Abstract
Main conclusion
Developmental and organ-specific expression of genes in dhurrin biosynthesis, bio-activation, and recycling offers dynamic metabolic responses optimizing growth and defence responses in Sorghum.
Abstract
Plant defence models evaluate the costs and benefits of resource investments at different stages in the life cycle. Poor understanding of the molecular regulation of defence deployment and remobilization hampers accuracy of the predictions. Cyanogenic glucosides, such as dhurrin are phytoanticipins that release hydrogen cyanide upon bio-activation. In this study, RNA-seq was used to investigate the expression of genes involved in the biosynthesis, bio-activation and recycling of dhurrin in Sorghum bicolor. Genes involved in dhurrin biosynthesis were highly expressed in all young developing vegetative tissues (leaves, leaf sheath, roots, stems), tiller buds and imbibing seeds and showed gene specific peaks of expression in leaves during diel cycles. Genes involved in dhurrin bio-activation were expressed early in organ development with organ-specific expression patterns. Genes involved in recycling were expressed at similar levels in the different organ during development, although post-floral initiation when nutrients are remobilized for grain filling, expression of GSTL1 decreased > tenfold in leaves and NITB2 increased > tenfold in stems. Results are consistent with the establishment of a pre-emptive defence in young tissues and regulated recycling related to organ senescence and increased demand for nitrogen during grain filling. This detailed characterization of the transcriptional regulation of dhurrin biosynthesis, bioactivation and remobilization genes during organ and plant development will aid elucidation of gene regulatory networks and signalling pathways that modulate gene expression and dhurrin levels. In-depth knowledge of dhurrin metabolism could improve the yield, nitrogen use efficiency and stress resilience of Sorghum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ontogenetic trajectories in defence have been described in hundreds of species of plants. Typically, defence compound levels are high in young plants, with overall concentration decreasing as plants mature (e.g. Gleadow and Woodrow 2002). Such changes may be adaptive, providing protection for the meristems and newly formed organs. When evaluating possible trade-offs between costs and benefits of investment in biotic defences, there are a number of other considerations that could moderate and/or off-set the cost of production (Neilson et al. 2013; Gleadow and Møller 2014; Blomstedt et al. 2018): (1) Are there developmental stage dependent metabolic constraints? (2) Are light or mineral nutrients limiting? (3) How effective are the defence compounds produced in deterring herbivores? and (4) Could these compounds have other potentially beneficial functions? Allocation models of investment provide mechanistic explanations of why costs of defence change as plants mature (Boege and Marquis 2005) and under different environmental conditions (Gleadow et al. 1998) but do not address the underlying molecular regulatory mechanisms.
Cyanogenic glucosides are present in over 2500 plant species and are known to be effective in herbivore defence (Tattersall et al. 2001a; Gleadow and Woodrow 2002; Ballhorn et al. 2006). Cyanogenic glucosides are classified as phytoanticipins, i.e. the defensive agent is released when required from a constitutive progenitor. In this case, a hydrogen cyanide ‘bomb’ is detonated when the cells in which they are stored are disrupted, e.g. by mastication, mixing the components with specific hydrolyzing enzymes (Gleadow and Møller 2014). As with many chemical defence systems, the costs of deployment in terms of a growth sacrifice are small, and usually only detectable under resource-limiting conditions, if at all (Blomstedt et al. 2018; Simon et al. 2010; Sohail et al. 2020; Gershenzon 1989). This lack of apparent cost could be because the costs of production are tiny (Gershenzon 1989) or off-set by other metabolic or physiological benefits (Neilson et al. 2013).
Dhurrin, the cyanogenic glucoside present in Sorghum bicolor, has been widely studied and established as a model for testing defence theories (Miller et al. 2014; Blomstedt et al. 2018; Gleadow et al. 2016). While the biosynthetic pathway is well characterised (Halkier and Møller 1989; Kahn et al. 1997; Møller and Conn 1979), little is known about the transcriptional regulation of genes in the pathway (Busk and Møller 2002). The concentration of dhurrin in sorghum is dependent on the genotype (Emendack et al. 2018; Hayes et al. 2015; Burke et al. 2013), tissue type (O’Donnell et al. 2013), plant age (Blomstedt et al. 2018), stage of development (Miller et al. 2014) and growing conditions (O’Donnell et al. 2013; Gleadow et al. 2016; Rosati et al. 2019). Information on pathway gene expression in organs, tissues and cells during development and under different environmental conditions is needed to provide information for gene regulatory network analysis and to elucidate the signalling pathways that regulate expression of the genes involved in dhurrin synthesis, transport, hydrolysis and re-mobilization.
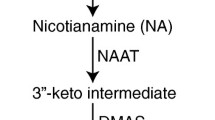
Dhurrin ((S)-p-hydroxymandelonitrile-β-D-glucopyranoside) is biosynthesized from tyrosine in a process catalysed by the sequential action of two cytochrome P450s (CYP79A1 and CYP71E1) and a UDP-glucosyltransferase (UGT85B1) with the NADPH-dependent cytochrome P450 oxidoreductase (POR) serving as an electron donor to the P450s (Fig. 1) (Kahn et al. 1997; Bak et al. 1998; Møller and Conn 1979; Jones et al. 1999). These enzymes form a metabolon which facilitates channelled conversion of the intermediates into dhurrin formation and prevents release of toxic intermediates and undesired metabolic cross-talk (Jørgensen et al. 2005; Kristensen et al. 2005; Nielsen et al. 2008; Laursen et al. 2015). The genes encoding the enzymes in this metabolon (except POR) are clustered on chromosome 1 together with SbMATE2 and a glutathione S-transferase (GST) (Darbani et al. 2016; Hayes et al. 2016; Takos et al. 2011). Genes involved in dhurrin biosynthesis located in the cluster are co-expressed and protein production is regulated in part at the transcriptional level (Takos and Rook 2012; Darbani et al. 2016; Busk and Møller 2002). In S. bicolor, dhurrin synthesis is highly active in young leaf and stem tissues during vegetative plant development (Busk and Møller 2002).
Overview of the genes involved in dhurrin biosynthesis, recycling, transport, bioactivation and in prevention of auto-toxicity. a Dhurrin is synthesised from tyrosine by the sequential action of three key enzymes. b Bioactivation of dhurrin to release a hydrogen cyanide bomb occurs upon tissue disruption by the action of specific β-glucosidases (DHR) and an α-hydroxynitrile lyase (HNL). c Auto-toxicity is prevented by β-cyanoalanine synthase and nitrilases retrieving the nitrogen of the hydrogen cyanide for production of amino acids and ammonia. d Endogenous recycling pathway mediated by glutathione transferases and a heteromeric NIT4A/NIT4B2 nitrilase resulting in formation of ammonia and p-hydroxyphenylacetic acid without the release of HCN. e Dhurrin synthesis occurs in the cytosol and two potential genes, SbMATE2 and SbCGTR1, may be involved in dhurrin transport to sites of storage within the cell or to other parts of the plant. Abbreviations: Asn asparagine; Asp aspartic acid; DHR dhurrinase; NH3, ammonia
Dhurrin may also be recycled in a glutathione transferase-catalysed process proceeding without concomitant release of HCN (Pičmanová et al. 2015; Bjarnholt et al. 2018; Jenrich et al. 2007). In the recycling process, the glucose moiety of dhurrin is replaced by glutathione, either spontaneously or potentially by a glutathione transferase (e.g. GST1 within the biosynthesis cluster). The glutathione conjugate is subsequently cleaved by GSTs of the lambda class (GSTL1/2) resulting in the production of p-hydroxyphenylacetonitrile, which through the action of the NIT4A/NIT4B2 heterodimer is converted into p-hydroxyphenylacetic acid and ammonium. The possibility of recycling without the release of toxic intermediates increases the likelihood that dhurrin could have other roles, depending on stage of plant development and environmental conditions (Gleadow and Møller 2014; Bjarnholt et al. 2018).
The sub-cellular storage site of dhurrin has previously been reported to be the vacuole (Saunders and Conn 1978) but more recent data points to the cytosol as the storage site, maybe in the form of a biocondensate (Møller and Laursen 2021; Heraud et al. 2018; Knudsen et al. 2018, 2020). Proper storage of dhurrin may involve transporters. In this context, dhurrin may be transported to the vacuole or other vesicle enclosed compartments by SbMATE2, a transporter of the multidrug and toxic compound extrusion (MATE) family (Darbani et al. 2016). In addition, a member of the nitrate and peptide family (NPF) of transporters has been identified in cassava, another well-studied cyanogenic plant, to exhibit high substrate specificity for linamarin, the main cyanogenic glucoside in that species [MeCGTR1; (Jørgensen et al. 2017)].
Upon cell destruction and exerting its function as part of a two-component defence system, dhurrin is hydrolysed and the hydrogen cyanide is rapidly released with co-production of stoichiometric amounts of p-hydroxybenzaldehyde. This process is catalysed by specific β—glucosidases (dhurrinases) and α-hydroxynitrilases (HNLs). Two dhurrinases (DHR1 and DHR2) have been identified that exhibit tissue-specific expression (Hösel et al. 1987; Cicek and Esen 1998). Sequence analysis has identified two additional dhurrinase-like genes and all four DHR genes are co-located (within ~ 630 kb) on chromosome 8 (Hayes et al. 2015, 2016; Krothapalli et al. 2013). The prevalence of two or more differentially expressed β-glucosidase isoenzymes in most plants suggests that the two-component defence system exemplified by cyanogenic glucosides is spatially and/or temporally regulated (Morant et al. 2008b). The α-hydroxynitrile intermediate formed is converted to hydrogen cyanide and p-hydroxybenzaldehyde via the action of α-hydroxynitrile lyase (HNL), although at pH > 6, the dissociation of the α-hydroxynitrile intermediate also proceeds non-enzymatically (Poulton 1990). To avoid auto-toxicity, the released hydrogen cyanide can be detoxified and reincorporated into primary metabolism in a process mediated by β-cyanoalanine synthase (CAS) and the nitrilases NIT4A, NIT4B1 or NIT4B2 (Fig. 1).
