Abstract
Main conclusion
Panax notoginseng PnMYB2 is a transcriptional activator of primary and secondary cell wall formation by promoting the PCW-specific gene CesA3 and key lignin biosynthetic gene CCoAOMT1 , respectively.
Abstract
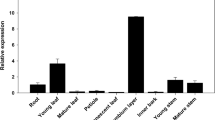
R2R3-MYB transcription factors play important roles in regulation secondary cell wall (SCW) formation. However, there are few reports on the functions of MYB transcription factors which involved in both primary cell wall (PCW) and SCW formation. Here, we isolated an R2R3-MYB transcription factor, PnMYB2, from Panax notoginseng roots which are widely used in Chinese traditional medicines and contain abundant cellulose and lignin. The expression pattern of PnMYB2 was similar to the accumulation pattern of cellulose and lignin contents in different organs. PnMYB2 localized in the nucleus and may function as a transcriptional activator. Overexpression of PnMYB2 in Arabidopsis thaliana enhanced cellulose and lignin biosynthesis, and remarkably increased thickness of PCW and SCW in the stem of transgenic plants compared with wild-type plants. The expression levels of genes associated with PCW-specific cellulose synthase (CesA) genes and key SCW-specific lignin biosynthetic genes were significantly increased in PnMYB2-overexpressing plants compared to the wild type plants. Furthermore, yeast one-hybrid, dual-luciferase reporter assays and electrophoretic mobility shift assays (EMSA) results verified that PnMYB2 could bind and activate the promoters of AtCesA3 and PnCesA3, which are the PCW-specific cellulose biosynthetic genes, and AtCCoAOMT1 and PnCCoAOMT1, which are the key lignin biosynthetic genes. These results demonstrated the central role of PnMYB2 in PCW-specific cellulose formation and SCW-specific lignin biosynthesis.







Similar content being viewed by others
Data availability
All data analyzed or generated in our study are included in this article and its supplementary information flies.
Abbreviations
- 4CL:
-
4-Coumarate-CoA ligase
- C3H:
-
P-coumarate 3-hydroxylase
- CCoAOMT1:
-
Caffeoyl-CoA 3-O-methyltransferase 1
- CesA:
-
Cellulose synthase
- COMT:
-
Caffeic acid O-methyltransferase
- EMSA:
-
Electrophoretic mobility shift assay
- HCT:
-
Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase
- PCW:
-
Primary cell wall
- SCW:
-
Secondary cell wall
References
Barrière Y, Courtial A, Soler M, Pettenati J (2015) Toward the identification genes underlying maize QTLs for lignin content, focusing on colocalizations with lignin biosynthetic genes and their regulatory MYB and NAC transcription factors. Mol Breeding 35:87–110. https://doi.org/10.1007/s11032-015-0275-8
Bo J, Zhao X, Lu W, Li G, Luo K (2019) The R2R3 MYB transcription factor MYB189 negatively regulates secondary cell wall biosynthesis in Populus. Tree Physiol 39:1187–1200. https://doi.org/10.1093/treephys/tpz040
Bomal C, Duval I, Giguère I, Fortin E, Caron S, Stewart D, Boyle B, Séguin A, MacKay JJ (2014) Opposite action of R2R3-MYBs from different subgroups on key genes of the shikimate and monolignol pathways in spruce. J Exp Bot 65:495–508. https://doi.org/10.1093/jxb/ert398
Bonawitz N, Kim J, Tobimatsu Y, Ciesielski P, Anderson N, Ximenes E, Maeda J, Ralph J, Donohoe B, Ladisch M, Chapple C (2014) Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509:376–380. https://doi.org/10.1038/nature13084
Endler A, Schneider R, Kesten C, Lampugnani E, Persson S (2016) The cellulose synthase companion proteins act non-redundantly with cellulose synthase interacting1/POM2 and cellulose synthase 6. Plant Signal Behav 11:4. https://doi.org/10.1080/15592324.2015.1135281
