Abstract
Our studies conducted on reptilian limb muscle development revealed, for the first time, early forelimb muscle differentiation at the morphological and molecular level. Sand lizard skeletal muscle differentiation in the early forelimb bud was investigated by light, confocal, and transmission electron microscopy as well as western blot. The early forelimb bud, filled with mesenchymal cells, is surrounded by monolayer epithelium cells. The immunocytochemical analysis revealed the presence of Pax3- and Lbx-positive cells in the vicinity of the ventro-lateral lip (VLL) of the dermomyotome, suggesting that VLL is the source of limb muscle progenitor cells. Furthermore, Pax3- and Lbx-positive cells were observed in the dorsal and ventral myogenic pools of the forelimb bud. Skeletal muscle development in the early limb bud is asynchronous, which is manifested by the presence of myogenic cells in different stages of differentiation: multinucleated myotubes with well-developed contractile apparatus, myoblasts, and mitotically active premyoblasts. The western blot analysis revealed the presence of MyoD and Myf5 proteins in all investigated developmental stages. The MyoD western blot analysis showed two bands corresponding to monomeric (mMyoD) and dimeric (dMyoD) fractions. Two separate bands were also detected in the case of Myf5. The observed bands were related to non-phosphorylated (Myf5) and phosphorylated (pMyf5) fractions of Myf5. Our investigations on sand lizard forelimb myogenesis showed that the pattern of muscle differentiation in the early forelimb bud shares many features with rodents and chicks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vertebrate limb muscle differentiation and growth are one of the best known developmental processes, and this knowledge is obtained from chick and mouse embryonic studies. The appearance of buds along the lateral body wall is the first step of limb development (Martin 1998). Both forelimbs and hind limbs develop from the lateral mesodermal plate and grow into three main axes controlled by different signaling centers. The differentiation along the proximal–distal axis (from girdles to digits) is controlled by the apical ectodermal ridge (AER). The primary function of the AER is to maintain the distal cells in the mesenchymal core of the limb bud mitotically active and undifferentiated, both which are necessary for further limb growth (Kengaku et al. 1998; Kawakami et al. 2001). The posteriorly placed zone of polarizing activity (ZPA) is the site of sonic hedgehog (SHH) expression responsible for the anterior–posterior (from the 1st to the 5th digit) axis (Aono and Ide 1988). The third dorsal–ventral axis is regulated by wingless 7a (WNT7a) signaling from the ectoderm (Church and Francis-West 2002; Zeller et al. 2009; Butterfield et al. 2010).
The limb buds are composed of undifferentiated mesenchymal cells covered by an ectodermal cell layer. Mesenchymal cells of limb buds express one of the T-box transcription factor (TBX) family genes: TBX5 or TBX4 in the forelimb and hind limb respectively (Gibson-Brown et al. 1996). The mentioned transcription factors upregulate fibroblast growth factor 10 (Fgf10) expression in mesenchymal cells. FGF10 induces Fgf8 expression in the overlying ectoderm (Ohuchi et al. 1997; Xu et al. 1998; Sekine et al. 1999; Ng et al. 2002; Agarwal et al. 2003; Minguillon et al. 2012; Nishimoto et al. 2015). The interactions between the mesenchyme and the ectoderm play a crucial role in the maintenance of mesenchymal cells’ proliferative activity and limb growth (Dealy et al. 1997)
In tetrapods, three parts of the limb can be distinguished: the stylopodium (arm/thigh), the zeugopodium (forearm/crus), and the autopodium (hand/foot). All three regions are present in quadrupedal tetrapods, with a number of individual bone elements varying between species (Petit et al. 2017).
In vertebrates, somites are the source of skeletal muscle precursors. During embryogenesis, somites differentiate into the dermomyotome, myotome, and sclerotome. Subsequently, the dermomyotome can be divided into epaxial and hypaxial parts with the ventro-lateral (VLL) and dorso-medial (DML) lips. The epaxial dermomyotome gives rise to the back musculature, whereas the limb skeletal muscles are formed by the cells derived from the hypaxial part of the dermomyotome. It has been shown that in mouse muscle progenitor cells express paired homeobox transcription factor 3 (Pax3) and paired homeobox transcription factor 7 (Pax7) proteins (Relaix et al. 2005). The mesenchymal cells located in limb buds differentiate into numerous tissues (including skeletal muscle tissue). The VLL is the source of limb muscle progenitor cells. The process of limb muscle differentiation shares similarities with trunk muscle development (premyoblasts divide mitotically, myoblasts withdraw from cell cycle and fuse to form multinucleated myotubes which differentiate into mature muscle fibers, MRF regulatory control) (reviewed by Bentzinger et al. 2012). The VLL cells undergo epithelial–mesenchymal transition, which leads to their migration into the limb bud. Simultaneously, migrating progenitor cells are characterized by proliferative activity, which results in an increase of the cell population and allows differentiation into striated muscles to begin (Christ and Ordahl 1995; Gros and Tabin 2014; Christ and Brand-Saberi 2002).
Hypaxial cell deepithelialization and migration are genetically regulated by numerous proteins, e.g., Pax3 (the marker of progenitor muscle cells detected in the dermomyotome and migrating cells) or tyrosine–protein kinase Met/c-Met (detected in progenitor muscle cells deepithelialized from the hypaxial dermomyotome) (Bober et al. 1994; Goulding et al. 1994; Williams and Ordhal 1994; Daston et al. 1996; Bladt et al. 1995). It is well known that the presence of ladybird homeobox transcription factor 1 (Lbx1) in migrating cells is unequivocal evidence of the migration of progenitor muscle cells (Jagla et al. 1995; Gross et al. 2000). In chicken and mammals, two Lbx genes were found, whereas zebrafish has three Lbx genes. In contrast, Xenopus tropicalis and lamprey have just one Lbx gene (Wotton et al. 2008, 2015). Recent studies revealed that the expression pattern of these genes in vertebrates is different during development. In mammals, Lbx2 is expressed in the urogenital system, eye, and brain, whereas the two chicken and three zebrafish genes are co-expressed in migratory muscle precursors (Chen et al. 1999). It is hypothesized that Lbx genes in vertebrates are the result of the genome duplication during evolution. However, it is controversial if the genome duplication occurred before or after the gnathostome–agnathan divergence (reviewed by Wotton et al. 2015)
The muscle progenitor cells migrate into the dorsal and ventral parts of limb buds and form myogenic pools. As limb bud growth proceeds, the population of mesenchymal cells splits into three subpopulations (stylopodium, zeugopodium, autopodium) along the long axis of the future limb. The differentiation of limb muscles shows a proximal–distal gradient, which is correlated with limb growth (Christ and Brand-Saberi 2002).