Sorghum bicolor (L.) Moench is one of the world’s leading cereal crops and a valuable forage crop, particularly in the semi-arid tropics. Photoperiod-sensitive sorghum is also a good source of biomass for bioenergy production due to the high productivity of this C4 grass and the crop’s drought tolerance, allowing production on marginal annual cropland. Bioenergy sorghum hybrids have been developed and under field conditions have shown good greenhouse gas mitigation potential (Olson et al. 2012; Rooney et al. 2007; Gill et al. 2014; Mullet et al. 2014; McKinley et al. 2018b). Improvements in biotic stress resilience and nitrogen use efficiency would benefit all types of sorghum crops.
The availability of sequenced and annotated sorghum genomes (Paterson et al. 2009; Mace et al. 2013; McKinley et al. 2018a; Cooper et al. 2019) presents new opportunities for improving S. bicolor for grain, forage and bioenergy production (McCormick et al. 2018; Kebrom et al. 2017, 2020; McKinley et al. 2018a, 2018b). In this study, transcriptome analysis aided by RNA-seq was used to compare the expression of the key genes involved in sorghum dhurrin biosynthesis, bio-activation, recycling, and transport in the juvenile and adult-phase vegetative tissues and from floral initiation until anthesis. Diel expression of genes in the dhurrin pathway was characterized over two light and one dark cycle in leaves of adult-phase vegetative plants.
Materials and methods
Plant material, growing conditions and sampling for RNA-seq study
RNA-seq data were derived primarily from S. bicolor BTx623, an important public breeding line and the genotype used for the first sorghum reference genome sequence (McCormick et al. 2018; Paterson et al. 2009). Tissue and development-dependent analyses of RNA-seq profiles related to dhurrin biosynthesis, bio-activation and detoxification, recycling and transport were carried out using the methodologies described in McCormick et al. (2018). For the diel experiment, BTx623 plants were grown in a growth chamber (300 μmol, 30 °C day/23 °C night, 50% relative humidity) and entrained to a 14 h photoperiod for 39 day (adult v-phase) (Murphy et al. 2011). Fully expanded adult leaf blades were harvested every 3 h, with the first time-point (0) within 10 min after lights were turned on in the morning and continuing every 3 h for 36 h. RNA-seq data were likewise derived from the photoperiod-sensitive bioenergy sorghum genotype R.07020 (Kebrom et al. 2017; Casto et al. 2018) and from the bioenergy sorghum hybrid TX08001 (Kebrom and Mullet 2016). RNA-seq data were generated from roots of the bioenergy hybrid TX08001 grown to 120 DAE in a Westwood silt loam soil under field conditions (see Fig. 2 for tissue locations) and from the tiller buds of leaves of the 100 M cultivar grown in growth chambers, as described above. 100 M is a photoperiod-sensitive line (Ma1Ma2Ma3Ma4Ma5ma6) that is part of a panel of genotypes that vary in flowering time (Quinby 1974). Sorghum genotypes that are photoperiod-sensitive (i.e., 100 M, R07020, TX08001) exhibit delayed flowering when grown in long days (> 12.4 h day lengths), whereas photoperiod-insensitive genotypes such as BTx623 flower in about 65–70 days in both long and short days. The genotypes selected for this study represent breeding lines or hybrids used in production of grain sorghum (i.e., BTx623) and bioenergy sorghum (i.e., TX08001, R07020) that have been the subject of prior transcriptome analysis. Seed for all genotypes was obtained from the Sorghum Breeding Lab at Texas A&M University in College Station, TX.

taken from the high bioenergy cultivar TX08001. DAE—days after emergence. The colour of the square or oval denoting the tissue samples indicates the different tissue selected and corresponds to the expression data in tables; Green—nodal roots Table 2a; Yellow square—root Table 2b; Red—stem Table 3a; Yellow circle—Table 4; Blue—leaves; Table 5
Schematic showing the developmental stages of Sorghum bicolor BTx623 and the tissues selected for RNA-seq analysis. The nodal root tissue is shown on this figure, but the tissue was
RNA extraction, template preparation and sequencing
The RNA-seq data reported on in this study were generated from multiple experiments conducted over several years focused on characterizing organ, tissue and developmental patterns of gene expression in sorghum (Kebrom et al. 2017, 2020; McKinley et al. 2016, 2018a; McCormick et al. 2018; Casto et al. 2018). Following tissue harvest, three biological replicates of each tissue were stored at − 80 °C until RNA extraction. Tissues were then pulverized by mortar and pestle and total RNA was extracted from ~ 100 mg of frozen tissue solubilized with 1 mL of Trizol reagent. RNA was isolated and purified using the Direct-zol RNA Miniprep kit (Zymo Research, Irvine CA). Samples were then aliquoted into 96-well plates and shipped to the Joint Genome Institute for sequencing using an automated sequencing pipeline to a depth of 30–50 M reads per sample (McCormick et al. 2018).
RNA-seq analysis
For all RNA-seq experiments, reads were aligned to the S. bicolor V3.1 genome using the HISAT2 aligner (Daehwan et al. 2015; McCormick et al. 2018). Transcriptome assembly and Transcripts Per Million Reads (TPM) normalization were performed using StringTie version 1.3 (Pertea et al. 2015). Analysis and visualization of gene expression data were conducted using TPM normalized data. The prepDE.py script was used to convert nucleotide coverage data from StringTie into reads that are compatible with differential expression statistical packages that use conventional raw reads. Differential expression analysis (DE) was calculated using the edgeR package to determine the fold change (FC) between samples, and the false discovery rate (FDR)—adjusted p values. The fold change displayed in the tables is the absolute value of the fold change between the maximum and minimum expression values. The FDR-adjusted p value also corresponds to the maximum and minimum expression values for each gene analysed. In some cases, zero expression at one time point when compared to expression at another time-point led to a very high differential in expression values. To minimize this effect, in instances with very high DE values, samples with < 1 TPM were set to a TPM = 1 and the fold change was recalculated as FC = maxTPM/1 (McKinley et al. 2016). These instances are indicated with the “ > ” sign in front of the fold change values since the actual fold change is assumed to be some value greater than the value displayed. Functional analysis of gene families utilized protein family annotations (pfam) as well as gene functional annotations generated by DOE-JGI for the S. bicolor V3.1 genome available through Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Sbicolor) (McCormick et al. 2018).
Basis for selection of genes for analysis
Transcript levels of genes with an identified role in dhurrin biosynthesis, hydrogen cyanide bio-activation and detoxification, recycling and transport (Fig. 1) were measured (Gleadow and Møller 2014; Darbani et al. 2016; Hayes et al. 2015; Nielsen et al. 2016; Akbudak et al. 2019; Bjarnholt et al. 2018). Additional details are available in Supplementary Information (Methods S1).
Phylogenetic analysis of dhurrin pathway genes
The cytochrome P450s, CYP79A1 and CYP71E1 have been well characterised (Fig. 1) (Gleadow and Møller 2014; Bak et al. 2000; Tattersall et al. 2001b; Thorsøe et al. 2005; Kahn et al. 1999). In this study, phylogenetic analysis was done to check or expand the cytochrome P450s (PF0067) and the glycoside hydrolases (PF00232). Genes related to either CYP79A1 or DHR1 were initially identified by selecting genes that encode proteins that possess the respective pfam domains present in the relevant gene. The protein sequences encoded by these genes were obtained from Biomart, hosted by the Phytozome database (https://phytozome.jgi.doe.gov/biomart). Multiple sequence alignment and generation of phylogenetic trees using the neighbor-joining method were performed using Clustal Omega using default settings (Madeira et al. 2019). Phylogenetic trees were further rendered using the ggtree and Treeio R packages (Yu et al. 2017). Additional putative DHR encoding genes were identified based on shared membership within a clade containing the query genes of interest. Sequence alignment of DHR1 (Sobic.008G079800.1) with other members of the glycoside hydrolases identified DHR2 (Sobic.008G08400.1), DHR-like3 (Sobic.008G08100) and DHR-like4 (Sobic.008G080600) as the most closely related genes in the sorghum genome with alignment occurring across the entire length of the DHR1 sequence and very low E-values (set to 0 in Phytozome) (Supplementary Fig. S1). The genes in clades adjacent to the DHR-clade aligned with higher E-values and with low sequence alignment over the N-terminal 75 amino acids of DHR1. Phylogenetic trees are given in supplementary Figure S2 and S3. GenBank accession numbers are listed in Table S7.
Plant material and growing conditions for dhurrin analysis
Seeds of S. bicolor BTx623 were germinated and grown in pots under controlled greenhouse conditions at Monash University, Melbourne, Australia (coordinates: 37°54′36″S 145°08′02″E) from December 2019–February 2020 with day/night temperatures of 28 ± 2 °C and 18 ± 2 °C, respectively, and an average photoperiod of 14 h (average photosynthetic photon flux density of 430 ± 80 μmol quanta m–2 s–1). Leaf and nodal root tissue were sampled for dhurrin quantification. The leaves selected were: (1) the last fully emerged and unfurled leaf, classified as older; and (2) a single younger, unfurled leaf positioned two leaves immediately above the last fully unfurled leaf. In plants that had bolted, this younger leaf was the leaf penultimate to the flag leaf. Nodal roots were divided into four sections at different distances from the root tip and their hydrogen cyanide potential analysed (Supplementary Information Fig. S4).
The concentration of the cyanogenic glucoside dhurrin was determined spectrophotometrically from the hydrogen cyanide potential (HCNp) using the method of Woodrow et al. (2002). The hydrogen cyanide potential (HCNp) is used as a proxy for the dhurrin content, with each mg of HCN being equivalent to 11.5 mg of dhurrin in the plant tissue. Free hydrogen cyanide released as a result of ethylene biosynthesis was assumed to be negligible. Leaf discs were sampled from two leaves (young and old) on four individual S. bicolor BTx623 plants: two plants 47-day post sowing and two plants 51-day post sowing that had undergone floral transition and stem bolting. Three leaf discs from each sampled leaf were placed in duplicate vials in 0.1 M citrate buffer (pH 5.0) with 300 µL of β-glucosidase (almond emulsin, G4511, Sigma). A duplicate set of analyses were performed without the addition of exogenous β-glucosidase. The HCN released was captured as NaCN in a 1 M NaOH solution and measured via a colorimetric assay (Woodrow et al. 2002). The leaf discs were subsequently dried and weighed to calculate HCNp mg g−1 dry mass. Mean tissue hydrogen cyanide potentials were compared using ANOVA in Microsoft Excel.