Fang L, Xu X, Li J, Zheng F, Li M, Yan J, Li Y, Zhang X, Li L, Ma G, Zhang A, Lv F, Wu K (2020) Transcriptome analysis provides insights into the non-methylated lignin synthesis in Paphiopedilum armeniacum seed. BMC Genomics 21:524–537. https://doi.org/10.1186/s12864-020-06931-1
Fornalé S, Sonbol F, Capellades M, Encina A, Tourino S, Fuguet E, Torres J, Rovira P, Ruel K, Puigdomenech P, Rigau J, Caparrós D (2009) ZmMYB31 & ZmMYB42: two maize R2R3-MYB transcription factors having complementary roles in the lignin and phenylpropanoid metabolism regulation. New Biotechnol 25:279–280. https://doi.org/10.1016/j.nbt.2009.06.632
Geng P, Zhang S, Liu J, Zhao C, Wu J, Cao Y, Fu C, Han X, He H, Zhao Q (2019) MYB20, MYB42, MYB43 and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol 182:1273–1283. https://doi.org/10.1104/pp.19.01070
Hamann T, Osborne E, Youngs H, Misson J, Nussaume L, Somerville C (2004) Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11:279–286. https://doi.org/10.1186/1471-2229-10-282
Hao Y, Song Q, Chen H, Zou F, Wei W, Kang X, Ma B, Zhang W, Zhang J, Chen S (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232:1033–1043. https://doi.org/10.1007/s00425-010-1238-2
Huang J, Chen F, Guo Y, Gan X, Yang M, Zeng W, Persson S, Li J, Xu W (2021) GhMYB7 promotes secondary wall cellulose deposition in cotton fibers by regulating GhCesA gene expression through three distinct cis-elements. New Phytol 232:1718–1737. https://doi.org/10.1111/nph.17612
Legay S, Sivadon P, Blervacq A, Pavy N, Baghdady A, Tremblay L, Levasseur C, Ladouce N, Lapierre C, Séguin A, Hawkins S, Mackay J, Pettenati J (2010) EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol 188:774–786. https://doi.org/10.1111/j.1469-8137.2010.03432.x
Li C, Wang X, Ran L, Tian Q, Fan D, Luo K (2015) PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant Cell Physiol 56:2436–2446. https://doi.org/10.1093/pcp/pcv157
Li C, Ma X, Yu H, Fu Y, Luo K (2018) Ectopic expression of PtoMYB74 in Poplar and Arabidopsis promotes secondary cell wall formation. Front Plant Sci 9:1262–1275. https://doi.org/10.3389/fpls.2018.01262
Li L, He Y, Zhang Z, Shi Y, Zhang X, Xu X, Wu J, Tang S (2021) OsNAC109 regulates senescence, growth and development by altering the expression of senescence- and phytohormone-associated genes in rice. Plant Mol Biol 105:637–654. https://doi.org/10.1007/s11103-021-01118-y
Liao P, Shi Y, Li Z, Chen Q, Xu T, Cui X, Guan H, Guo L, Yang Y (2019) Impaired terpenoid backbone biosynthesis reduces saponin accumulation in Panax notoginseng under Cd stress. Funct Plant Biol 46:56–68. https://doi.org/10.1071/FP18003
Liu Y (2010) Investigation of a KNAT7-BLH-OFP transcription factor complex involved in regulation of secondary cell wall biosynthesis in Arabidopsis thaliana. Acta Agron Sin 1:40–51. https://doi.org/10.1007/s00249-010-0592-0
Michelle S, Michele H, Haowen X, David H, Ramón O, Arthur S, Lawrence S (2013) Enzymatic saccharification of shrub willow genotypes with differing biomass composition for biofuel production. Front Plant Sci 4:57–65. https://doi.org/10.3389/fpls.2013.00057
Ng T (2010) Pharmacological activity of sanchi ginseng (Panax notoginseng). J Pharm Pharmacol 58:1007–1019. https://doi.org/10.1211/jpp.58.8.0001
Noda S, Yamaguchi T, Hattor T, Yamaguchi M, Suzuki S, Umezawa T (2015) The expression of a rice secondary wall-specific cellulose synthase gene, OsCesA7, is directly regulated by a rice transcription factor, OsMYB58/63. Planta 243:589–600. https://doi.org/10.1007/s00425-015-2343-z
Preston J, Wheeler J, Heazlewood J, Li S, Parish R (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40:975–995. https://doi.org/10.1111/j.1365-313X.2004.02280.x
