Abstract
Main conclusion
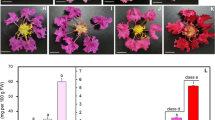
SmANS deletion leads to white flower mutation in Salvia miltiorrhiza.
Abstract
SmANS deletion leads to white flower mutation in Salvia miltiorrhiza. Abstract Salvia miltiorrhiza is an essential traditional Chinese medicine (TCM) with purple flowers, and S. miltiorrhiza Bge. f. alba is a unique intraspecific variation with white flowers. The molecular mechanism of flower color formation in S. miltiorrhiza will provide vital information for the variation and evolution. Here, we performed HPLC, transcriptomic, and re-sequencing analyses of purple-flowered S. miltiorrhiza line ‘Zihua105’ (ZH105) and white-flowered S. miltiorrhiza Bge. f. alba line ‘Baihua18’ (BH18). Delphinidin was the most anthocyanidin in ZH105, which become the main different between ZH105 vs. BH18 flowers. Transcriptome analysis revealed 299 differentially expressed genes (DEGs). SmANS, the anthocyanidin synthase gene in the down-stream anthocyanin biosynthesis pathway, was significantly expressed in ZH105 corollas, suggesting it might play a key role in white petal formation. Whole-genome re-sequencing revealed that a 6.75 kb segment located on chromosome 5, which contains the complete sequence of the SmANS genes, was lost in BH18 and another S. miltiorrhiza Bge. f. alba line. In contrast, the other five purple-flowered S. miltiorrhiza lines both possessed this segment. Further molecular marker identification also confirmed that wild S. miltiorrhiza Bge. f. alba lines lost regions that contained a complete or important part of SmANS sequences. Subsequently, the research showed that the deletion mutant of SmANS genes resulted in the natural white flower color variant of S. miltiorrhiza.






Similar content being viewed by others
Data availability
The RNA-seq data of ZH105 and BH18 in the GenomeWarehouse in BIG Data Center under Project numbers PRJCA****. The re-sequencing data of six lines were also available under Project numbers PRJCA****, which are accessible at https://bigd.big.ac.cn/gwh.
Abbreviations
- ANS:
-
Anthocyanidin synthase
- DEGs:
-
Differentially expressed genes
- DFR:
-
Dihydroflavonol 4-reductase
References
Aizza LCB, Sawaya CHFSA, Dornelas MC (2019) Identification of anthocyanins in the corona of two species of Passiflora and their hybrid by ultra-high performance chromatography with electrospray ionization tandem mass spectrometry (UHPLC–ESI–MS/MS). Biochem Syst Ecol 85:60–67. https://doi.org/10.1016/j.bse.2019.05.003
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol. 11: 106. http://genomebiology.com/2010/11/10/R106.
Ben-Simhon Z, Judeinstein S, Trainin T, Harel-Beja R, Bar-Ya’akov I, Borochov-Neori H, Holland D (2015) A “white” anthocyanin-less pomegranate (Punica granatum L.) caused by an insertion in the coding region of the leucoanthocyanidin dioxygenase (LDOX; ANS) gene. PLoS ONE 10(11):e0142777. https://doi.org/10.1371/journal.pone.0142777
Britten RJ, Rowen L, Williams J, Cameron A (2003) Majority of divergence between closely related DNA samples is due to Indels. Proc Natl Acad Sci USA 100(8):4661–4665. https://doi.org/10.1073/pnas.0330964100
Ghosh S, Chan CK (2016) Analysis of RNA-seq data using Tophat and Cufflinks. Methods Mol Biol 1374:339–361. https://doi.org/10.1007/978-1-4939-3167-5_18
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083. https://doi.org/10.1105/tpc.7.7.1071