The expression of the myogenic regulatory factor (MRF) family starts when the progenitor muscle cells are localized in limb buds. The superficial progenitor muscle cells express Myf5, whereas MyoD-positive cells localize in deeper layers of the limb bud (Patel et al. 2002). Limb bud myogenesis starts when the MyoD-positive cells fuse with each other, leading to primary muscle fiber formation. The primary muscle fibers are believed to be a scaffold for the secondary muscle fibers and every future muscle. Their main functions are the maintenance of the type, shape, and localization of mature muscles (Stockdale 1992, 1997; Christ and Brand-Saberi 2002).
Limb myogenesis in rodents and chicks has been investigated in detail at the morphological and molecular levels. Despite some molecular differences in MRF expression during limb muscle development between rat, mouse, and chick (in rat, MyoD occurs first, whereas, in mouse and chick, Myf5 is the first transcription factor observed in early limb bud), studies showed that the pattern of muscle differentiation in the limb is evolutionarily conserved (Christ and Brand-Saberi 2002; Duprez 2002; Francis-West et al. 2003; Murphy and Kardon 2011; Lee et al. 2013). So far, reptilian limb muscle development has not been studied at a detailed molecular level. The aim of our studies was to demonstrate reptilian early forelimb muscle differentiation at the morphological and molecular levels.
Materials and methods
Study animals
Gravid females of the sand lizard, Lacerta agilis, were caught in Poland in the vicinity of Wrocław at the beginning of June 2016. All of the specimens used in the experiments were captured according to the Polish regulations concerning the protection of wild species (Journal of Laws 1991, No. 114 Item 492; Journal of Laws 200, No. 66 Item 802; Journal of Laws 2004, No. 112 Item 1183; Journal of Laws 2015, No. 133 Item 266). The Department of Animal Developmental Biology of the University of Wrocław obtained approvals from the Local Ethics Commission in Wrocław (77/2013) and the Polish Ministry of Environment (Ref. No.: WPN.6401.51.2016.IW.1). The animals were kept in vivaria in an open area, in conditions similar to those in the wild (similar temperature, ventilation, humidity, differentiated bedding, hiding places) until the eggs were laid, and then they were released into their native area. The sand lizard eggs (n = 33) after oviposition were carefully collected and were placed inside plastic boxes filled with moistened perlite (at 100% humidity) with ventilation holes. Holes in the bottom and top of containers ensured air circulation. The eggs were prevented from desiccating by moistening perlite with water (twice a day) which ensured constant humidity in containers. The eggs were incubated at 30 °C, reflecting the seasonal ambient temperatures in the wild. The developmental stages of embryos were estimated using the developmental table published by Peter (1904).
The collected embryos were anesthetized with tricaine methanesulfonate (MS-222; 500 μg/g of body weight) (Conroy et al. 2009), before being decapitated and dissected for further analysis (Journal of Laws 2015, No. 133 Item 266).
Light and transmission electron microscopy
For light and electron microscopic examination, the embryonic body wall with limb buds (including differentiated muscle tissue) was fixed in modified Karnovsky fixative (1% paraformaldehyde [PFA] and 1% glutaraldehyde, in a 0.1 M phosphate buffer pH 7.2) for 24 h at 4 °C. The material was repeatedly rinsed with the same buffer and was postfixed for 2 h in a 1:1 mixture of osmium tetroxide-potassium ferricyanide (OsO4-K3Fe(CN)6). Following rinsing in the phosphate buffer, the material was dehydrated, first in a graded alcohol series and then in acetone, and was then embedded in epoxy resin Epon 812 (Sigma-Aldrich) (Luft 1961). The Epon blocks were cut on Leica Ultracut UCT (Leica, Wetzlar, Germany). Semithin sections (0.6 μm) were collected on glass slides and were stained with methylene blue in a 1% borax solution. The stained material was examined under an Olympus BX60 light microscope (Olympus). The ultrathin sections were collected on 200-mesh copper grids and were stained with uranyl acetate and lead citrate according to the standard protocol (Reynolds 1963), before being examined under the transmission electron microscope (TEM), Zeiss EM 900 (Carl Zeiss AG, Oberkochen, Germany; 80 kV).
Immunofluorescence analysis
After the dissection and fixation of embryos (4% PFA in phosphate buffer saline, PBS for 45 min at room temperature), the samples were transferred to 30% sucrose in PBS for overnight incubation at 4 °C. Next, samples were embedded in optimal cutting temperature medium (OCT) and placed in a cryomold and frozen. The samples were cut into 10 μm sections in a cryostat (Leica) at − 24 °C and were placed on SuperFrost Plus slides and subjected to immunofluorescence staining.
Standard immunofluorescence reactions were carried out on tissue cryosections described in our previous paper (Lewandowski et al. 2017). The following primary antibodies were used: rabbit polyclonal anti-phospho-histone H3 (pSer10) (Sigma-Aldrich) at dilution of 1:200 in PBST, mouse monoclonal anti-Pax3 (Developmental Studies Hybridoma Bank) at dilution of 1:50 in PBST, mouse monoclonal anti-Pax7 (Developmental Studies Hybridoma Bank) at dilution of 1:50 in PBST, mouse monoclonal anti-Lbx2 (Abcam) at dilution of 1:200 in PBST. Additionally, the following secondary antibodies were used: goat anti-mouse IgG-FITC conjugated (Sigma-Aldrich) at dilution of 1:50 in PBST, goat anti-rabbit IgG TRITC conjugated (Sigma-Aldrich) at dilution of 1:50 in PBST, donkey anti-mouse IgG Cy5 conjugated (Jackson ImmunoResearch) at dilution of 1:100 in PBST, donkey anti-rabbit IgG Cy5 conjugated (Jackson ImmunoResearch) at dilution of 1:100 in PBST, donkey anti-rabbit IgG Cy3 conjugated (Jackson ImmunoResearch) at dilution of 1:100 in PBST. For the F-actin identification, Alexa Fluor 488–conjugated phalloidin and Alexa Fluor 546–conjugated phalloidin (Molecular Probes) were used at a dilution of 1:80 in PBS. The DNA was stained with 4,6-diamidino-2-phenylindole (DAPI; 0.2 μg/ml). For the imaging, an Olympus FluoView FV1000 confocal laser scanning microscope (Olympus) was used. The images were recorded by employing the Plan-Apochromat × 10, × 20, or × 40 objectives. Brightness and contrast adjustments were performed in the FV10-ASW_Viewer or in ImageJ.