Rashmi S, Voesenek V, Ronald P (2011) Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses. Crit Rev Plant Sci 30:548–562. https://doi.org/10.1080/07352689.2011.615706
Ray T, Xiao H, Ingles J, Greenblatt J (1993) Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J Biol Chem 268:2284–2287. https://doi.org/10.1016/S0021-9258(18)53769-8
Riazunnisa K, Padmavathi L, Scheibe R, Raghavendra S (2007) Preparation of Arabidopsis mesophyll protoplasts with high rates of photosynthesis. Physiol Plant 129:679–686. https://doi.org/10.1111/j.1399-3054.2007.00867.x
Royo JN, Gómez E, Barrero C, Muñiz LM, Sanz Y, Hueros G (2009) Transcriptional activation of the maize endosperm transfer cell-specific gene BETL1 by ZmMRP-1 is enhanced by two C2H2 zinc finger-containing proteins. Planta 230:807–818. https://doi.org/10.1007/s00425-009-0987-2
Sena J, Lachance D, Duval I, Nguyen T, Séguin A (2019) Functional analysis of the PgCesA3 white spruce cellulose synthase gene promoter in secondary xylem. Front Plant Sci 10:626–639. https://doi.org/10.3389/fpls.2019.00626
Shi Y, Pu R, Guo L, Man J, Shang B, Ou X, Dai C, Liu P, Cui X, Ye Y (2020) Formula fertilization of nitrogen and potassium fertilizers reduces cadmium accumulation in Panax notoginseng. Arch Agron Soil Sci 66:343–357. https://doi.org/10.1080/03650340.2019.1616176
Shi J, Zhang Q, Yan X, Zhang D, Zhou Q, Shen Y, Anupol N, Wang X, Bao M, Larkin R, Luo H, Ning G (2021) A conservative pathway for coordination of cell wall biosynthesis and cell cycle progression in plants. Plant J 106:630–648. https://doi.org/10.1111/tpj.15187
Takenaka Y, Watanabe Y, Schuetz M, Unda F, Hill JL, Phookaew P, Yoneda A, Mansfield SD, Samuels L, Ohtani M, Demuraa T (2018) Patterned deposition of xylan and lignin is independent from the secondary wall cellulose of Arabidopsis xylem vessels. Plant Cell 30:2663–2676. https://doi.org/10.1105/tpc.18.00292
Tian Q, Wang X, Li C, Lu W, Yang L, Jiang Y, Luo K, Wu K (2013) Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS One 8:76369–76379. https://doi.org/10.1371/journal.pone.0076369
Wei K, Zhao Y, Zhou H, Jiang C, Zhang B, Zhou Y, Song X, Lu M (2019) PagMYB216 is involved in the regulation of cellulose synthesis in Populus. Mol Breeding 39:39–65. https://doi.org/10.1007/s11032-019-0970-y
Xi W, Song D, Sun J, Shen J, Li L (2017) Formation of wood secondary cell wall may involve two type cellulose synthase complexes in Populus. Plant Mol Biol 93:419–429. https://doi.org/10.1007/s11103-016-0570-8
Yang L, Zhao X, Ran L, Li C, Fan D, Luo K (2017) PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci Rep 24:41209–44121. https://doi.org/10.1038/srep41209
Yang Q, Li J, Sun J, Cui X (2022) Comparative transcriptomic and proteomic analyses to determine the lignin synthesis pathway involved in the fungal stress response in Panax notoginseng. Physiol Mol Plant P 119: 101814. https://doi.org/10.21203/rs.3.rs-361945/v1
Zhang J, Ge H, Zang C, Li X, Grierson D, Chen K, Yin X (2016) EjODO1, a MYB transcription factor, regulating lignin biosynthesis in developing loquat (Eriobotrya japonica) fruit. Front Plant Sci 7:1360–1369. https://doi.org/10.3389/fpls.2016.01360
Zhong R, Mccarthy R, Lee C, Ye Z (2011) Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 157:1452–1468. https://doi.org/10.1104/pp.111.181354
Zhou J, Lee C, Zhong R, Ye Z (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21:248–266. https://doi.org/10.1105/tpc.108.063321
Acknowledgements
This work was supported by Beijing Science and Technology Planning Project (Z201100005420005).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Human participants and/or animals
Research did not involve human and /or animal subjects.
