responsive element. Development 124:2007–2014
Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jonsson B, Schluter D, Bell MA, Kingsley DM (2010) Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327:302–305. https://doi.org/10.1126/science.1182213
Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML (1995) Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev 9:663–674
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Ferretti E, Cambronero F, Tumpel S, Longobardi E, Wiedemann LM, Blasi F, Krumlauf R (2005) Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol Cell Biol 25:8541–8552. https://doi.org/10.1128/MCB.25.19.8541-8552.2005
Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F (2000) Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 127:155–166
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279. https://doi.org/10.1093/nar/gkh458
Gavalas A, Davenne M, Lumsden A, Chambon P, Rijli FM (1997) Role of Hoxa-2 in axon pathfinding and rostral hindbrain patterning. Development 124:3693–3702
Gehring WJ, Affolter M, Bürglin T (1994) Homeodomain proteins. Annu Rev Biochem 63:487–526
Gehrke AR, Shubin NH (2016) Cis-regulatory programs in the development and evolution of vertebrate paired appendages. Semin Cell Dev Biol 57:31–39. https://doi.org/10.1016/j.semcdb.2016.01.015
Gendron-Maguire M, Mallo M, Zhang M, Gridley T (1993) Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell 75:1317–1331
Grammatopoulos GA, Bell E, Toole L, Lumsden A, Tucker AS (2000) Homeotic transformation of branchial arch identity after Hoxa2 overexpression. Development 127:5355–5365
Hamilton H, Saarman N, Short G, Sellas AB, Moore B, Hoang T (2017) Molecular phylogeny and patterns of diversification in Syngnathid fishes. Mol Phylogenetics Evol 17:388–403
Herald ES (1959) From pipefish to seahorse - a study of phylogenetic relationships. Proc Calif Acad Sci 29(13):465–473
Hoegg S, Boore JL, Kuehl JV, Meyer A (2007) Comparative phylogenomic analyses of teleost fish Hox gene clusters: lessons from the cichlid fish Astatotilapia burtoni. BMC Genomics 8:317. https://doi.org/10.1186/1471-2164-8-317
Hoekstra HE, Coyne JA (2007) The locus of evolution: evo devo and the genetics of adaptation. Evolution: International Journal of Organic Evolution 61:995–1016
Holland PW (2013) Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol 2:31–45. https://doi.org/10.1002/wdev.78
Hunter MP, Prince VE (2002) Zebrafish hox paralogue group 2 genes function redundantly as selector genes to pattern the second pharyngeal arch. Dev Biol 247:367–389. https://doi.org/10.1006/dbio.2002.0701
Infante CR, Mihala AG, Park S, Wang JS, Johnson KK, Lauderdale JD, Menke DB (2015) Shared enhancer activity in the limbs and phallus and functional divergence of a limb-genital cis-regulatory element in snakes. Dev Cell 35:107–119. https://doi.org/10.1016/j.devcel.2015.09.003
Kiecker C, Lumsden A (2005) Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci 6:553–564. https://doi.org/10.1038/nrn1702
Kimmel CB, Small CM, Knope ML (2017) A rich diversity of opercle bone shape among teleost fishes. PLoS One 12:e0188888. https://doi.org/10.1371/journal.pone.0188888
Kitazawa T et al (2015) Distinct effects of Hoxa2 overexpression in cranial neural crest populations reveal that the mammalian hyomandibular-ceratohyal boundary maps within the styloid process. Dev Biol 402:162–174. https://doi.org/10.1016/j.ydbio.2015.04.007
Knoepfler PS, Lu Q, Kamps MP (1996) Pbx-1 Hox heterodimers bind DNA on inseparable half-sites that permit intrinsic DNA binding specificity of the Hox partner at nucleotides 3′ to a TAAT motif. Nucleic Acids Res 24:2288–2294
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152–D157. https://doi.org/10.1093/nar/gkq1027
Krumlauf R (1994) Hox genes in vertebrate development. Cell 78:191–201