SDS-PAGE electrophoresis and western blot
The decapitated embryos and limb bud lysates were prepared, separated by SDS-PAGE, and analyzed by Western blot as described previously (Lewandowski et al. 2017). The membranes with separated and transferred proteins were incubated with the following primary antibodies: mouse monoclonal anti-Pax3 (Developmental Studies Hybridoma Bank) at dilution of 1:100, mouse monoclonal anti-Lbx2 (Abcam) at dilution of 1:200, rat monoclonal anti-α-actinin (Babraham Bioscience Technologies) at dilution of 1:250, mouse monoclonal anti-MyoD (Santa Cruz Biotechnology) at dilution of 1:200, rabbit polyclonal anti-Myf5 (GeneTex) at dilution of 1:200. Additionally, secondary antibodies were used: donkey anti-mouse IgG HRP conjugated (Jackson ImmunoResearch) at dilution of1:10000, donkey anti-rabbit IgG HRP conjugated (Jackson ImmunoResearch) at dilution of 1:10000, donkey anti-rat HRP conjugated (Jackson ImmunoResearch) at dilution of 1:10000. Membranes were then detected and documented with a chemiluminescent method using the Bio-Rad imaging system. The protein content in the Pax3, Lbx, MyoD, and Myf5 bands was then normalized according to the α-actinin content in each lane.
LC-MS analysis
Homogenized samples were prepared as described by Mroczek et al. (2017). The limb bud lysates were separated by SDS-PAGE and stained by modified silver staining (Shevchenko et al. 1996). A stained portion of the gel (size of cutout bands corresponded to the size of bands detected in western blot technique) was cut out. Samples of proteins were sent for the identification by an LC-MS method in the Mass Spectrometry Laboratory, IBB PAS and searched with MASCOT (Matrix Science) against homemade protein database containing 538 entries including known Lbx sequences, downloaded 18.04.2019 from UniProt.
Densitometric analysis of MyoD and Myf5 fractions
The ratio of different fractions of the tested proteins (monomeric vs. dimeric in the case of MyoD and non-phosphorylated vs. phosphorylated in the case of Myf5) was determined via densitometric measurement of the signal generated by a western blot analysis. Imaging software (Image Lab 6.0; Bio-Rad) was used to compare the signal generated by the bands detected on the membranes.
Statistical analyses were carried out using MS Excel. Student’s t test was used for comparisons of the percentage amounts (n = 5 for MyoD; n = 3 for Myf5). The results are reported in the graph, and p < 0.05 is considered statistically significant.
Results
The sand lizard limb bud myogenesis was investigated at stages 21–23 by the use of a light microscope, confocal microscope, TEM, and Western blot.
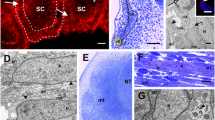
During the early stage (stage 23) of sand lizard embryogenesis, the forelimb bud, surrounded by a monolayer of epithelial cells, is filled with mesenchymal cells (Fig. 1a). Mononucleated mesenchymal cells are irregularly shaped. Their cytoplasm, with a large nucleus, is rich in mitochondria and rough endoplasmic reticulum (RER) (Fig. 1b). The immunodetection of the Pax3 protein (a marker of muscle progenitor cells) revealed that this protein is present in the epithelial dermomyotome, in the VLL, DML, and in the myotome (Fig. 1c). At this stage of myogenesis, Pax3-positive muscle precursor cells are also observed in the dorsal muscle mass of the forelimb (Fig. 1c). The presence of Pax3 protein during trunk muscle myogenesis was confirmed by western blot analysis (stages 22–32) (Fig. 1d). No Pax7- (a marker of muscle progenitor and satellite cells) positive cells are detected in the L. agilis forelimb bud (stage 23), while in the L. agilis trunk Pax7-positive cells in the myotomes were previously observed at the same stage of embryogenesis (Rupik et al. 2012; Lewandowski, unpublished).
Limb bud muscle differentiation. a Stage 23. Cross section through embryo. Dermomyotome (yellow, dashed line), ventro-lateral (vll), and dorso-medial (dml) lips of dermomyotome. Note mesenchymal cells (white arrows) in developing limb bud (lb). Transverse, semithin section, methylene blue staining, light microscope. Scale bar: 100 μm. Inset: Magnification of region marked by white, dashed frame. Myotome (red, dotted line), ventro-lateral (vll) lip of dermomyotome (yellow, dashed line), limb bud (lb). Transverse, semithin section, methylene blue staining, light microscope. Scale bar: 20 μm. b Stage 23. Ultrastructure of forelimb bud myogenic pool filled with mesenchymal cells. Nuclei (N), nucleoli (Nu), RER (black arrowheads), mitochondria (blue arrowheads). Transverse, ultrathin section. Scale bar: 1 μm. c Stage 23. Immunodetection of Pax3 protein (green) in dermomyotome (white, dotted line) and dorsal forelimb bud (lb). Nuclei (blue), F-actin (red), ventro-lateral (vll), and dorso-medial (dml) lips of dermomyotome, neural tube (NT). Transverse, cryosection, confocal microscope. Scale bar: 100 μm. Inset: Pax3-positive cells (green) in dorsal muscle mass of forelimb bud (lb). Transverse, cryosection, confocal microscope. Scale bar: 100 μm. d Western blot analysis of Pax3 protein expression in myotome during successive developmental stages (stages 22–32). Pax3 is marked together with the α-actinin band used as a loading control
To demonstrate the presence of migratory muscle progenitor cells, immunodetection of Lbx protein was carried out in the trunk (myotome). In the myotome (stage 23), Lbx-positive cells were observed in the vicinity of the VLL (Fig. 2a). Lbx-positive cells were mitotically active, which was confirmed by the detection of phosphorylated histone H3 (Fig. 2b). Moreover, western blot analysis revealed that Lbx protein is present in trunk muscles in all investigated stages (stages 22–32) (Fig. 2c).