Statistical analysis
In all cases, RNA-seq sample data from each individual experiment shown are the mean of three biological replicates. The three biological replicates of juvenile BTx623 stems were composed of tissue from 5 plants per biological replicate to obtain enough tissue for RNA extraction. Similarly, the three biological replicates of the 100 M tiller bud samples were composed of six axillary buds per sample. For differential expression calculations, FDR-adjusted p values were calculated using the edgeR package. Error bars shown in Figs. 3, 4, and 5 are standard error of the mean (SEM). As stated previously, the significance between tissue hydrogen cyanide potentials was determined by calculating an F-statistic (ANOVA). Phylogenetic trees were constructed using the neighbor-joining method in Clustal Omega with default settings.
Hydrogen cyanide potential (HCNp) of nodal roots of S. bicolor BTx623 sampled at different distances from the tip, corresponding to the regions analysed by RNA-seq: section A—Meristematic region of root tip; Section B—Elongation zone; Section C—Mature zone 1; Section D—Mature zone 2, adjacent to stem. See supplementary figure (Fig. S4) for images of the roots used in this experiment
Diurnal expression of components of the dhurrin biosynthesis, bioactivation and recycling pathways and putative membrane transporter in leaves of BTx623 during vegetative growth. The day length during the experiment was 14 h/10 h, light/dark. White regions of the plots represent lights-on and grey-shaded regions represent lights-off during the experiment. Expression values are the means of three biological replicates (± 1 SEM). Data have been normalized as transcripts per million. The full data set for all genes in this is given Supplementary information (Table S6)
Results
Data on the expression of genes involved in dhurrin biosynthesis (CYP79A1, CYP71E1, UGT85B1 and POR), bioactivation (DHR1, DHR2, DHR-like3, DHR-like4; HNL), prevention of auto-toxicity (CAS, NIT) and endogenous recycling (GST lambda candidates and NIT4A/NIT4B2) during organ and plant development are presented. The nitrilases NIT4A, NIT4B2 and NIT4B1 are involved in detoxification as well as endogenous dhurrin recycling (Jenrich et al. 2007). We also analysed the expression of SbMATE2, which has been shown to transport dhurrin (Darbani et al. 2016) and a candidate nitrate/peptide family (NPF) transporter (SbCGTR1) that is the closest sorghum homolog of a transporter identified in cassava, MeCGTR1, specific for the cyanogenic glucoside, linamarin (Jørgensen et al. 2017). There were four key observations: (1) Expression of the genes involved in dhurrin biosynthesis, except POR, varied significantly during organ and plant development with highest expression in the active meristematic growth zones and juvenile tissues; (2) Expression of genes involved in dhurrin bio-activation was tissue dependent, with DHR1 preferentially expressed in roots and stems, DHR2 in leaf blades, DHR-like3 in stem growing zones and DHR-like4 in young leaf: leaf sheath collar tissue; (3) The nitrilases, NIT4A and NIT4B2, which form heterodimers specifically involved in the endogenous recycling of dhurrin, were expressed at higher levels in mature tissue of plants entering the reproductive stage; (4) Expression of dhurrin pathway genes varied during a diel cycle in leaves, with a peak of expression of the biosynthetic genes (CYP79A1, CYP71E1, UGT85B1 including POR) occurring 3 h after onset of plant illumination at dawn and shortly after the onset of the dark period, whilst SbMATE2 decreased at night and expression of DHR2, specific for bio-activation in the leaves, showed only small changes in expression amplitude during diel cycles. For clarity, data in tables have been limited to genes that showed significant variation in expression during organ or plant development or a diel cycle. Expanded tables that include genes with low or undetectable expression can be found in the Supplementary Information Tables S1–S6.
Transcriptomic analysis of tissue from different organs and developmental stages
Dhurrin pathway gene expression increases during seed imbibition
Transcript levels of the dhurrin biosynthetic pathway genes CYP79A1, CYP71E1 and UGT85B1 were very low in dry seed, consistent with prior results (Nielsen et al. 2016). Transcript levels of CYP79A1, CYP71E1 and UGT85B1 increased in seeds imbibed for 24 h, especially the key gene CYP79A1, consistent with prior data showing rapid accumulation of dhurrin in emerging seedlings (Table 1) (Heraud et al. 2018; Halkier and Møller 1989; Busk and Møller 2002). Transcript levels of the DHR1 increased with seed imbibition (Table 1). There was no change in the expression of hydroxynitrile lyase (HNL) in imbibed seed but the expression of CAS C1 increased 15-fold (Table 1). The recycling genes NIT4A/B2 both increased in expression during imbibition. A small increase in expression of NIT4B1 involved in hydrogen cyanide detoxification is also observed (Supplementary information Table S1). The expression of the SbMATE2 dhurrin transporter and the putative cyanogenic glucoside transporter, SbCGTR1, also increased following seed imbibition.
Dhurrin pathway genes are expressed during tiller bud out growth
Tiller buds formed in the first leaf axil grow steadily in size for 6–8 day post germination prior to becoming dormant or entering into a phase of rapid leaf outgrowth (Kebrom and Mullet 2016). Dhurrin pathway genes involved in biosynthesis were highly expressed in tiller buds of the first leaf axil of 100 M at 8 day after germination (Table 7b; Supplementary information Table S1). DHR2, a gene differentially expressed in leaves, was expressed at very high levels in tiller buds, DHR1 and DHR-like3 at low levels, and DHR-like4 expression was low or undetectable (Supplementary information Table S1). HNL and CAS are also expressed at high levels. In the recycling pathway, three GST genes are highly expressed including two transcripts derived from GST1 (Table S1). However, these GSTs are likely to be involved in functions other than recycling as GSTL1, GSTL2 and NIT4A/4B2, which have been identified in dhurrin recycling, show low or no expression (Table S1). Expression of the SbMATE2 dhurrin transporter was also detected in tiller buds.
Dhurrin pathway gene expression during root and plant development
Nodal roots grow out from the stem and extend into the soil profile through tip growth. Expression of genes in the dhurrin pathway during nodal root development was analysed by collecting tissue from TX08001 nodal roots that had recently become embedded in the soil but prior to lateral root proliferation. Three regions of the developing nodal root were collected; ~ 2 cm of nodal root tissue that spans the root tip-growing zone, the adjacent ~ 2 cm of root tissue that may span a portion of the zone of elongation and 4 cm of more fully differentiated root tissue (Table 2a; Fig. 2 green boxes). Expression of CYP79A1, CYP71E1 and UGT85B1 was highest in root tissue that spans the root tip meristem/growing zone (Fig. 2a, Meristematic). Expression of these genes gradually decreased during maturation of nodal root tissues. DHR1 was expressed at very high levels in the root tip-growing zone but at very low levels in older root tissue. DHR-like 3 was expressed at low levels in the root tip-growing zone but not in more mature root tissues and expression of DHR2 and DHR-like4 was not detected in nodal roots. Expression of the genes involved in dhurrin recycling was 2–4-fold higher in root tissue just above the root tip meristematic tissue (Table 2a). Taken together, the results indicate that dhurrin pathway genes involved in biosynthesis, bio-activation and detoxification/recycling are regulated in very different ways during nodal root development although all pathway genes are highly expressed in nodal root tip-growing zones.
Expression of genes in the dhurrin pathway was examined in roots of S. bicolor BTx623 at four stages of plant development (Table 2b; Fig. 2). At all stages of plant development, roots were collected from the portion of the soil profile where root proliferation was occurring as indicated by the presence of root tips. Expression of CYP79A1, CYP71E1 and UGT85B1 was high at all stages of plant development indicating that dhurrin biosynthesis occurs in proliferating root systems throughout plant development. DHR1 was expressed at very high levels and DHR-like3 at low levels throughout development whereas DHR2 and DHR-like4 expression was very low or not detected. In general, expression of genes involved in dhurrin biosynthesis, detoxification/recycling and transport was high in proliferating roots throughout plant development (Table 2b).
Expression of dhurrin pathway genes during stem development
Dhurrin pathway gene expression during early stages of stem internode development in the vegetative phase of R.07020 is shown in Table 3a. Nascent internodes of R.07020 produced by the shoot apical meristem increase in size through cell division, followed by a phase of elongation, growth cessation, and further differentiation (Kebrom et al. 2017). These four stages of internode development occur sequentially in the top four visible internodes of R.07020 (Table 3a, see Table S3a for a diagram of internode development). Expression of genes in the dhurrin pathway in the top four visible internodes was characterized to better understand dhurrin pathway gene expression during stem and internode development. CYP79A1 and CYP71E1 expression was high in internode 1 and 2, followed by a decrease in expression of ~ 2–3-fold in internodes 3 and 4. UGT85B1 and POR expression was high and uniform in internodes 1–4 (Table 3a, Supplementary Table S3a). Among the dhurrinase genes, DHR1 was highly expressed in internode 1 followed by a ~ 50-fold decrease in expression during this phase of internode development. DHR-like3 expression was also highest in internode 1 and decreased steadily to ~ 100-fold lower levels in internode 4. DHR2 was expressed at relatively low levels that did not change with development and DHR-like4 expression was very low or undetectable in developing R.07020 internodes (Table 3a). HNL expression was low in all tissue sampled (Supplementary Table S3a), whilst CAS C1 showed high and uniform expression across all internodes (Table 3a). GSTL1 and NITB2 expression increased markedly with internode development (Table 3a), whilst the expression of GSTL2 was uniformly high (Supplementary Table S3a). Expression of the putative transporter SbCGTR1 increased approximately 17-fold during this phase of internode development.