Kurosawa G, Takamatsu N, Takahashi M, Sumitomo M, Sanaka E, Yamada K, Nishii K, Matsuda M, Asakawa S, Ishiguro H, Miura K, Kurosawa Y, Shimizu N, Kohara Y, Hori H (2006) Organization and structure of hox gene loci in medaka genome and comparison with those of pufferfish and zebrafish genomes. Gene 370:75–82. https://doi.org/10.1016/j.gene.2005.11.015
Le Pabic P, Scemama JL, Stellwag EJ (2010) Role of Hox PG2 genes in Nile tilapia pharyngeal arch specification: implications for gnathostome pharyngeal arch evolution. Evol Dev 12:45–60. https://doi.org/10.1111/j.1525-142X.2009.00390.x
Lee AP, Koh EG, Tay A, Brenner S, Venkatesh B (2006) Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proc Natl Acad Sci U S A 103:6994–6999. https://doi.org/10.1073/pnas.0601492103
Leysen H, Jouk P, Brunain M, Christiaens J, Adriaens D (2010) Cranial architecture of tube-snouted gasterosteiformes (Syngnathus rostellatus and Hippocampus capensis). J Morphol 271:255–270. https://doi.org/10.1002/jmor.10795
Lin Q, Fan S, Zhang Y, Xu M, Zhang H, Yang Y, Lee AP, Woltering JM, Ravi V, Gunter HM, Luo W, Gao Z, Lim ZW, Qin G, Schneider RF, Wang X, Xiong P, Li G, Wang K, Min J, Zhang C, Qiu Y, Bai J, He W, Bian C, Zhang X, Shan D, Qu H, Sun Y, Gao Q, Huang L, Shi Q, Meyer A, Venkatesh B (2016) The seahorse genome and the evolution of its specialized morphology. Nature 540:395–399. https://doi.org/10.1038/nature20595
Lin Q, Qiu Y, Gu R, Xu M, Li J, Bian C, Zhang H, Qin G, Zhang Y, Luo W, Chen J, You X, Fan M, Sun M, Xu P, Venkatesh B, Xu J, Fu H, Shi Q (2017) Draft genome of the lined seahorse. Hippocampus erectus Gigascience 6:1–6. https://doi.org/10.1093/gigascience/gix030
Lumsden A (2004) Segmentation and compartition in the early avian hindbrain. Mech Dev 121:1081–1088. https://doi.org/10.1016/j.mod.2004.04.018
Lumsden A, Krumlauf R (1996) Patterning the vertebrate neuraxis. Science 274:1109–1115
Maconochie M, Krishnamurthy R, Nonchev S, Meier P, Manzanares M, Mitchell PJ, Krumlauf R (1999) Regulation of Hoxa2 in cranial neural crest cells involves members of the AP-2 family. Development 126:1483–1494
Maconochie MK, Nonchev S, Manzanares M, Marshall H, Krumlauf R (2001) Differences in Krox20-dependent regulation of Hoxa2 and Hoxb2 during hindbrain development. Dev Biol 233:468–481. https://doi.org/10.1006/dbio.2001.0197
Manzanares M, Bel-Vialar S, Ariza-McNaughton L, Ferretti E, Marshall H, Maconochie MM, Blasi F, Krumlauf R (2001) Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involve auto- and cross-regulatory mechanisms. Development 128:3595–3607
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047. https://doi.org/10.1093/bioinformatics/16.11.1046
McEllin JA, Alexander TB, Tumpel S, Wiedemann LM, Krumlauf R (2016) Analyses of fugu hoxa2 genes provide evidence for subfunctionalization of neural crest cell and rhombomere cis-regulatory modules during vertebrate evolution. Dev Biol 409:530–542. https://doi.org/10.1016/j.ydbio.2015.11.006
McGinnis W, Krumlauf R (1992) Homeobox genes and axial patterning. Cell 68:283–302
Minoux M, Rijli FM (2010) Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 137:2605–2621. https://doi.org/10.1242/dev.040048
Neutens C, Adriaens D, Christiaens J, Kegel B, Dierick M, Boistel R (2014) Grasping convergent evolution in syngnathids: a unique tale of tails. J Anat 224:710–723. https://doi.org/10.1111/joa.12181
Nonchev S et al (1996a) The conserved role of Krox-20 in directing Hox gene expression during vertebrate hindbrain segmentation. Proc Natl Acad Sci U S A 93:9339–9345
Nonchev S et al (1996b) Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development 122:543–554
Parker HJ, Bronner ME, Krumlauf R (2014) A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature 514:490–493. https://doi.org/10.1038/nature13723