Expression of Lbx protein in myotome. a Stage 23. Immunodetection of Lbx-positive muscle progenitor cells (red) in the myotome (yellow, dashed line) and the vicinity of ventro-lateral lip (vll) of dermomyotome (white, dotted line). Nuclei (blue), F-actin (green). Transverse, cryosection, confocal microscope. Scale bar: 10 μm. b Stage 23. Immunodetection of mitotically active migrating muscle progenitor cells (white arrowheads) in the myotome. Lbx (yellow), phosphorylated histone H3 (red), nuclei (blue). Transverse, cryosection, confocal microscope. Scale bar: 10 μm. c Western blot analysis of Lbx protein expression during successive developmental stages (stages 22–32). Lbx is marked together with the α-actinin band used as a loading control
At stage 24, in the forelimb, muscle mass mononucleated post-mitotic cells are observed in the proximal part, whereas mitotically active cells occupied a distal part of the developing forelimb (Fig. 3a). Their mitotic activity was confirmed by the detection of phosphorylated histone H3 (Fig. 3c). TEM analysis showed mitotically active premyoblasts and elongated post-mitotic myoblasts with centrally located large nuclei. The cytoplasm of myoblasts revealed the presence of numerous mitochondria and RER (Fig. 3b).
Myogenic pool in the forelimb. a Stage 24. Structure of forelimb bud (lb) with post-mitotic cells (white arrows) localized in proximal (Pr) part of the limb bud, mitotically active cells (circled) are in the distal (Di) part of the limb bud. Longitudinal, semithin section, methylene blue staining, light microscope. Scale bar: 20 μm. b Stage 24. Ultrastructure of forelimb bud. Elongated myoblasts (mb), nuclei (N), nucleoli (Nu), RER (black arrows), mitochondria (blue arrows). Longitudinal, ultrathin section. Scale bar: 1 μm. Inset: Ultrastructure of interphase (left) and mitotic (right) premyoblasts (pmb). Longitudinal, ultrathin section. Scale bar: 1 μm. c Stage 24. Immunodetection of phosphorylated histone H3 (red). Nuclei (blue), F-actin (green). Transverse, cryosection, confocal microscope. Scale bar: 5 μm
In the forelimb bud myogenic pool (stage 28), mononucleated myotubes are accompanied by premyoblasts. The characteristic feature of mononucleated myotubes at this developmental stage is irregularly arranged myofibrils. The myotube sarcoplasm contains numerous mitochondria, glycogen granules, and RER (Fig. 4a, b).
Structure of forelimb bud muscles. a Stage 28. Structure of forelimb bud (lb). Mononucleated cells (black arrows), elongated myoblasts (white arrows), premyoblasts (circled), proximal (Pr), distal (Di). Longitudinal, semithin section, methylene blue staining, light microscope. Scale bar: 20 μm. b Stage 28. Ultrastructure of forelimb bud myogenic pool. Nuclei (N), mitochondria (blue arrowheads), myotube (MT), myofibrils (MF), RER (black arrowheads), glycogen (red arrowheads). Longitudinal, ultrathin section. Scale bar: 1 μm. Inset: Ultrastructure of premyoblast. Nucleus (N), nucleolus (Nu), mitochondria (blue arrowheads). Longitudinal, ultrathin section. Scale bar: 1 μm. c Stage 32. Ultrastructure of myotube (MT) in the myogenic pool. Nucleus (N), mitochondria (blue arrowheads), myofibrils (MF), glycogen (red arrowheads), Z lines (white arrows). Longitudinal, ultrathin section. Scale bar: 2 μm. d Stage 32. Ultrastructure of forelimb myogenic pool filled with cells at different stages of differentiation: myotube (MT) and premyoblast (circled). Nucleus (N), mitochondria (blue arrowheads), myofibrils (MF), glycogen (red arrowheads). Longitudinal, ultrathin section. Scale bar: 2 μm
Western blot analysis of the forelimb bud revealed the presence of Pax3 protein in all studied stages (stages 21–32) (Fig. 5a, b), which was confirmed by immunocytodetection of Lbx and Pax3 proteins in the dorsal and ventral limb bud (Fig. 5c–e).
Expression of Lbx and Pax3 proteins in forelimb bud. a, b Western blot analysis of Lbx (a) and Pax3 (b) proteins expression during successive developmental stages (Stages 21–32). Lbx and Pax3 proteins are marked together with α-actinin bands used as a loading control. c, d Stage 30. Immunodetection of Lbx (white arrowheads and red) protein in ventral (c) and dorsal (d) part of forelimb bud. Nuclei (blue), F-actin (green), ventral (V), dorsal (D), proximal (Pr), distal (Di). Longitudinal, cryosection, confocal microscope. Scale bar: 50 μm. e Stage 30. Immunodetection of Pax3 (green) protein in ventral (V) and dorsal (D) part of forelimb bud. Nuclei (blue), F-actin (red), proximal (Pr), distal (Di). Longitudinal, cryosection, confocal microscope. Scale bar: 50 μm
To confirm the presence of Lbx protein in limb buds, peptide identification from silver-stained protein gel samples was performed. The comparison of obtained masses of peptides and their fragments with the NCBI protein sequences database was carried out using the MASCOT program. Obtained results revealed the presence of three peptides corresponding to Xenopus laevis Lbx1 (Fig. 1a, b; suppl.).
As limb bud muscle differentiation proceeded (stage 32), cells at different stages of differentiation can be observed: mitotic active premyoblasts, myoblasts, and multinucleated myotubes with well-developed contractile apparatus (Fig. 4d). The ultrastructure of myotube sarcoplasm shows that sarcomeres form characteristic repetitive units of light and dark bands with pronounced Z lines (Fig. 4c).
Our research revealed the presence of MyoD and Myf5 proteins in all investigated developmental stages (stages 22–32) (Fig. 6). The Western blot analysis of MyoD protein showed two bands corresponding to monomeric (mMyoD) and dimeric (dMyoD) fractions of MyoD (Fig. 6a). Since the intensity of detected bands was different in distinct developmental stages, we decided to estimate the ratio of monomeric and dimeric MyoD fractions in each investigated developmental stage. The conducted analysis showed statistically significant differences between the levels of mMyoD and dMyoD at stage 32 (p < 0.05) (Fig. 6a).