Expression of dhurrin pathway genes in stems during plant development was examined using stem tissues derived from BTx623 (Table 3b; Fig. 2 red boxes). The stems of juvenile-phase seedlings were ~ 2 mm in length 8 d after emergence (8 DAE) and 1 cm in length at 24 DAE, a short time after transition from the juvenile to the vegetative phase. Genes encoding the first three steps in dhurrin biosynthesis (CYP79A1, CYP71E1, UGT85B1) were expressed at relatively high levels in stems of juvenile seedlings (8, 24 DAE) prior to the onset of internode elongation (Table 3b). Expressions of dhurrin pathway genes were also analysed in stem internodes that were elongating at floral initiation and in non-growing internodes at anthesis and grain maturity. CYP79A1, CYP71E1 and UGT85B1 transcript levels were lower in elongating and fully elongated internodes of plants at floral initiation and still lower at anthesis or grain maturity (Table 3b). POR was expressed at similar levels in stems at all stages of plant development (Supplementary information Table S3b). DHR1 and DHR2 were expressed at high levels in growing stems of juvenile and vegetative plants. Expression levels of both dhurrinase genes were lower at later stages of plant development (Table 3b). DHR-like3 was expressed at a low but constant level in stems through to floral initiation, whereas DHR-like4 expression in stems was consistently low or not detected (Table 3b). Expression of HNL, CAS C1, GSTL1 and GSTL2 was also highest in juvenile tissue and decreased with plant development, in a pattern similar to the dhurrinase genes. In the recycling pathway, NIT4A/4B2 expression increased during plant development with maximum expression in plant stems at anthesis (65 DAE). Expression of the SbMATE2 dhurrin transporter was highest in stems of juvenile and vegetative plants and ~ fourfold lower at later stages of plant development. The SbCGTR1 putative transporter showed maximum expression in the stems at floral initiation (44 DAE).
Expression of dhurrin pathway genes in leaf sheaths
Information on dhurrin pathway gene expression during leaf sheath development was collected from elongating and fully elongated leaf sheaths of TX08001 during vegetative development. Variation in expression at later stages of plant development was obtained by analysis of fully expanded leaf sheaths of BTx623 at floral initiation, anthesis and grain maturity (Table 4; Fig. 2). CYP79A1, CYP71E1 and UGT85B1 were expressed at relatively high levels in growing leaf sheaths of TX08001. However, expression of these genes was reduced to low levels in fully expanded leaf sheaths of TX08001 and non-growing leaf sheaths of BTx623 at floral initiation, anthesis, and grain maturity. POR expression showed only small differences in expression during leaf sheath and plant development (Supplementary information Table S4). Expression of DHR1 and DHR2 was very low in all leaf sheath tissue sampled and DHR-like3 was expressed at a low level in growing leaf sheaths of TX08001 (Table 4). However, expression of DHR-like4 was high and expression of HNL was very high in the growing sheath of TX08001, decreasing ~ 50-fold in expanded sheath tissue of TX08001 (Table 4). In BTx623, HNL expression was highest in leaf sheath tissue at floral initiation followed by 54-fold lower expression at grain maturity. CAS C1 expression in both TX08001 and BTx623 increased during leaf sheath development followed by a decrease in BTx623 at grain maturity. The GSTL1 and GSTL2 genes, involved in dhurrin recycling, were highly expressed in the growing sheath of TX08001, but at lower levels in BTx623 leaf sheaths post floral initiation. NIT4A, NIT4B1 and NIT4B2 were expressed in all leaf sheath tissues with up to fivefold variation depending on stage of plant development. SbMATE2 dhurrin transporter gene expression was also relatively high in growing leaf sheaths of TX08001, lower in fully expanded leaf sheath tissue, and very low in BTx623 from floral initiation through to grain maturity. The SbCGTR1 putative transporter is highly expressed in leaf sheath tissue with ~ threefold variation in expression during development (Table 4).
Expression of dhurrin pathway genes in leaves during plant development
Expression of dhurrin pathway genes was quantified in green leaf blades collected from BTx623 during the juvenile phase (8 DAE), early vegetative phase (24 DAE), at floral initiation, anthesis and at grain maturity. The 24 DAE RNA was extracted from a bulk sample of expanded leaf blades and growing leaves located in the whorl. CYP79A1, CYP71E1 and UGT85B1 expression was highest in leaves collected from plants at 8 and 24 DAE and very low in fully expanded leaves of plants at anthesis and grain maturity (Table 5). POR was expressed at relatively high levels in leaves until anthesis and at a lower level at grain maturity (Supplementary Table S5). DHR2 was expressed at very high levels in leaf blades except at grain maturity. DHR1 and DHR-like3 expression was very low in leaf blades (Table 5). DHR-like4 showed low levels of expression that increased to a peak at anthesis. HNL was expressed in young juvenile and vegetative-phase leaf blades then decreased to undetected levels at anthesis and grain maturity. CAS C1 increased in expression up to the floral initiation stage, then decreased to approximately half in leaves of plants at grain maturity. In the endogenous recycling pathway, the GSTL2 expression was approximately twice that of GSTL1, with both genes expressed at higher but uniform levels up to the floral initiation stage then decreasing to low levels at grain maturity (Table 5). Expression of NIT4A/4B2 was high in leaf tissues with maximal expression at grain maturity, suggesting recycling may be higher at that stage of plant development to aid remobilization of stored N and C. The SbMATE2 dhurrin transporter gene was expressed at low levels in leaves at all stages of development and very low levels at anthesis and grain maturity (Supplementary Table S5). Expression of the SbMATE2 genes in leaves was also low relative to the expression of this gene in stems and roots. The SbCGTR1 putative transporter gene also showed relatively low expression in leaves with peak expression in vegetative-phase leaves.
Expression of dhurrin pathway genes in a developing phytomer of field grown TX08001
The expression of dhurrin pathway genes in TX08001 under field conditions was investigated 60 days after plant emergence during vegetative-phase growth (Table 6). Dhurrin pathway expression was examined in tissues collected from a single phytomer that contained a fully expanded leaf blade/sheath and a partially elongated internode. Expression of genes involved in dhurrin biosynthesis was high in the leaf sheath collar, but much lower in the leaf blade. In the leaf sheath, these genes were expressed at high levels in the sheath collar and base and lower in the middle of the leaf sheath. Expression of genes involved in dhurrin biosynthesis was highest in the pulvinus and basal growing zone tissues of the internode and lower in the nodal plexus. Expression of genes encoding DHR1 and DHR-like3 was low in leaf blades and sheaths, low in the nodal plexus and upper portions of the internode, with higher expression in the lower tissues of the internode and pulvinus. DHR2 expression was low in most tissues of the phytomer. DHR-like4 expression was high in the leaf collar and very low in all other tissues of the phytomer. Expression of HNL was high in the leaf blade, very high in the leaf sheath and very low in stem tissues.
Coordinated expression of genes encoding dhurrinase and dhurrin biosynthesis
Three genes involved in dhurrin biosynthesis are clustered on chromosome 1 (CYP79A1, CYP71E1, UGT85B1) and genes encoding dhurrinases are clustered on chromosome 8 (Table 7b. The three genes involved in dhurrin biosynthesis have been previously reported to be co-expressed and co-expression was also observed in this study (Table 7b). Co-expression analysis of DHR-gene expression showed that DHR1 and DHR-like3 are also co-expressed (correlation = 0.91). DHR1 and DHR-like3 are differentially expressed in roots and stems and show higher expression in growing zones compared to older non-growing tissues (Table 7a). However, the ratio of expression of DHR1/DHR-like3 can vary from ~ 42 in the root growing zone to ~ 1.8 in the stem internode growing zone indicating that DHR-like3 plays a more significant role in stem growing zones and DHR1 in roots (Table 7a). In contrast, DHR2 expression is not correlated with other DHR genes with highest expression in tiller buds, mature leaf blades and lower expression in the stem apex, possibly associated with nascent leaf formation (Table 7a, b). High DHR2 expression was observed in vegetative-phase young leaves of BTx623 grown in growth chambers (~ 2861 TPM), however, DHR2 expression was low in leaves of TX08001 grown in the field (2 TPM) (Table 7a). The basis of this difference is unclear but may be due to differences in stage of leaf development, genotype, and environmental factors. DHR-like4 expression was also highly specific to the leaf collar of young leaves and low expression in the upper portion of the leaf sheath (Table 7a). Genes involved in dhurrin biosynthesis and dhurrinases were often co-expressed in organ growing zones, but not in fully developed organs (Table 7b). For example, in fully elongated non-growing (NG) stem internodes and roots, genes involved in dhurrin biosynthesis were highly expressed when genes encoding dhurrinases were expressed at very low levels (Table 7b). In contrast, expression of DHR2 was very high in non-growing leaves of BTx623 at anthesis although expression of genes involved in dhurrin biosynthesis was very low. This temporal difference in expression could help separate synthesis of dhurrin and proteins that are involved in bio-activation, or it could reflect other non-defence functions associated with DHR2.
HCNp and β-glucosidase activity in tissues of roots, shoots and tiller buds
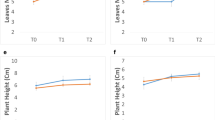
Hydrogen cyanide potential (HCNp) in nodal roots of BTx623 equivalent to the stages in Table 2a was highest in the portion of the root that spans the growing zone and lower in the fully elongated root tissue (Fig. 3). We also measured HCNp in young developing leaves and older fully developed leaves of BTx623 at or shortly after floral initiation. To confirm the activity of DHR2 in these leaves, we compared the amount of hydrogen cyanide released with and without the addition of exogenous β-glucosidase in four individual plants. Greater amounts (~ two-fold) of evolved HCN were observed upon addition of exogenous β-glucosidase. The analysis showed higher HCN production in young growing leaves in plants 2, 3 and 4 compared to older fully expanded leaves (Fig. 4).