Parker HJ, Bronner ME, Krumlauf R (2016) The vertebrate Hox gene regulatory network for hindbrain segmentation: evolution and diversification: coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. Bioessays 38:526–538. https://doi.org/10.1002/bies.201600010
Parker HJ, Bronner ME, Krumlauf R (2019a) An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates. Dev Biol 453:19–33. https://doi.org/10.1016/j.ydbio.2019.05.001
Parker HJ, de Kumar B, Green SA, Prummel KD, Hess C, Kaufman CK, Mosimann C, Wiedemann LM, Bronner ME, Krumlauf R (2019b) A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nat Commun 10:1189. https://doi.org/10.1038/s41467-019-09197-8
Parker HJ, Krumlauf R (2020) A Hox gene regulatory network for hindbrain segmentation. Curr Top Dev Biol 139:169–203. https://doi.org/10.1016/bs.ctdb.2020.03.001
Pascual-Anaya J, Sato I, Sugahara F, Higuchi S, Paps J, Ren Y, Takagi W, Ruiz-Villalba A, Ota KG, Wang W, Kuratani S (2018) Hagfish and lamprey Hox genes reveal conservation of temporal colinearity in vertebrates. Nat Ecol Evol 2:859–866. https://doi.org/10.1038/s41559-018-0526-2
Pasqualetti M, Ori M, Nardi I, Rijli FM (2000) Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development 127:5367–5378
Prince V, Lumsden A (1994) Hoxa-2 expression in normal and transposed rhombomeres: independent regulation in the neural tube and neural crest. Development 120:911–923
Raff RA (2012) The shape of life: genes, development, and the evolution of animal form. University of Chicago Press, Chicago
Ravi V, Lam K, Tay BH, Tay A, Brenner S, Venkatesh B (2009) Elephant shark (Callorhinchus milii) provides insights into the evolution of Hox gene clusters in gnathostomes. Proc Natl Acad Sci U S A 106:16327–16332. https://doi.org/10.1073/pnas.0907914106
Rebeiz M, Tsiantis M (2017) Enhancer evolution and the origins of morphological novelty. Curr Opin Genet Dev 45:115–123. https://doi.org/10.1016/j.gde.2017.04.006
Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, Chambon P (1993) A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 75:1333–1349
Santagati F, Minoux M, Ren SY, Rijli FM (2005) Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development 132:4927–4936. https://doi.org/10.1242/dev.02078
Santagati F, Rijli FM (2003) Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci 4:806–818. https://doi.org/10.1038/nrn1221
Scott MP (1992) Vertebrate homeobox gene nomenclature. Cell 71:551–553
Small CM, Bassham S, Catchen J, Amores A, Fuiten AM, Brown RS, Jones AG, Cresko WA (2016) The genome of the Gulf pipefish enables understanding of evolutionary innovations. Genome Biol 17:258. https://doi.org/10.1186/s13059-016-1126-6
Stern DL (2000) Perspective: evolutionary developmental biology and the problem of variation. Evolution 54:1079–1091
Stern DL, Frankel N (2013) The structure and evolution of cis-regulatory regions: the shavenbaby story. Philos Trans R Soc B-Biol Sci 368:20130028. https://doi.org/10.1098/rstb.2013.0028
Teske PR, Beheregaray LB (2009) Evolution of seahorses’ upright posture was linked to Oligocene expansion of seagrass habitats. Biol Lett 5:521–523. https://doi.org/10.1098/rsbl.2009.0152
Thisse C, Thisse B (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3:59–69
Torresen OK et al (2017) An improved genome assembly uncovers prolific tandem repeats in Atlantic cod. BMC Genomics 18:95. https://doi.org/10.1186/s12864-016-3448-x
Trainor PA, Krumlauf R (2000) Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci 1:116–124. https://doi.org/10.1038/35039056
Trainor PA, Krumlauf R (2001) Hox genes, neural crest cells and branchial arch patterning. Curr Opin Cell Biol 13:698–705
Tumpel S, Cambronero F, Ferretti E, Blasi F, Wiedemann LM, Krumlauf R (2007) Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev Biol 302:646–660. https://doi.org/10.1016/j.ydbio.2006.10.029