Expression of MyoD and Myf5 fractions during forelimb bud development. a Percentage amounts of different MyoD fractions (mMyoD, monomeric; dMyoD, dimeric) during successive developmental stages (stages 22–32). The analysis was based on densitometric measurement of Western blot detected bands. Statistically significant difference is indicated with an asterisk (p < 0.05; Student’s t test; n = 5). Western blot analysis of MyoD protein expression during successive developmental stages (stages 22–32). Note the presence of two bands corresponding to monomeric (MyoD) and dimeric (dMyoD) fraction of MyoD. MyoD is marked together with the α-actinin band used as a loading control. b Percentage amounts of different Myf5 fractions (Myf5, non-phosphorylated; pMyf5, phosphorylated) during successive developmental stages (stages 22–32). The analysis was based on densitometric measurement of Western blot detected bands. Statistically significant differences are indicated with asterisks (p < 0.05; Student’s t test; n = 3). Western blot analysis of Myf5 protein expression during successive developmental stages (stages 22–32). Note the presence of two bands corresponding to non-phosphorylated (Myf5) and phosphorylated (pMyf5) fractions of Myf5. Myf5 is marked together with α-actinin band used as a loading control
We also observed two separate bands during Myf5 Western blot analysis (Fig. 6b). The observed bands are related to non-phosphorylated (Myf5) and phosphorylated (pMyf5) fractions of Myf5 (Fig. 6b). Since the intensity of detected bands was different in distinct developmental stages, we decided to estimate the ratio of non-phosphorylated and phosphorylated Myf5 fractions. The conducted analysis showed statistically significant differences between Myf5 and pMyf5 at stages 24 and 28 (Fig. 6b).
Discussion
Structure of the early limb bud
Our investigations on sand lizard forelimb myogenesis showed that the pattern of muscle differentiation in the early forelimb bud shares many features with rodents and chicks. In sand lizard embryos, forelimb buds are surrounded by a monolayer of epithelial cells and present a core of mesenchymal cells. The myogenesis of the limb bud initially starts in the myogenic pool divided into the dorsal and ventral muscle mass. The process of muscle differentiation in the limb bud is asynchronous. In the myogenic pool, mitotically active progenitor muscle cells (premyoblasts), mononucleated myoblasts, mononucleated myotubes, and multinucleated muscle fibers are observed. As in rodents and chicks, sand lizard mononucleated myoblasts elongate and differentiate into mononucleated myotubes accompanied by mononucleated cells (premyoblasts or myoblasts). Lee et al. (2013) compared mononucleated myotubes observed in the early rodent and chick limb bud to mononucleated, so-called founder cells (FCs), found during Drosophila melanogaster somatic muscle development. In the fruit fly, FCs initiate myogenesis by fusion with non-specific cells, called fusion-competent myoblasts (FCMs). It is noteworthy that muscle morphology, localization in each hemisegment, innervation, and the connection site to the exoskeleton in Drosophila depend on the identity of FC genes (Baylies et al. 1998; Dobi et al. 2015). In contrast to vertebrate multi-fiber muscles, Drosophila muscles are mono-fiber. Our knowledge of mononucleated myotube gene identity in mammals and chicks is still very poor. It has been reported that the mouse adult limb muscles contain a different proportion of slow and fast fibers, but despite this they do not show notable differences in gene expression (Schafer and Braun 1999; Gross et al. 2000). Our ultrastructural studies on sand lizard muscles showed that not all myoblasts elongate and form multinucleated myotubes; some of them behave like FCMs observed in Drosophila myogenesis. These data show that muscle differentiation in a phylogenetically distant group of organisms may share unexpected similar features. To find some specific markers of mononucleated myotubes, detailed investigations at a molecular level should be carried out.
Pattern expression of Pax3, Pax7, and Lbx proteins during early limb bud development
During chick and mammal limb myogenesis, Pax3 protein is essential for the survival of muscle progenitor cells and for their migration from VLLs into limb buds, whereas Pax7 is responsible for the specification of adult satellite cells at later developmental stages (Buckingham 2007). In the mentioned organisms, Pax3-positive progenitor cells were observed initially in the VLL of the dermomyotome and then in the dorsal and ventral muscle mass of the early forelimb bud (reviewed by Bentzinger et al. 2012; Zammitt 2017). Our investigation on sand lizard also showed Pax3-positive cells in the VLL and in the myogenic pool of the early forelimb bud. Sand lizard migrating cells also expressed Lbx protein. Based on studies carried out on mice, it was shown that Lbx1 and Pax3 are co-expressed in all migrating hypaxial muscle precursors. The authors assume that Lbx1 regulates their migration (Gross et al. 2000; Masselink et al. 2017). Previous studies demonstrated that in mice there exist two Lbx genes: Lbx1 and Lbx2. Lbx1 is expressed in hypaxial migratory progenitor cells, whereas Lbx2 is observed in the central nervous and genitourinary systems (Chen et al. 1999). In Danio rerio, the presence of three Lbx genes were confirmed: Lbx2, Lbx1a, Lbx1b, Lbx2 is expressed in fin buds and in the ventral part of the somite. In Drosophila, Lbx protein is engaged in the establishment of leg morphological and functional features (Maqbool et al. 2006). It was evidenced that in D. rerio Morpholino knockdown of Lbx2 suppresses MyoD expression in fin buds. Of interest, the Lbx2 gene is also implicated in myofibrillogenesis in both trunk and fin bud muscles (Ochi and Westerfield 2009). In our studies, to identify migratory muscle progenitors (MMPs), we used a commercial anti-Lbx2 antibody. LC-MS analysis showed the presence of three peptides corresponding to Xenopus laevis Lbx1. It is worthy to note that only Lbx1 was found in veiled chameleon (Chamaeleo calyptratus) embryo transcriptome at the early limb bud stage (Pinto et al. 2019). Based on those results, our LC-MS analysis, and amino acids sequence comparison (epitope of anti-Lbx2 antibody vs. Anolis carolinensis vs. Pogona vitticeps) we assume that used anti-Lbx2 antibody recognizes the Lbx1 protein in Lacerta agilis. However, the presence of Lbx2 protein in Lacerta agilis cannot be excluded.
Our studies showed for the first time the presence of Lbx protein during sand lizard limb bud myogenesis. We strongly believe that the mentioned protein is engaged in the acquisition of the migratory potential of limb muscle progenitor cells, but further studies are necessary.