Expression of dhurrin pathway genes in leaves during a diel cycle
Expression of dhurrin pathway genes in young leaves during a diel cycle was investigated in vegetative-phase BTx623 plants. Tissue sampling started within 10 min after dawn (time 0) and samples were collected every 3 h, including a 10-h night period and four time points into the light period of day 2. Transcript abundance of biosynthesis pathway genes CYP79A1, CYP71E1 and UGT85B1 peaked in the morning (3HL and 27HL) and 2 h after the onset of dark (15HL) (Fig. 5). In the morning, UGT85B1 expression was lower than CYP79A1 and CYP71E1, but in the evening, UGT85B1 expression was similar to or higher than CYP79A1/CYP71E1. POR expression varied ~ 50-fold during a diel cycle with highest expression at dawn followed by a decrease in transcript levels to low levels from 9HL until the following dawn. DHR2 transcript levels were high throughout the day–night cycle. By contrast, expression of HNL was highest during the day peaking at 9HL followed by a decrease in the late afternoon and at night (Fig. 5). NIT4B2 expression increased at dawn and stayed high for the first 6 h of the day then decreased in the evening. Expression of the SbMATE2 dhurrin transporter increased in the morning and remained high during the day and for 1 h after darkness, then decreased to low levels for the duration of the night, increasing the following dawn.
Discussion
A detailed understanding of gene expression in plant organs/tissues during development and in response to variation in nutrient status and environmental constraints facilitates the identification of signalling pathways, transcription factors and gene regulatory networks that modulate gene expression. In this study, RNA-seq was used to characterize the expression of genes involved in the biosynthesis, recycling and bio-activation of dhurrin during organ and plant development in sorghum. Broadly, we found that expression of genes involved in dhurrin biosynthesis was highest in growing tiller buds, imbibed seed, and the growing zones of leaves, roots, and stems. The bio-activation genes showed organ- and tissue-specific expression whereas expression of the recycling genes was maintained or increased during the reproductive phase. The expression of genes involved in dhurrin biosynthesis and recycling in leaves varied during diel cycles. If the function of dhurrin is for herbivore defence, then synthesis and bio-activation would be expected to vary with the stage of development and within different tissues, with the highest deployment in juvenile or meristematic tissues following a typical ontological defence trajectory (Boege and Marquis 2005). The expression of genes in the dhurrin pathway was complex and varied with plant development in a way that is consistent with multiple roles in defence and metabolism, including serving as a source of reduced nitrogen during grain filling and germination. Instances of differential expression of biosynthesis and bio-activation genes, and the tissue-specific expression of DHR1, DHR2 and DHR-like4 indicates the involvement of complex regulatory systems. If the multiple and changing roles for dhurrin over the life of the sorghum plant—herbivore defence, mitigation of abiotic stresses, nitrogen storage and recycling—are found to be a general feature of other cyanogenic plants, this may explain why cyanogenic plants are so common, and why humans appear to have selected for higher expression of this pathway during domestication.
High transcript levels of biosynthesis genes in young tissues indicate a defence function
The three genes involved in dhurrin biosynthesis that are clustered on chromosome 1 (CYP79A1, CYPE1, UGT85B1) are generally co-expressed in developing organs and during plant development. These genes were highly expressed in growing zones of leaves, stems (shoot apex, base of elongating internodes), roots and young developing tiller buds and imbibing seeds. Expression extended into zones of elongation and early maturation of developing roots and stem internodes with gradual decreases in expression of these genes as the organ or tissue became fully differentiated. High expression was also observed in leaf collars and the stem pulvinus, tissues adjacent to elongating leaf blades and stem internodes. These tissues may be prioritized for protection or sources of dhurrin transported into developing leaf blades and internodes. High rate of dhurrin synthesis in young developing organs and growing zones is consistent with elevated priority of deployment of protection. Very low expression of genes involved in dhurrin biosynthesis was observed in older fully developed organs of vegetative-phase adult plants (i.e., leaf blades of TX08001 in the field) and during the reproductive phase, especially from anthesis and grain maturity (i.e., leaf blades, leaf sheath and stems of BTx623). Down-regulation of dhurrin biosynthesis in vegetative organs during the reproductive phase of development would reduce competition for nitrogen needed for grain filling.
Previous studies have consistently found young sorghum plants and immature tissues of older plants to have much higher concentrations of cyanogenic glucosides (Halkier and Møller 1989; Miller et al. 2014; Blomstedt et al. 2018; Sohail et al. 2020; Cowan et al. 2021). Here we determined that the HCNp in roots was highest in a region immediately adjacent to the root tip that spans the growing zone, with a significantly lower concentration in the older tissue of the maturation zone (Table 2b; Fig. 3). During root development, transcript levels were correlated with accumulation of dhurrin (Fig. 3), supporting the earlier observation of Busk and Møller (2002) that dhurrin biosynthesis is transcriptionally regulated.
In leaf blades, genes involved in dhurrin biosynthesis are also expressed at higher levels when they are growing (Table 5). Dhurrin concentrations in the leaves are significant in older leaves (Fig. 4), even though expression of genes involved in de novo synthesis is very low in these older tissues (Tables 5, 7a). This is likely due to the presence of stably stored dhurrin synthesized during leaf development (Knudsen et al. 2020) or could be due transport of dhurrin into fully developed portions of leaf blades from the base of the leaf blade or leaf collars (see below). Similar expression patterns of genes encoding biosynthesis of the cyanogenic glucoside linamarin in the rubber tree, Hevea brasiliensis were obtained upon analysis of transcriptome prepared from leaves of different age classes (Fang et al. 2016).
The first step in the biosynthesis pathway, the conversion of tyrosine to (E)-p-hydroxyphenylacetaldoxime is catalysed by CYP79A1 and is considered a rate limiting step (Busk and Møller 2002). However, if this step proceeds faster than the subsequent formation of p-hydroxymandelonitrile catalysed by CYP71E1, then the intermediate E-oxime would accumulate. Oximes with the E-configuration have high biological activity compared with Z-oximes exhibiting strong insecticidal, fungicidal, and herbicidal activity (Sørensen et al. 2018). They inhibit mitochondrial oxidases and thereby promote lipid peroxidation and the production of toxic reactive oxygen species (Sakurada et al. 2009; Møller 2010). Upon introduction of CYP79A1 and CYP71E1 into Arabidopsis, the plants do not develop properly due to auto-toxicity related to p-hydroxymandelonitrile accumulation or the associated liberation of hydrogen cyanide. Upon introduction of the entire dhurrin pathway, the glucosylating activity of the introduced UGT85B1 efficiently converts p-hydroxymandelonitrile into dhurrin and the wild-type phenotype is restored (Tattersall et al. 2001b). In the transcriptomes here analysed, we observe co-expression of CYP79A1, CYP71E1 and UGT85B1 in a tissue although not always at equal transcript levels at the time of day the tissue was collected. In leaves, CYP79A1 and CYP71E1 were more highly expressed than UGT85B1 in the morning when most samples were collected, whereas UGT85B1 expression was higher in the evening. A similar pattern is seen for equivalent genes in barley (Knoch et al. 2016).
Bio-activation: tissue-specific dhurrinase and HNL expression allow HCN release to be finely regulation
Phylogenetic analysis of DHR1 and DHR2 identified two additional genes that encoded closely related proteins we named DHR-like3 and DHR-like4. All four genes are located in close proximity on sorghum chromosome 8. In our current study, we characterized the expression of DHR1, DHR2 and the two DHR-like genes (Table 7a, b). Follow-up studies will be required to determine if DHR-like3 and DHR-like4 catalyse dhurrin hydrolysis. Ganjewala et al. (2010) noted that expression of DHRs is often localized to specific organs. Here, we confirm that the two previously identified DHRs (DHR1 and DHR2) show a high degree of tissue specificity with DHR1 differentially expressed in roots > shoot apex/stem intercalary meristem/pulvinus and DHR2 in leaf blades (Tables 2, 5, 6). DHR1 is highly expressed in imbibed seeds undergoing root outgrowth and DHR2 is highly expressed in developing tiller buds associated with nascent leaf growth. Both DHR1 and DHR2 are highly expressed in stem tissue that includes the shoot apex of juvenile and vegetative stems (24 DAE) that have not started to elongate (Table 3a). Krothapalli et al. (2013) also found that DHR1 was differentially expressed in root tissue and the mesocotyl. DHR2 was expressed at high levels in young developing leaves and fully developed leaves (anthesis) of BTx623 grown in growth chambers/greenhouse (Table 5). However, DHR2 expression in leaves of field grown TX08001 was much lower indicating growing conditions modulate DHR2 expression (Table 6). Overall, expression of the dhurrinase-like genes DHR-like3 and DHR-like4 was low relative to DHR1 and DHR2, with two notable exceptions: DHR-like3 was expressed in the stem shoot apex and stem intercalary meristematic regions (Tables 3, 6) and DHR-like4 was highly expressed in leaf collars with lower expression in upper portion of leaf sheaths of the bioenergy cultivar TX08001 (Tables 4, 6). DHR1 and DHR-like3 expression was correlated, although DHR1 is more highly expressed in roots and DHR-like3 showed highest expression in the shoot apex and stem intercalary meristems. These genes are adjacent on chromosome 8, possibly the result of a recent gene duplication. It is interesting that expression of DHR-like4 is highly expressed in leaf collars and not correlated with the expression of any of the other members of this gene family. This is typical of gene paralogs that diverge and acquire unique patterns of expression that in this case may help protect the leaf collar. DHR1, DHR2 and HNL were expressed at high levels in young tissue (roots, stem, sheath, tiller buds), decreasing with plant age (Tables 2, 3, 4; Table S1). DHR-like3 expression also decreased with organ maturity, consistent with our observations that dhurrin metabolism is associated with early organ growth and development. Follow-up studies will be required to confirm that DHR-like3 and DHR-like4 encode dhurrinases and to better understand the activity of these enzymes.