Informed consent
All authors have read and approved submission of this work.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2022_3891_MOESM4_ESM.pptx
Supplementary file4 (PPTX 17006 KB) Expression level of PnMYB2 and thickness of cell wall in PnMYB2 overexpression plants and wild-type plants. a The expression level of PnMYB2 in WT and PnMYB2 OE plants by RT-PCR analysis. b The expression levels of PnMYB2 in WT and PnMYB2 OE plants by qRT-PCR analysis. c PCW and SCW thickness of xylem fiber in vascular bundle in the WT and PnMYB2 OE plants. d Thickness of SCW in the WT and PnMYB2 OE plants. e Vessel cell wall thickness in the WT and PnMYB2 OE plants. Statistical analysis was performed with one-way analyses of variance (ANOVA) with Duncan’s multiple range tests to separate means. Error bars represent SD of three replicates; significance **P < 0.01, *P < 0.05
425_2022_3891_MOESM5_ESM.pptx
Supplementary file5 (PPTX 10631 KB) Functional analysis of different expression gene and identification of MYB transcription factors involved in cell wall biosynthesis. a KEGG pathway analysis of different expression gene from WT and PnMYB2 OE plants. b GO functional categories of different expression gene from WT and PnMYB2 OE plants. c Heatmap of AtMYB20/42/43/85 from the WT and PnMYB2 OE plants. d Expression levels of AtMYB20/42/43/85 in WT and PnMYB2 OE plant stems. Statistical analysis was performed with one-way analyses of variance (ANOVA) with Duncan’s multiple range tests to separate means. Error bars represent SD of three replicates; significance **P < 0.01, *P < 0.05
425_2022_3891_MOESM6_ESM.pptx
Supplementary file6 (PPTX 22380 KB) PnCesA3 sequence analysis. a Multiple sequence alignment of PnCesA3 and AtCesA3. The alignment was made with DNAMAN program. Black shading indicates amino acid identities, blue shading indicate amino acids with different similarities. b Phylogenetic analysis of PnCesA3 and the other CesA protein from A. thaliana. Multiple sequence alignments of CesA sequences were performed using ClustalW, and the phylogenetic tree was constructed using MEGA6 with the neighbor-joining (NJ) method and 1000 bootstrap replicates
425_2022_3891_MOESM7_ESM.pptx
Supplementary file7 (PPTX 23984 KB) PnCCoAOMT1 sequence analysis. a Multiple sequence alignment of PnCCoAOMT1 and AtCCoAOMT1 protein. The alignment was made with DNAMAN program. Black shading indicates amino acid identities, blue shading indicate amino acids with different similarities. The red line indicates conserved site. b Phylogenetic analysis of PnCCoAOMT1 and the other CCoAOMT from A. thaliana or Nicotiana tabacum. Multiple sequence alignments of CCoAOMT sequences were performed using ClustalW, and the phylogenetic tree was constructed using MEGA6 with the neighbor-joining (NJ) method and 1000 bootstrap replicates
Rights and permissions
About this article
Cite this article
Shi, Y., Man, J., Huang, Y. et al. Overexpression of PnMYB2 from Panax notoginseng induces cellulose and lignin biosynthesis during cell wall formation. Planta 255, 107 (2022). https://doi.org/10.1007/s00425-022-03891-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-03891-6