Our previous research revealed the presence of Pax7-positive cells in myotomal muscles during early stages (stage 24) of sand lizard development (Rupik et al. 2012). It is noteworthy that we did not observe Pax7-positive cells in analyzed stages of limb bud development. Similar results were obtained by Lee et al. (2013) in the early rat forelimb. Pax7 expression was not observed in the limb muscle mass, whereas this protein was clearly present in the dorsal neural tube and the dermomyotome. During later developmental stages, Pax7 protein was detected in the central and basal part of the forelimb bud, then Pax7-positive cells were observed in the dorsal and ventral limb bud mass. The spatiotemporal expression of Pax3 and Pax7 proteins was also noted in the mouse limb, in which Pax3 expression precedes the expression of Pax7 (Relaix et al. 2004). These data strongly confirm the hypothesis that Pax3 protein plays a crucial role in primary myogenesis, whereas Pax7 is required later for the maintenance of satellite cells (Tajbakhsh et al. 1997; Seale et al. 2000). Based on our results, we suggest that the Pax3/Pax7 pattern expression during limb bud muscle development is conserved in amniotes.
Expression of MyoD and Myf-5 proteins during early limb bud development
It is commonly known that Myf5 and MyoD are involved in the shift from proliferating myoblasts to multinucleated muscle fibers capable of contractions. It has been suggested that the activation of MyoD and Myf5 is controlled in developing myoblasts by their degradation (Thayer et al. 1989), modification (Lindon et al. 1998), signaling (Vaidya et al. 1989; Li et al. 1992), or interference with heterodimerization (Benezra et al. 1990).
Western blot analysis of sand lizard limb buds revealed the presence of MyoD and Myf5 proteins in all investigated developmental stages (stages 22–32). It has been demonstrated that in chicks and mice during early developmental stages of limb buds Myf5 is the first MRF protein expressed. As myogenesis proceeds, Myf5 was no longer detectable, but MyoD was observed. In contrast to chicks and mice, during rat limb bud development, MyoD is the first MRF protein detectable (Lee et al. 2013; Mok et al. 2015). Although our studies revealed the presence of MyoD and Myf5 proteins in all analyzed stages, we cannot exclude spatiotemporal pattern expression of the mentioned proteins because of asynchronous myogenesis in sand lizard limb buds, i.e., in the limb bud myogenic pool the muscle cells are at different stages of differentiation: mitotically active premyoblasts, myoblasts, and myotubes were observed.
It is commonly accepted that MyoD induces the myogenic program only as homo- or heterodimers with other helix–loop–helix transcription factors such as E12 or E47 (Maleki et al. 1997). Sand lizard limb bud Western blot analysis revealed the presence of two bands of MyoD protein corresponding to monomeric (mMyoD) and dimeric (dMyoD) fractions. Statistical analysis showed significant differences between the levels of mMyoD and dMyoD at a later developmental stage (stage 32). A higher level of mMyoD suggests a decrease in the number of undifferentiated cells in the limb bud myogenic pool.
It has been reported that Myf5 is a “determination” factor expressed in proliferating myoblasts (Braun et al. 1989). Lindon et al. (1998) found that Myf5 is downregulated in cells undergoing differentiation. The authors suggested that the Myf5 level in proliferative cells is strongly regulated by cell cycle–associated events that include the degradation of this factor during mitosis. Several studies revealed that the cell cycle depends on the Myf5 level and its phosphorylation (Lindon et al. 1998; Song et al. 1998; Kitzmann et al. 1998; 1999; Tintignac et al. 2000). Previous data suggested that Myf5 undergoes phosphorylation by a mitosis-specific kinase(s), and that this modified Myf5 is highly unstable in mitotic cells and is rapidly degraded, probably by a 26-S proteasome-dependent mechanism (Lindon et al. 1998). Based on the results from Ohya et al. (2006), Myf5 in turtles undergoes alternative splicing (twelve nucleotides deletion comparing it with the sequences of other vertebrate Myf5 genes). However, the authors suggest that the mentioned deletion is only turtle lineage–specific feature. The sand lizard limb bud western blot analysis revealed two separate bands (~ 30 kDa) of Myf5 protein. We assume that the observed bands correspond to non-phosphorylated (Myf5) and phosphorylated (pMyf5) fractions of Myf5. The conducted analysis showed statistically significant differences between Myf5 and pMyf5 at stages 24 and 28. Therefore, the limb bud myogenesis is asynchronous; both non-phosphorylated and phosphorylated fractions of Myf5 could be observed. However, it is not excluded that separated bands were represented proteins translated on two Myf5 isoforms, future studies will clarify this.
It is now clearly established that limb myogenesis shares many similar features among all vertebrates. During limb myogenesis, mononucleated myoblasts elongate and differentiate into mononucleated myotubes accompanied by mononucleated cells (premyoblasts or myoblasts). Similar to other tetrapods, the VLLs of the dermomyotome are a source of limb muscle progenitor cells. Our studies on the sand lizard for the first time revealed the presence of Lbx2 protein in progenitor muscle cells migrating to the limb bud. Despite the fact that in all vertebrates genetic control of muscle fiber differentiation includes the same transcription factors (Pax3/7, MyoD, Myf5), the known differences in myogenic genetic control, observed in mouse, chick, rat, and sand lizard, only concern their spatiotemporal expression.