Tumpel S, Cambronero F, Wiedemann LM, Krumlauf R (2006) Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes). Proc Natl Acad Sci U S A 103:5419–5424. https://doi.org/10.1073/pnas.0600993103
Tumpel S, Wiedemann LM, Krumlauf R (2009) Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol 88:103–137. https://doi.org/10.1016/S0070-2153(09)88004-6
Wilkins AS (2002) The evolution of developmental pathways. Sinauer Associates Inc, Sunderland
Wilson NG, Rouse GW (2010) Convergent camouflage and the non-monophyly of ‘seadragons’ (Syngnathidae: Teleostei): suggestions for a revised taxonomy of syngnathids. Zool Scr 39:551–558. https://doi.org/10.1111/j.1463-6409.2010.00449.x
Wray GA (2007) The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8:206–216. https://doi.org/10.1038/nrg2063
Yasuike M, Fujiwara A, Nakamura Y, Iwasaki Y, Nishiki I, Sugaya T, Shimizu A, Sano M, Kobayashi T, Ototake M (2016) A functional genomics tool for the Pacific bluefin tuna: development of a 44K oligonucleotide microarray from whole-genome sequencing data for global transcriptome analysis. Gene 576:603–609. https://doi.org/10.1016/j.gene.2015.10.023
You X et al (2014) Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat Commun 5:5594. https://doi.org/10.1038/ncomms6594
Acknowledgements
We wish to thank S. Bassham, C. Small, M. Currey, A. Amores, Y.L. Yan, and J. Poslethwait for providing feedback on the study design and results, as well as other members of the Cresko and Postlethwait labs; H. Mason-Jones and E. Rose for help with obtaining Gulf pipefish embryos; A. Jones and A. Anderson for sending robust ghost pipefish, messmate pipefish, and dwarf seahorse tissue samples; and A. Bentley for sending bluestripe pipefish and sculptured pipefish tissue samples from the KU fish tissue collection.
Funding
This work would not have been possible without multiple sources of generous support: grant from the National Science Foundation Doctoral Dissertation Improvement Grant in the Directorate for Biological Sciences (DEB-1701854 to W. A. Cresko and A. M. Fuiten), fellowships to A. M. Fuiten from the National Institutes of Health for both the Developmental Biology Training Grant Fellow (T32 HD007348) and the Genetics Training Grant Fellow (T32 GM007413), and from University of Oregon UO Under-Represented Minority Fellow.
Author information
Authors and Affiliations
Contributions
Allison Fuiten and Bill Cresko designed this study. Allison Fuiten performed experiments and data analysis. Interpretation of results was by Allison Fuiten and Bill Cresko. Allison Fuiten wrote the original draft, and Bill Cresko reviewed and edited the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The care and use of animals were in accordance under the IACUC protocol (assurance number A-3009-01).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Mark Q. Martindale
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplemental Figure 1
Evolution of Hox complex. a) Evolutionary timing of Hox complex duplications are denoted on the animal phylogeny based on (Carroll et al. 2013), with updates from (Pascual-Anaya et al. 2018; Ravi et al. 2009). Dashed arrow indicates current uncertainty where the second vertebrate Hox cluster duplication occurred relative to agnathans. b) A cartoon of the previously reported Hox clusters in Gulf pipefish, lined seahorse and tiger tail seahorse with boxes representing genes and circles representing microRNAs arranged along chromosome segments oriented left to right 5′ to 3′. The hollow box represents the hoxa7a pseudogene as described in Small et al. 2016. (EPS 3390 kb)
Supplemental Figure 2