Many cyanogenic species encode two or more dhurrin β-glucosidase isoenzymes (Hösel et al. 1987). The genome of Lotus japonicus harbours four genes encoding β-glucosidases able to hydrolyze aliphatic cyanogenic glucosides. These four genes are differentially expressed; BGD2 and BGD4 are expressed in the shoot, BGD3 in the keel and the enclosed reproductive organs of the flowers, whilst BGD7 is expressed in roots (Morant et al. 2008a; Lai et al. 2014; Takos et al. 2011). The confined expression of the individual dhurrinases to specific tissues observed here may be related to the presence of particular cell types in the relevant organs. Kojima et al. (1979), for example, found the DHR in leaves of sorghum to be specific to mesophyll cells, and possibly even the chloroplast (Kojima et al. 1979; Thayer and Conn 1981). Elevated expression of dhurrinase could occur in the root cortex requiring root cortical cell specific expression. RAMAN, Laser dissection/qPCR, in situ hybridization and micro-extraction technologies will be useful for defining where root dhurrinase is transcribed and accumulates (Heraud et al. 2018; Montini et al. 2020).
Upon cell disruption dhurrin is hydrolysed by dhurrinases (β–glucosidases) and hydrogen cyanide is rapidly released from dhurrin through the action of α-hydroxynitrilases (HNLs) with co-production of stoichiometric amounts of p-hydroxybenzaldehyde. High expression of HNL is leaf blades and the leaf sheath is consistent with a role of this enzyme in defence since these organs are exposed to pests. In contrast, HNL expression was low in stems even though genes involved in dhurrin biosynthesis were expressed at high levels. One could posit that since the stem is enclosed by the leaf sheath that has high expression of HNL, there is less need for rapid HNL-mediated release of HCN by stem tissue. Low HNL levels in stems may increase flux through the recycling pathway or facilitate dhurrin transport without turnover.
Cyanogenic glucosides are usually considered to be phytoanticipins, that is, they are synthesised and stored and only release the toxic HCN when mixed with specific hydrolytic enzymes (β-glucosidase and α-hydroxynitrile lyase). The rate of this release is important for the effectiveness of the system in deterring herbivores (Krothapalli et al. 2013). Ballhorn et al. (2006), for example, found a strong correlation between the activity of β-glucosidase and deterrence of herbivores in lima bean (Phaseolus lunatus). One might expect synchronous expression of biosynthesis and bio-activation genes, even though they are clustered on two different chromosomes as found by Fang et al. (2016) for Hevea brasiliensis. High expression of genes involved in dhurrin biosynthesis and organ-specific DHRs was observed in regions of organ cell division (i.e., root tips, shoot/stem apex, internode 1, intercalary meristems). However, there were notable exceptions, such as high DHR2 expression in fully expanded leaf blades despite biosynthetic gene expression decreasing to very low levels (Tables 5, 7). Similarly, DHR1 expression decreased to low levels in root tissues just above the root tip whereas expression of the three genes involved in dhurrin biosynthesis remained elevated. This difference may allow plants to restrict synthesis of DHR1 to specific cell types generated in the meristematic zone, but continue to accumulate dhurrin for sequestration in vacuoles in elongating and fully elongated tissues. These differences in the timing of gene expression during organ biogenesis will need to be accounted for in gene regulatory network analysis.
The transport of specialized metabolites, such as dhurrin, is important to enable optimisation of the production costs and to ensure the compounds are deposited where they are most useful. MATE and NPF transporters have been implicated in relocation of various secondary metabolites, including dhurrin, linamarin and glucosinolates (Darbani et al. 2016; Nour-Eldin et al. 2012; Jørgensen et al. 2017). In our study, expression of SbMATE2 increased in imbibing seed and decreased > tenfold in the leaf sheath post floral initiation. However, in most other organs and stages of development expression of this gene was relatively high and consistent. Darbani et al. (2016) showed that SbMATE2 specifically transports dhurrin across the vacuolar membrane and it may be that SbMATE2 is involved in the transport of dhurrin for defence and remobilization depending on the stage of organ development. We also analysed the expression of SbCGTR1, a NPF transporter identified based on its high (72%) amino acid sequence similarity to AtGTR1 of Arabidopsis (Nour-Eldin et al. 2012) and 75% sequence similarity to MeCGTR1 of cassava (Jørgensen et al. 2017). The expression of SbCGTR1 increases with internode development but otherwise was similar in organs and stages of plant development. Regulation of transport and re-allocation of dhurrin between tissue types in preparation for remobilisation and/or recycling of reduced nitrogen at later developmental stages could occur at a post-transcriptional level.
Increase in recycling genes in maturing plants and with shift to reproductive stage is consistent with dhurrin as a mobilizable source of nitrogen
As the sorghum plants transitioned into the reproductive phase, a substantial decrease in the expression of the biosynthetic genes was observed concomitant with relatively small but consistent increases in expression of selective recycling genes. For example, NITB2 expression increased ~ 127-fold during vegetative internode development (Table 3a) and increased ~ 21-fold in stems during the transition from the vegetative phase to anthesis (Table 3b). Potentially, this could orchestrate the mobilization of dhurrin-bound nitrogen for plant growth or to support seed production (Tables 2a, 4, 5). The high expression of genes involved in dhurrin biosynthesis in stems during internode biogenesis, low expression of HNL in internodes, and up-regulation of NITB2 in stems post-floral initiation are consistent with dhurrin remobilization. It is clear that the recycling pathway, where dhurrin is metabolized without the release of toxic hydrogen cyanide affords the possibility of repurposing cyanogenic glucosides (Bjarnholt et al 2018). The expression of recycling genes, particularly around floral initiation, anthesis and grain maturity, may reflect the increased demand for nitrogen for seed set and filling. Root tissue just above root tips had a high dhurrin content although lower content in older more mature root tissue. The same root zone showed high expression of genes involved in dhurrin recycling, suggesting that the dhurrin present was subject to continuous endogenous recycling to provide nitrogen and carbon for growth uses (Fig. 4; Table 2b). In the rubber tree (Hevea brasiliensis), cyanogenic glucosides have been reported as being a mobilizable source of nitrogen and glucose used to build up the biosynthetic machinery required for latex production (Kongsawadworakul et al. 2009). The increased expression of CAS and NIT4A in mature rubber leaves observed by Fang et al. (2016) supports this hypothesis.
Diel effects show dhurrin concentrations are environmentally plastic
We detected significant diel variation in the transcript levels of the biosynthesis genes. CYP79A1 and CYP71E1 showed a peak of expression 3 h after initiation of the light period concomitant with a small increase in UGT85B1 expression. The transcript levels diminished during the light period followed by a small rise in expression about 1 h after onset of the dark period and a larger increase in UGT85B1 expression. POR expression was highest at the beginning of the light period and the following 3 h whereas HNL expression peaked at 9HL and decreased at night. Expression of DHR2 was high and showed only slight changes during diel cycles. Diel variation in gene expression can affect protein levels on a daily basis if protein stability is low, or more often, adjust levels of protein over a longer time frame. The oscillations may indicate signalling that connects expression of genes in the dhurrin pathway to the nitrogen demands of primary metabolism. Diurnal variation in synthesis of cyanogenic glucosides has previously been reported in cassava (Schmidt et al. 2018). In cassava, the transcripts encoding the biosynthetic genes disappeared at the beginning of the light period closely accompanied by disappearance of the biosynthetic enzymes and rapid recycling of the cyanogenic glucosides. It is possible that strong photosynthetic carbon fixation happening during the light period in the tropical and subtropical regions where cassava is grown may be balanced by a supply of reduced nitrogen in the form of ammonium ions derived from recycling of cyanogenic glucosides. The operation of a similar process in sorghum involving recycling of dhurrin is supported by the high transcript level of HNL and the NIT4B2 during the light period. Like cassava, sorghum is grown in geographic regions with high light irradiation levels during the day. The recycling of dhurrin in the light period may serve to balance the nitrogen demands in primary metabolism when carbon fixation rates are high.
Function and adaptive significance of cyanogenic glucosides
Three distinct expression patterns of the cyanogenic glucoside biosynthetic pathway genes in sorghum were observed in this study: (1) high expression of biosynthesis genes in young developing tissues and organs; (2) a diel pattern in leaves, with enhanced expression of the biosynthetic genes in the period immediately following dawn; (3) an increase in cyanogenic glucoside recycling genes post floral initiation. These phases align with ontological development and are consistent with the hypothesis that cyanogenic glucosides serve multiple functions in plants—herbivore defence, mitigation of stress, UV scavengers, nitrogen storage and recycling—arising from different evolutionary pressures at different stages of plant development and in different tissues (Gleadow and Møller 2014). We have developed a schematic model linking expression levels, plant developmental stage and function (Fig. 6). Different patterns of expression in different tissues support the notion of multiple roles. In addition to the biosynthesis and recycling pathways, the presence of multiple nitrilases, GSTs and DHRs, some with tissue-specific expression, increases the options for additional flexibility in phenotypic expression in space and time.
Model linking differences in gene expression of cyanogenesis-related genes at different stages of plant development with proposed functions of dhurrin. When plants are young, and in young tissues dhurrin is primarily functioning in herbivore defence. During vegetative growth, dhurrin has multiple roles mediated by environmental plasticity in response to light and abiotic stress. After anthesis, dhurrin becomes a
High expression of bio-activation and biosynthetic genes, but not recycling genes, indicates a primary role in defence in the early stage of organ and plant development consistent with the very high concentration of dhurrin observed in these tissues and consistent with ontogenetic trajectories observed in sorghum (Miller et al. 2014; O'Donnell et al. 2013). This pattern supports the optimal allocation theory, which hypothesises that plants will invest resources in defence of the most valuable and most vulnerable parts of the plant (McKey 1974). Similar ontogenetic and spatial patterns have been observed in many species for a wide range of chemical defence systems (Barton and Boege 2017). As organs and plants mature (Phase 2), there is less value in investing in defence per se (Boege and Marquis 2005). In sorghum, such investments are plastic, with increased allocation of resources to dhurrin in plants experiencing stress (e.g. Rosati et al. 2019; O'Donnell et al. 2013; Gleadow et al. 2016)), or in the case presented here, transcript level variation guided by the diel cycle (Fig. 5). Such changes in expression that modulates pathway deployment are likely related to the availability of energy to reduce nitrogen in the leaves, or to mitigate stress by reducing reactive oxygen species. Once sorghum plants enter the reproductive phase (Phase 3), herbivore defence becomes less important and grain yield becomes the main evolutionary driver. In accordance with these demands, dhurrin accumulated in the early phases of grain development is re-cycled during the subsequent maturation period. As a result, only negligible residual dhurrin amounts are present in the mature grain (Nielsen et al. 2016). A similar link has been seen in Nicotiana attenuata (Diezel et al. 2011). Indeed, lack of nitrogen is an important factor in limiting grain growth in cereals, including sorghum. The decrease in expression of genes involved in dhurrin biosynthesis and an increase in the expression of recycling genes that we observed in plants during the reproductive phase supports the hypothesis that sorghum remobilises dhurrin to provide nitrogen for the developing grain. This could be one reason why stay green varieties of sorghum have higher levels of dhurrin (Hayes et al. 2015, 2016) and may help explain why domesticated sorghum is much more cyanogenic than its wild relatives (Myrans et al. 2021; Cowan et al. 2021).