References
Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG (2003) Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130:623–633
Aono H, Ide H (1988) A gradient of responsiveness to the growth-promoting activity of ZPA (zone of polarizing activity) in the chick limb bud. Development 124:4547–4556
Baylies MK, Bate M, Ruiz Gomez M (1998) Myogenesis: a view from Drosophila. Cell 93(6):921–927
Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein ID: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49–59
Bentzinger CF, Wang YX, Rudnicki MA (2012) Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol 4(2). https://doi.org/10.1101/cshperspect.a008342
Bladt F, Reithmacher D, Isenmann S, Aguzzi A, Birchmeier C (1995) Essential role for the c-met receptor in the migration of myogenic precursor cells in to the limb bud. Nature 376:768–771
Bober E, Franz T, Arnolod H-H, Gruss P, Tremblay P (1994) Pax-3 is required for the development of the limb muscles: a possible role for the migration of dermomyotomal muscle precursor cells. Development 120:603–612
Braun T, Bober E, Buschhausen-Denker G, Kohtz S, Grzeschik KH, Arnold HH, Kotz S (1989) Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J 8(12):3617–3625
Buckingham M (2007) Skeletal muscle progenitor cells anf the role of Pax genes. C R Biol 330(6–7):530–533
Butterfield NC, McGlinn E, Wicking C (2010) The molecular regulation of vertebrate limb patterning. Curr Top Dev Biol 90:319–341
Chen F, Liu KC, Epstein JA (1999) Lbx2, a novel murine homeobox gene related to the Drosophila ladybird genes is expressed in the developing urogenital system, eye and brain. Mech Dev 84:181–184
Christ B, Brand-Saberi B (2002) Limb muscle development. Int J Dev Biol 46:905–914
Christ B, Ordahl CP (1995) Early stages of chick somite development. Anat Embryol 191:381–396
Church VL, Francis-West P (2002) Wnt signaling during limb development. Int J Dev Biol 46:927–936
Conroy CJ, Papenfuss T, Parker J, Hahn N (2009) Use of tricaine methanesulfonate (MS-222) for euthanasia of reptiles. J Am Assoc Lab Anim 48:28–32
Daston G, Lamar E, Olivier M, Goulding M (1996) Pax-3 is necessary for the migration but not differentiation of limb muscle precursors in mouse. Development 122:1017–1027
Dealy CN, Seghatoleslami D, Ferrari D, Kosher RA (1997) FGF-stimulated outgrowth and proliferation of limb mesoderm is dependent of syndecan-3. Dev Biol 184:343–350
Dobi KC, Schulman VK, Baylies MK (2015) Specification of the somatic musculature in Drosophila. Wiley Interdiscip Rev Dev Biol 4(4):357–375
Duprez D (2002) Signals regulating muscle formation in the limb during embryonic development. Int J Dev Biol 46(7):915–925
Francis-West PH, Antoni L, Anakwe K (2003) Regulation of myogenic differentiation in the developing limb bud. J Anat 202(1):69–81
Gibson-Brown JJ, Agulnik SI, Chapman DL, Alexiou M, Garvey N, Silver LM, Papaioannou VE (1996) Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech Dev 56:93–101
Goulding M, Lumsden A, Paquette AJ (1994) Regulation of Pax-3 expression the dermomyotome and its role in muscle development. Development 120:957–356
Gros J, Tabin CJ (2014) Vertebrate limb bud formation is initiated by localized epithelial to mesenchymal transition. Nature 343(6176):1253–1256
Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M (2000) Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127:413–424
Jagla K, Dolle P, Mettei MG, Jagla T, Schurbaur B, Drentan G, Bellard F, Bellard M (1995) Mouse Lbx1 and human LBX1 define a novel mammalian homeobox gene family related to the Drosophila lady bird genes. Mech Dev 23:345–356
Kawakami Y, Capdevila J, Buscher D, Itoh T, Rodriguez-Esteban C, Izpisua Belmonte JC (2001) WNT signals control FGF-dependent limb initiation and AER induction in chick embryo. Cell 104:891–900
Kengaku M, Capdevila J, Rodriguez-Esteban C, de la Pena J, Johnson RL, Belmonte JC, Tabin CJ (1998) Distinct WNT pathways regulating AER formation dorsoventral polarity in the chick limb bud. Science 280:1274–1277
Kitzmann M, Carnac G, Vandromme M, Priming M, Lamb NJ, Fernandez A (1998) The muscle regulatory factors MyoD and myf-5 undergo distinct cell-specific expression an muscle cells. J Cell Biol 142:1447–1459
Kitzmann M, Vandromme M, Schaeffer V, Carnac G, Labbe JC, Lamb N, Fernandez A (1999) cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: role in modulating MyoD half-life activity. Mol Cell Biol 19:3167–3176
Lee ASJ, Harris J, Bate M, Vijayraghavan K, Fisher L, Tajbakhsh S, Duxson M (2013) Initiation of primary myogenesis in amniote limb muscles. Dev Dyn 242:1043–1055
Lewandowski D, Dubińska-Magiera M, Posyniak E, Rupik W, Daczewska M (2017) Does the grass snake (Natrix natrix) (Squamata: Serpentes: Natricinae) fit the amniote-specific model of myogenesis? Protoplasma 254(4):1507–1516
Li L, Zhou J, James G, Heller-Harrison R, Czech MP, Olson EN (1992) FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell 71:1181–1194
Lindon C, Montarras D, Pinset C (1998) Cell cycle-regulated expression of the muscle determination factor Myf5 in proliferating myoblasts. J Cell Biol 140:111–118
Luft JH (1961) Improvements in epoxy resin embedding methods. J Biophys Biochem Cytol 9(2):409–414
Maleki SJ, Royer CA, Hutlburt BK (1997) MyoD-E12 heterodimers and MyoD-MyoD homodimers are equally stable. Biochemistry-US 36(22):6762–6767
Maqbool T, Soler C, Jagla T, Daczewska M, Lodha N, Palliyil S, VijayRaghavan K, Jagla K (2006) Shaping leg muscles in Drosophila: role of ladybird, a conservation regulator of appendicular myogenesis. PLoS One 1:e122
Martin GR (1998) The roles of FGFs in the early development of vertebrate limbs. Genes Dev 12:1571–1586
Masselink W, Masaki M, Sieiro D, Marcelle C, Currie PD (2017) Phosphorylation of Lbx1 controls lateral myoblast migration into the limb. Dev Boil 430:302–309
Minguillon C, Nishimoto S, Wood S, Vendrell E, Gibson-Brown JJ, Logan MP (2012) Hox genes regulate the onset of Tbx5 expression in the forelimb. Development 139:3180–3188
Mok GF, Mohammed RH, Sweetman D (2015) Expression of myogenic regulatory factors in chicken embryos during somite and limb development. J Anat 227:352–360
Mroczek S, Chlebowska J, Kuliński TM, Gewartowska O, Gruchota J, Cysewski D, Liudkovska V, Borsuk E, Nowis D, Dziembowski A (2017) The non-canonical poly(A) polymerase FAM46C acts as an onco-suppressor in multiple myeloma. Nat Commun 8:619. https://doi.org/10.1038/s41467-017-00578-5
Murphy M, Kardon G (2011) Origin of vertebrate limb muscle: the role of progenitor and myoblast populations. Curr Top Dev Biol 96:1–31
Ng JK, Kawakami Y, Buscher D, Raya A, Itoh T, Koth CM, Rodriguez-Esteban C, Rodriguez-Leon J, Garrity DM, Fishman MC, Izpisua Belmonte JC (2002) The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development 129:5161–5170