VISTA plots for the HoxB clusters with threespine stickleback HoxBa set as reference sequence. Exons are highlighted in blue, CNE in pink, microRNAs are in the blue boxes. Shuffle LAGAN alignment was used with gray lines indicate stretches of continuous sequence. The reference, Gac, is the threespine stickleback; Tru, fugu; Ola, medaka; Tor, tuna; Ssc, pipefish; Hco, tiger tail seahorse; Her, lined seahorse; Bpe, mudskipper; Gmo, cod; Dre, zebrafish; Loc, spotted gar; Mmu, mouse; Hsa, human. Syngnathid specific peak losses are in red boxes. (EPS 1333 kb)
Supplemental Figure 3
VISTA plots for the HoxB clusters with threespine stickleback HoxBb set as reference sequence. Exons are highlighted in blue, CNE in pink, microRNAs are in the blue boxes. Shuffle LAGAN alignment was used with gray lines indicate stretches of continuous sequence. The reference, Gac, is the threespine stickleback; Tru, fugu; Ola, medaka; Tor, tuna; Ssc, pipefish; Hco, tiger tail seahorse; Her, lined seahorse; Bpe, mudskipper; Gmo, cod; Dre, zebrafish; Loc, spotted gar; Mmu, mouse; Hsa, human. Syngnathid specific peak losses are in red boxes. (EPS 2281 kb)
Supplemental Figure 4
VISTA plots for the HoxC clusters. Exons are highlighted in blue, CNE in pink. Gac, is the threespine stickleback; Tru, fugu; Ola, medaka; Tor, tuna; Ssc, pipefish; Hco, tiger tail seahorse; Her, lined seahorse; Bpe, mudskipper; Gmo, cod; Dre, zebrafish; Loc, spotted gar; Mmu, mouse; Hsa, human. Syngnathid specific peak losses are in red boxes. a) Gac is set as the reference with Shuffle-LAGAN alignment used. b) Ola is set as the reference with Shuffle-LAGAN alignment used. c) Tru is set as the reference with Shuffle-LAGAN alignment used. d) Gac is set as the reference with LAGAN alignment used. e) Ola is set as the reference with LAGAN alignment used. f) Tru is set as the reference with LAGAN alignment used. (EPS 5785 kb)
Supplemental Figure 5
VISTA plots for the HoxC clusters. Exons are highlighted in blue, CNE in pink. Gac, is the threespine stickleback; Tru, fugu; Ola, medaka; Tor, tuna; Ssc, pipefish; Hco, tiger tail seahorse; Her, lined seahorse; Bpe, mudskipper; Gmo, cod; Dre, zebrafish; Loc, spotted gar; Mmu, mouse; Hsa, human. Syngnathid specific peak losses are in red boxes. a) Gac is set as the reference with Shuffle-LAGAN alignment used. b) Ola is set as the reference with Shuffle-LAGAN alignment used. c) Tru is set as the reference with Shuffle-LAGAN alignment used. d) Gac is set as the reference with LAGAN alignment used. e) Ola is set as the reference with LAGAN alignment used. f) Tru is set as the reference with LAGAN alignment used. (EPS 5534 kb)
Supplemental Figure 6
A conserved non-coding element is not detectable in the pipefish HoxAb cluster with different species set as the reference and mVISTA alignment algorithms. Exons are highlighted in blue, CNE in pink. Gac, is the threespine stickleback HoxAb sequence; Tru, fugu HoxAb sequence; Ola, medaka HoxAb sequence; Tor, tuna HoxAb sequence; Ssc, pipefish HoxAb sequence; Hco, tiger tail seahorse HoxAb sequence; Her, lined seahorse HoxAb sequence; Bpe, mudskipper HoxAb sequence; Gmo, cod HoxAb sequence; Dre, zebrafish HoxAb sequence; Loc, spotted gar HoxA sequence; Mmu, mouse HoxA sequence; Hsa, human HoxA sequence. Red arrows indicate missing CNE in syngnathid fish. a) Threespine stickleback (gac) is set as the reference with LAGAN alignment used. b) Fugu (tru) is set as the reference with LAGAN alignment used. c) Medaka (ola) is set as the reference with LAGAN alignment used. d) Fugu (tru) is set as the reference with Shuffle-LAGAN alignment used. e) Medaka (ola) is set as the reference with Shuffle-LAGAN alignment used. (EPS 3593 kb)
Rights and permissions
About this article
Cite this article
Fuiten, A.M., Cresko, W.A. Evolutionary divergence of a Hoxa2b hindbrain enhancer in syngnathids mimics results of functional assays. Dev Genes Evol 231, 57–71 (2021). https://doi.org/10.1007/s00427-021-00676-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-021-00676-x