A plant’s investment in the synthesis and maintenance of specialised metabolites is often quantified by calculating the costs in terms of growth forgone or reduced yield. While this approach is excellent at explaining why costs of defence change as plants mature (Boege and Marquis 2005), it does not account for the fact the costs are sometimes not detectable (e.g. (Sohail et al. 2020)). Results support the conjecture that dhurrin is not purely synthesised for defence but is integrated with the primary metabolism in sorghum functioning as a mobilizable source of reduced nitrogen during grain filling and germination. Allowing for roles other than defence can accommodate such discrepancies, at least for cyanogenic glucosides, while leaving the general tenant of resource allocation theory intact (Neilson et al. 2013). Clearly, different functions are of adaptive significance at different stages of development. Thus, rather than viewing the allocation of resources through the lens of ‘costs’ and ‘benefits’, it may be more useful to view them as integrated to the benefit of plant development and fecundity.
Any increase in nitrogen use efficiency would both increase yields and reduce costs to farmers. A significant amount of the nitrogenous fertiliser used in agriculture is currently wasted through either run-off or is converted to nitrous oxide, a potent greenhouse gas. Optimisation of dhurrin pathway gene expression could contribute to a reduced need for nitrogen fertilisers. The work presented here provides a number of options for doing this. While simply removing dhurrin biosynthesis seems an attractive option to reduce nitrogen use in cyanogenic plants, the observation that the storage of high-value reduced nitrogen that can be remobilized for grain yield may mean this is counterproductive, at least for grain crops. However, if the remobilization of nitrogen from dhurrin could be more targeted in space and time, this could lead to the desired improvements particularly in biomass production with a reduced need for fertilisers.
Author contribution statement
JM and RMG conceived of the project using RNA-seq data provided by JM. BMc performed the bioinformatics. All authors analysed and interpreted the results and contributed to the writing of the manuscript.
Data availability
Expression data are available in the accompanying excel files. All of the raw data are available through Phytozome (Joint Genome Institute) https://genome.jgi.doe.gov/portal/.
Abbreviations
- HCNp:
-
Hydrogen cyanide potential
References
Akbudak MA, Filiz E, Uylas S (2019) Identification of O-acetylserine(thiol)lyase (OASTL) genes in sorghum (Sorghum bicolor) and gene expression analysis under cadmium stress. Mol Biol Rep 46(1):343–354. https://doi.org/10.1007/s11033-018-4477-0
Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA (1998) Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol 36(3):393–405
Bak S, Olsen CE, Halkier BA, Møller BL (2000) Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in dhurrin biosynthesis. Plant Physiol 123(4):1437–1448
Ballhorn D, Heil M, Lieberei R (2006) Phenotypic plasticity of cyanogenesis in lima bean Phaseolus lunatus—activity and activation of β-glucosidase. J Chem Ecol 32:261–275
Barton KE, Boege K (2017) Future directions in the ontogeny of plant defence: understanding the evolutionary causes and consequences. Ecol Lett 20:403–411
Bjarnholt N, Neilson EHJ, Crocoll C, Jørgensen K, Motawia MS, Olsen CE, Dixon DP, Edwards R, Møller BL (2018) Glutathione transferases catalyze recycling of auto-toxic cyanogenic glucosides in Sorghum. Plant J 94(6):1109–1125. https://doi.org/10.1111/tpj.13923
Blomstedt CK, Rosati VC, Lindberg Møller B, Gleadow R (2018) Counting the costs: nitrogen partitioning in Sorghum mutants. Funct Plant Biol 45(7):705–718. https://doi.org/10.1071/FP17227
Boege K, Marquis RJ (2005) Facing herbivory as you grow up: the ontogeny of resistance in plants. Trends Ecol Evol 20(8):441–448. https://doi.org/10.1016/j.tree.2005.05.001
Burke JJ, Chen J, Burow G, Mechref Y, Rosenow D, Payton P, Xin Z, Hayes CM (2013) Leaf dhurrin content is a quantitative measure of the level of pre- and postflowering drought tolerance in sorghum. Crop Sci 53:1056–1065. https://doi.org/10.2135/cropsci2012.09.0520
Busk PK, Møller BL (2002) Dhurrin synthesis in sorghum is regulated at the transcriptional level and induced by nitrogen fertilization in older plants. Plant Physiol 129:1222–1231
Casto AL, McKinley BA, Yu KMJ, Rooney WL, Mullet JE (2018) Sorghum stem aerenchyma formation is regulated by SbNAC_D during internode development. Plant Direct 2(11):e00085. https://doi.org/10.1002/pld3.85
Cicek M, Esen A (1998) Structure and expression of a dhurrinase (beta-glucosidase) from Sorghum. Plant Physiol (rockville) 116(4):1469–1478
Cooper EA, Brenton ZW, Flinn BS, Jenkins J, Shu S, Flowers D, Luo F, Wang Y, Xia P, Barry K, Daum C, Lipzen A, Yoshinaga Y, Schmutz J, Saski C, Vermerris W, Kresovich S (2019) A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: implications for the genetics of sugar metabolism. BMC Genomics 20(1):420. https://doi.org/10.1186/s12864-019-5734-x
Cowan M, Blomstedt CK, Møller BL, Henry R, Gleadow RM (2021) Variation in production of cyanogenic glucosides during early plant development: a comparison of wild and domesticated Sorghum. Phytochemistry 184:112645
Daehwan K, Langmead B, Salzberg S (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360
Darbani B, Motawia MS, Olsen CE, Nour-Eldin HH, Møller BL, Rook F (2016) The biosynthetic gene cluster for the cyanogenic glucoside dhurrin in Sorghum bicolor contains its co-expressed vacuolar MATE transporter. Sci Rep 6(1):37079. https://doi.org/10.1038/srep37079
Diezel C, Allmann S, Baldwin IT (2011) Mechanisms of optimal defense patterns in Nicotiana attenuata: flowering attenuates herbivory-elicited Ethylene and Jasmonate Signaling F. J Integr Plant Biol 53(12):971–983
Emendack Y, Burke J, Sanchez J, Laza HE, Hayes C (2018) Agro-morphological characterization of diverse sorghum lines for pre-and post-flowering drought tolerance. Aust J Crop Sci 12(1):135–150
Fang Y, Mei H, Zhou B, Xiao X, Yang M, Huang Y, Long X, Hu S, Tang C (2016) De novo transcriptome analysis reveals distinct defense mechanisms by young and mature leaves of Hevea brasiliensis (para rubber tree). Sci Rep 5:33151. https://doi.org/10.1038/srep33151
Ganjewala D, Kumar S, Devi SA, Ambika K (2010) Advances in cyanogenic glycosides biosynthesis and analyses in plants: a review. Acta Biologica Szegediensis 54:1–14
Gershenzon J (1989) The cost of plant chemical defense against herbivory: a biochemical perspective. In: Bernays EA (ed) Insect-plant interactions, vol 5. CRC Press, Bacon Raton, pp 105–173
Gill JR, Burks PS, Staggenborg SA, Odvody GN, Heiniger RW, Macoon B, Moore KJ, Barrett M, Rooney WL (2014) Yield results and stability analysis from the sorghum regional biomass feedstock trial. BioEnergy Res 7(3):1026–1034. https://doi.org/10.1007/s12155-014-9445-5
Gleadow RM, Møller BL (2014) Cyanogenic glycosides: synthesis, physiology, and phenotypic plasticity. Annu Rev Plant Biol 65(1):155–185. https://doi.org/10.1146/annurev-arplant-050213-040027
Gleadow RM, Woodrow IE (2002) Constraints on effectiveness of cyanogenic glycosides in herbivore defense. J Chem Ecol 28:1301–1313
Gleadow RM, Foley WJ, Woodrow IE (1998) Enhanced CO2 alters the relationship between photosynthesis and defence in cyanogenic Eucalyptus cladocalyx F. Muell. Plant Cell Environ 21:12–22
Gleadow RM, Ottman MJ, Kimball BA, Wall GW, Pinter PJ Jr, LaMorte RL, Leavitt SW (2016) Drought-induced changes in nitrogen partitioning between cyanide and nitrate in leaves and stems of Sorghum grown at elevated CO2 are age dependent. Field Crop Res 185:97–102. https://doi.org/10.1016/j.fcr.2015.10.010
Halkier BA, Møller BL (1989) Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L.) Moench and partial purification of the enzyme system involved. Plant Physiol 90(4):1552–1559
Hayes CM, Weers BD, Thakran M, Burow G, Xin Z, Emendack Y, Burke JJ, Rooney WL, Mullet JE (2016) Discovery of a dhurrin QTL in Sorghum: co-localization of dhurrin biosynthesis and a novel stay-green QTL. Crop Sci 56(1):104–112. https://doi.org/10.2135/cropsci2015.06.0379
Hayes CM, Burow GB, Brown PJ, Thurber C, Xin Z, Burke JJ (2015) Natural variation in synthesis and catabolism genes influences dhurrin content in sorghum. Plant Genome. https://doi.org/10.3835/plantgenome2014.09.0048
Heraud P, Cowan MF, Marzec KM, Møller BL, Blomstedt CK, Gleadow R (2018) Label-free Raman hyperspectral imaging analysis localizes the cyanogenic glucoside dhurrin to the cytoplasm in Sorghum cells. Sci Rep 8(1):2691. https://doi.org/10.1038/s41598-018-20928-7