Nishimoto S, Wilde SM, Wood S, Logan MP (2015) RA acts in a coherent feed-forward mechanism with Tbx5 to control limb bud induction and initiation. Cell Rep 12:879–891
Ochi H, Westerfield M (2009) Lbx2 regulates formation of myofibrils. BMC Dev Biol 9:13. https://doi.org/10.1186/1471-213X-9-13
Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M, Itoh N, Noji S (1997) The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development 124:2235–2244
Ohya YK, Usuda R, Kuraku S, Nagashima H, Kuratani S (2006) Unique features of Myf-5 in turtles: nucleotide deletion, alternative splicing, and unusual expression pattern. Evol Dev 8(5):415–423
Patel K, Christ B, Stockdale FE (2002) Control of muscle size during embryonic, fetal, and adult life. In: Brand-Saberi B (ed) Results and problems in cell differentiation vol. 38. Vertebrate myogenesis pp 163–186. Springer-Verlag, Berlin, Heidelberg
Peter K (1904) Normantafel zur Entwicklungsgeschichte der Zauneidechse (Lacerta agilis). In: Keibel F (ed) Normantafel zur Entwicklungsgeschichte der Wirbeltiere, vol 4. Gustav Fischer, Jena
Petit F, Sears KE, Ahituv N (2017) Limb development: a paradigm of gene regulation. Nature 18:245–258
Pinto BJ, Card DC, Castoe TA, Diaz RE Jr, Nielsen SV, Trainor PA, Gamble T (2019) The transcriptome of the veiled chameleon (Chamaeleo calyptratus): a source for studying the evolution and development of vertebrates. Dev Dynam. https://doi.org/10.1002/dvdy.20
Relaix F, Rocancourt D, Mansouri A, Buckingham M (2004) Divergent functions of murine Pax3 and Pax7 limb muscle development. Genes Dev 18(9):1088–1105
Relaix F, Rocancourt D, Mansouri A, Buckingham M (2005) A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 435:948–953
Reynolds ES (1963) The use of lead citrate at high pH as an electronopaque stain in electron microscopy. J Cell Biol 17(1):208–212
Rupik W, Swadźba E, Dubińska-Magiera M, Jędrzejowska I, Daczewska M (2012) Reptilian myotomal myogenesis-lessons from the sand lizard Lacerta agilis L. (Reptilia, Lacertidae). Zool (Jena) 115:330–338
Schafer K, Braun T (1999) Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat Genet 23(2):213–216
Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA (2000) Pax7 is required for the specification of myogenic satellite cells. Cell 102:777–786
Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S (1999) Fgf10 is essential for limb and lung formation. Nat Genet 21:138–141
Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858
Song A, Wang Q, Goebl MG, Harrington MA (1998) Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol Cell Biol 18:4994–4999
Stockdale FE (1992) Myogenic cell lineages. Dev Biol 154:284–298
Stockdale FE (1997) Mechanisms of formation of muscle fiber types. Cell Struct Funct 22:37–43
Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M (1997) Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89(1):127–138
Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H (1989) Positive autoregulation of the myogenic determination gene MyoD1. Cell 58:241–248
Tintignac LA, Leibovitch MP, Kitzmann M, Fernandez A, Ducommun B, Meijer L, Leibovitch SA (2000) Cyclin E-cdk2 phosphorylation promotes late G1-phase degradation of MyoD in muscle cells. Exp Cell Res 259:300–307
Vaidya TB, Rhodes SJ, Taparowsky EJ, Konieczny SF (1989) Fibroblast growth factor and transforming growth factor beta repress transcription of the myogenic regulatory gene MyoD1. Mol Cell Biol 9:3576–3579
Williams BA, Ordhal CP (1994) Pax-3 expression in segmental mesoderm marks early stages in myogenic specification. Development 120:785–796
Wotton KR, Weierud FK, Dietrich S, Lewis KE (2008) Comparative genomics of Lbx loci reveales conservation of identical Lbx ohnolgs in bony vertebrates. BMC Evol Biol 8:171. https://doi.org/10.1186/1471-2148-8-171
Wotton KR, Schubert FR, Dietrich S (2015) Hypaxial muscle: controversial classification and controversial? In: Brand-Saberi B (ed) Vertebrate myogenesis. Results Probl Cell Differ, vol 56. Springer, Berlin, Heidelberg, pp 25–48
Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C (1998) Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development 125:753–765
Zammitt PS (2017) Function of myogenic regulatory factors Myf5, MyoD, myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol 72:19–32
Zeller R, Lopez-Rios J, Zuniga A (2009) Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet 10:845–858
Acknowledgments
The authors would like to thank Katarzyna Pajer (Department of Animal Developmental Biology, University of Wroclaw) and Sylwia Nowak (Laboratory of Microscopic Techniques, University of Wroclaw) for their technical assistance and Anna Najbar (Department of Evolutionary Biology and Conservation of Vertebrates, University of Wroclaw) for her consultations in animal care. The LC-MS equipment was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. The equipment used was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland. We acknowledge the support of the Polish State Committee for Scientific Research, Project No. 1068/S/IBE/2017.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Angelika Stollewerk
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
LC-MS based identification of Lbx peptides from Lacerta agilis limb buds extract. A. The limb bud lysates from Lacerta agilis embryos (Stages 21 and 24) were separated by SDS-PAGE and stained by modified Silver Staining (Shevchenko et al. 1996). Stained portion of the gel were cut out (black box). The size of cutout bands corresponded to the size of bands detected in Western blot technique (see Fig. 5A). Samples of proteins were sent for the identification by an LC-MS method in the Mass Spectrometry Laboratory, IBB PAS and searched with MASCOT (Matrix Science). B. Protein sequence alignment of Xenopus laevis Lbx1 (Lbx1; accession number NP_001089192.1), epitope (Ep) recognized by commercially available antibody (mouse monoclonal anti-Lbx2; Abcam), identified peptide 1 (P1), identified peptide 2 (P2), and identified peptide 3 (P3). The comparison identified peptides with the NCBI protein sequences database was carried out using MASCOT program. Obtained results revealed the presence of three peptides corresponding to regions present in X. laevis Lbx1 amin acid sequence. Sequence alignment: ClustalW2, http://www.ebi.ac.uk (PNG 1377 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lewandowski, D., Dubińska-Magiera, M., Garbiec, A. et al. Primary myogenesis in the sand lizard (Lacerta agilis) limb bud. Dev Genes Evol 229, 147–159 (2019). https://doi.org/10.1007/s00427-019-00635-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-019-00635-7