Jia Y, Selva C, Zhang YJ, Li B, McFawn LA, Broughton S, Zhang XQ, Westcott S, Wang PH, Tan C, Angessa T, Xu YH, Whitford R, Li CD (2020) Uncovering the evolutionary origin of blue anthocyanins in cereal grains. Plant J 101(5):1057–1074. https://doi.org/10.1111/tpj.14557
Jiang T, Zhang M, Wen C, Xie X, Tian W, Wen S, Liu L (2020) Integrated metabolomics and transcriptomic analysis of the anthocyanin regulatory networks in Salvia miltiorrhiza Bge flowers. BMC Plant Biol 20:349
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242. https://doi.org/10.1016/j.tplants.2005.03.002
Kotepong P, Ketsa S, Van Doorn WG (2011) A white mutant of Malay apple fruit (Syzygium malaccense) lacks transcript expression and activity for the last enzyme of anthocyanin synthesis, and the normal expression of a MYB transcription factor. Func Plant Biol 38:75–86. https://doi.org/10.1071/FP10164
Kubo H, Nawa N, Lupsea SA (2007) Anthocyaninless1 gene of Arabidopsis thaliana encodes a UDP-glucose: flavonoid-3-O-glucosyltransferase. J Plant Res 120:445–449. https://doi.org/10.1007/s10265-006-0067-7
Larsen ES, Alfenito MR, Briggs WR, Walbot V (2003) A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and petunia An9. Plant Cell Rep 21:900–904. https://doi.org/10.1007/s00299-002-0545-x
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li JT, Dong JE, Liang ZS, Shu ZM, Yuan GW (2008) Distributional differences of fat-soluble compounds in the roots, stems and leaves of four Salvia plants. J Mol Cell Biol 41:44–52
Li N, Song Y, Li J, Hao R, Feng X, Li L (2021) Resequencing and transcriptomic analysis reveal differences in nitrite reductase in jujube fruit (Ziziphus jujube Mill.). Plant Methods 17:75. https://doi.org/10.1186/s13007-021-00776-9
Li J, Sun X, Zhou F, Li J, Wang S (1995) New resources of Salvia miltiorrhiza in Shandong Province. J Shandong Coll Tradit Chin Med 19(3):190–191
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) 1000 Genome project data processing subgroup the sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Li YL, Xin XM, Chang ZM, Shi RJ, Miao ZM, Ding J, Hao GP (2015) The endophytic fungi of Salvia miltiorrhiza Bge.f. alba are a potential source of natural antioxidants. Bot Stud 56:5. https://doi.org/10.1186/s40529-015-0086-6
Li H, Liu J, Pei T, Bai Z, Han R, Liang Z (2019) Overexpression of SmANS enhances anthocyanin accumulation and alters phenolic acids content in Salvia miltiorrhiza and Salvia miltiorrhiza Bge f. alba plantlets. Int J Mol Sci 20:2225. https://doi.org/10.3390/ijms20092225
Lao F, Giusti MM (2016) Quantification of Purple Corn (Zea mays L.) Anthocyanins using spectrophotometric and HPLC approaches: method comparison and correlation. Food Anal. Methods 9:1367–138. https://doi.org/10.1007/s12161-015-0318-0
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.2307/1592215
Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J 1:37–49. https://doi.org/10.1111/j.1365-313X.1991.00037.x
Nakatsuka T, Saito M, Sato-Ushiku Y, Yamada E, Nakasato T, Hoshi N, Fujiwara K (2012) Hikage Development of DNA markers that discriminate between white-and blue-flowers in Japanese gentian plants. EUPHYTICA 184(3):335–344
Pedersen BS, Quinlan AR (2018) Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34(5):867–868. https://doi.org/10.1093/bioinformatics/btx699
Peng YY, Wang KL, Cooney JM, Wang TC, Espley RV, Allan AC (2019) Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic Res 6:3. https://doi.org/10.1038/s41438-018-0076-4
Rafique MZ, Elisabete C, Ralf S, Luisa P, Lorena H, Antje F, Malnoy M, Martens S (2016) Nonsense mutation inside anthocyanidin synthase gene controls pigmentation in yellow raspberry (Rubus idaeus L.). Front Plant Sci 19(7):1892. https://doi.org/10.3389/fpls.2016.01892
Ran R, Zhou FQ, Wang T, Kong QY (2008) Comparison of tanshinone IIA in radix Salvia miltiorrhiza from Shandong. J Chin Med Mater 31(3):331–333
Roberts WR, Roalson EH (2017) Comparative transcriptome analyses of flower development in four species of Achimenes (Gesneriaceae). BMC Genomics 18:240. https://doi.org/10.1186/s12864-017-3623-8
Shang D, Zhao X (1986) Treatment of thromboangiitis obliterans by activating blood circulation and removing blood stasis - Clinical study of 144 cases. Shandong J Tradit Chin Med 2:14–15
Shimizu K, Ohnishi N, Morikawa N, Ishigami A, Otake S, Rabah IO, Sakata Y, Hashimoto F (2011) A 94-bp deletion of anthocyanidin synthase gene in acyanic flower lines of lisianthus [Eustoma grandiflorum (Raf.) Shinn.]. J Jpn Soc Hortic Sci 80(4):434–442. https://doi.org/10.2503/jjshs1.80.434
Shin DH, Choi MG, Kang CS, Park CS, Choi SB, Park YI (2016) Overexpressing the wheat dihydroflavonol 4-reductase gene TaDFR increases anthocyanin accumulation in an Arabidopsis dfr mutant. Genes Genom 38:333–340. https://doi.org/10.1007/s13258-015-0373-3
Song Z, Li X (2015) Expression profiles of rosmarinic acid biosynthesis genes in two Salvia miltiorrhiza lines with differing water-soluble phenolic contents. Ind Crops Prod 71:24–30. https://doi.org/10.1016/j.indcrop.2015.03.081
Song JY, Luo HM, Li CF, Sun C, Xu J, Chen SL (2013) Salvia miltiorrhiza as medicinal model plant. Acta Pharm Sin 48:1099–1106
Song Z, Wang J, Li X (2016) Expression profiles of genes involved in tanshinone biosynthesis of two Salvia miltiorrhiza genotypes with different tanshinone contents. J Genet 95:433–439. https://doi.org/10.1007/s12041-016-0621-6
Song Z, Lin C, Xing P, Fen Y, Jin H, Zhou C, Gu YQ, Wang J, Li X (2020) A high-quality reference genome sequence of Salvia miltiorrhiza provides insights into tanshinone synthesis in its red rhizomes. Plant Genome 13:e20041. https://doi.org/10.1002/tpg2.20041
Wu ZY, Li XW (1977) Flora of China, vol 66. Science Press, Beijing, pp 145–150. https://doi.org/10.3100/1043-4534-13.2.301
Yang YF, Hou S, Cui GH, Chen SL, Wei JH, Huang LQ (2010) Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol Biol Rep 37:507–513. https://doi.org/10.1007/s11033-009-9703-3
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:14. https://doi.org/10.1186/gb-2010-11-2-r14
Zhou XW, Li JY, Zhu YL, Ni S, Chen LJ, Feng XJ, Zhang YF, Li SQ, Zhu HG, Wen YG (2017) De novo assembly of the Camellia nitidissima transcriptome reveals key genes of flower pigment biosynthesis. Front Plant Sci 8:1545. https://doi.org/10.3389/fpls.2017.01545
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81872949), the Natural Science Foundation of Shandong province (ZR2019HM081), and Agricultural seed improvement project of Shandong Province (2021LZGC008).
Funding
National Natural Science Foundation of China, 81872949, Zhenqiao Song,Natural Science Foundation of Shandong Province,ZR2019
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no conflicts of interest in this work and have nothing to disclose.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lin, C., Xing, P., Jin, H. et al. Loss of anthocyanidin synthase gene is associated with white flowers of Salvia miltiorrhiza Bge. f. alba, a natural variant of S. miltiorrhiza. Planta 256, 15 (2022). https://doi.org/10.1007/s00425-022-03921-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03921-3