Hösel W, Tober I, Eklund SH, Conn EE (1987) Characterisation of betaglucosidases with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench seedlings. Arch Biochem Biophys 252:152–162
Jenrich R, Trompetter I, Bak S, Olsen CE, Møller BL, Piotrowski M (2007) Evolution of heteromeric nitrilase complexes in Poaceae with new functions in nitrile metabolism. Proc Natl Acad Sci 104(47):18848–18853. https://doi.org/10.1073/pnas.0709315104
Jones PR, Møller BL, Hoj PB (1999) The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor isolation, cloning, heterologous expression, and substrate specificity. J Biol Chem 274(50):35483–35491. https://doi.org/10.1074/jbc.274.50.35483
Jørgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Møller BL (2005) Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol 8:280–291
Jørgensen M, Xu D, Crocoll C, Ernst H, Ramírez D, Motawia M, Olsen C, Mirza O, Nour-Eldin H, Halkier B (2017) Origin and evolution of transporter substrate specificity within the NPF family. Elife 6:19466. https://doi.org/10.7554/eLife.19466
Kahn RA, Bak S, Svendsen I, Halkier BA, Møller BL (1997) Isolation and reconstitution of cytochrome P450ox and in vitro reconstitution of the entire biosynthetic pathway of the cyanogenic glucoside dhurrin from Sorghum. Plant Physiol 115(4):1661–1670
Kahn R, Fahrendorf T, Halkier B, Møller B (1999) Substrate specificity of the cytochrome P450 enzymes CYP79A1 and CYP71E1 involved in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Archives Biochem Biophys 363(1):9–18
Kebrom TH, Mullet JE (2016) Transcriptome profiling of tiller buds provides new insights into phyb regulation of tillering and indeterminate growth in Sorghum. Plant Physiol 170:2232–2250
Kebrom TH, McKinley B, Mullet JE (2017) Dynamics of gene expression during development and expansion of vegetative stem internodes of bioenergy sorghum. Biotechnol Biofuels 10(1):159. https://doi.org/10.1186/s13068-017-0848-3
Kebrom TH, McKinley BA, Mullet JE (2020) Shade signals alter the expression of circadian clock genes in newly-formed bioenergy sorghum internodes. Plant Direct 4(6):e00235. https://doi.org/10.1002/pld3.235
Knoch E, Motawie MS, Olsen CE, Møller BL, Lyngkjær MF (2016) Biosynthesis of the leucine derived a-, b- and c-hydroxynitrile glucosides in barley (Hordeum vulgare L.). Plant J 88:247–256
Knudsen CB, Gallage NJ, Hansen CC, Møller BL, Laursen T (2018) Dynamic metabolic solutions to the sessile life style of plants. Nat Prod Rep 35:1140–1155
Knudsen CB, Bavishi K, Viborg KM, Drew DP, Simonsen HT, Motawie MS, Møller BL, Laursen T (2020) Stabilization of the dhurrin biosynthetic enzymes using a natural deep eutectic solvent. Phytochemistry 170:112214
Kojima M, Poulton JE, Thayer SS, Conn EE (1979) Tissue distributions of dhurrin and of enzymes involved in its metabolism in leaves of Sorghum bicolor. Plant Physiol 63:1022–1028
Kongsawadworakul P, Viboonjun U, Romruensukharom P, Chantuma P, Ruderman S, Chrestin H (2009) The leaf, inner bark and latex cyanide potential of Hevea brasiliensis: evidence for involvement of cyanogenic glucosides in rubber yield. Phytochemistry 70:730–739
Kristensen C, Morant M, Olsen CE, Ekstrøm CT, Galbraith DW, Møller BL, Bak S (2005) Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc Natl Acad Sci USA 102(5):1779–1784
Krothapalli K, Buescher EM, Li X, Brown E, Chapple C, Dilkes BP, Tuinstra MR (2013) Forward genetics by genome sequencing reveals that rapid cyanide release deters insect herbivory of Sorghum bicolor. Genetics 195(2):309–318. https://doi.org/10.1534/genetics.113.149567
Lai D, Abou Hachem M, Robson F, Olsen CE, Wang TL, Møller BL, Takos AM, Rook F (2014) The evolutionary appearance of non-cyanogenic hydroxynitrile glucosides in the Lotus genus is accompanied by the substrate specialization of paralogous β–glucosidases resulting from a crucial amino acid substitution. Plant J 79(2):299–311. https://doi.org/10.1111/tpj.12561
Laursen T, Møller BL, Bassard J-E (2015) Plasticity of specialized metabolism as mediated by dynamic metabolons. Trends Plant Sci 20:20–32
Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L, Campbell BC, Hu W, Innes DJ, Han X, Cruickshank A, Dai C, Frère C, Zhang H, Hunt CH, Wang X, Shatte T, Wang M, Su Z, Li J, Lin X, Godwin ID, Jordan DR, Wang J (2013) Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun. https://doi.org/10.1038/ncomms3320
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47(W1):W636–W641. https://doi.org/10.1093/nar/gkz268
McCormick RF, Truong SK, Sreedasyam A, Jenkins J, Shu S, Sims D, Kennedy M, Amirebrahimi M, Weers BD, McKinley B, Mattison A, Morishige DT, Grimwood J, Schmutz J, Mullet JE (2018) The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J 93(2):338–354
McKey D (1974) Adaptive patterns in alkaloid physiology. Am Nat 108:305–320
McKinley B, Rooney W, Wilkerson C, Mullet J (2016) Dynamics of biomass partitioning, stem gene expression, cell wall biosynthesis, and sucrose accumulation during development of Sorghum bicolor. Plant J 88(4):662–680. https://doi.org/10.1111/tpj.13269
McKinley BA, Casto AL, Rooney WL, Mullet JE (2018a) Developmental dynamics of stem starch accumulation in Sorghum bicolor. Plant Direct 2:e00074. https://doi.org/10.1002/pld3.74
McKinley BA, Olson SN, Ritter KB, Herb DW, Karlen SD, Lu F, Ralph J, Rooney WL, Mullet JE (2018b) Variation in energy Sorghum hybrid TX08001 biomass composition and lignin chemistry during development under irrigated and nonirrigated field conditions. PLoS ONE 13(4):e0195863. https://doi.org/10.1371/journal.pone.0195863
Miller RE, Gleadow RM, Cavagnaro T (2014) Age versus stage: does ontogeny modify the effect of phosphorus and arbuscular mycorrhizas on above- and below-ground defence in forage sorghum? Plant Cell Environ 37:929–942
Møller BL (2010) Dynamic metabolons. Science 330:1328
Møller BL, Conn EE (1979) The biosynthesis of cyanogenic glucosides in higher plants N-Hydroxytyrosine as an intermediate in the biosynthesis of dhurrin by Sorghum bicolor (L.) Moench. J Biol Chem 254(17):8575–8583
Møller BL, Laursen T (2021) Metabolons and bio-condensates: the essence of plant plasticity and the key elements in development of green production systems. Adv Bot Res 97:185–223
Montini L, Crocoll C, Gleadow R, Motawia M, Janfelt C, Bjarnholt N (2020) Metabolite dynamics during Sorghum grain germination visualised by MALDI-mass spectrometry imaging. Plant Physiol 183:925–942
Morant AV, Bjarnholt N, Kragh ME, Kjaergaard CH, Jorgensen K, Paquette SM, Piotrowski M, Imberty A, Olsen CE, Møller BL, Bak S (2008a) The β-Glucosidases responsible for bioactivation of hydroxynitrile glucosides in Lotus japonicus. Plant Physiol 147(3):1072–1091. https://doi.org/10.1104/pp.107.109512
Morant AV, Jørgensen K, Kristensen C, Paquette SM, Sanchez-Perez R, Møller BL, Bak S (2008b) β-glucosidases as detonators of plant chemical defense. Phytochemistry 69:1795–1813
Mullet J, Morishige D, McCormick R, Truong S, Hilley J, McKinley B, Anderson R, Olson SN, Rooney W (2014) Energy Sorghum—a genetic model for the design of C4 grass bioenergy crops. J Exp Bot 65(13):3479–3489. https://doi.org/10.1093/jxb/eru229
Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE (2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic lowering in sorghum. Proc Natl Acad Sci USA 108(16):469–474. https://doi.org/10.1073/pnas.1106212108
Myrans H, Vandegeer RK, Henry RJ, Gleadow RM (2021) Nitrogen availability and allocation in sorghum and its wild relatives: divergent roles for cyanogenic glucosides. J Plant Physiol JPLPH 258:153393
Neilson EH, Goodger JQD, Woodrow IE, Møller BL (2013) Plant chemical defense: at what cost? Trends Plant Sci 18(5):250–258
Nielsen KA, Tattersall DB, Jones PR, Møller BL (2008) Metabolon formation in dhurrin biosynthesis. Phytochemistry 69:88–98
Nielsen LJ, Stuart P, Pičmanová M, Rasmussen S, Olsen CE, Harholt J, Møller BL, Bjarnholt N (2016) Dhurrin metabolism in the developing grain of Sorghum bicolor (L.) Moench investigated by metabolite profiling and novel clustering analyses of time-resolved transcriptomic data. BMC Genomics 17:1021
Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jørgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488(7412):531–534. https://doi.org/10.1038/nature11285
O’Donnell NH, Møller BL, Neale AD, Hamill JD, Blomstedt CK, Gleadow RM (2013) Effects of PEG-induced osmotic stress on growth and dhurrin levels of forage Sorghum. Plant Physiol Biochem 73:83–92
Olson SN, Ritter K, Rooney W, Kemanian A, McCarl BA, Zhang Y, Hall S, Packer D, Mullet J (2012) High biomass yield energy Sorghum: developing a genetic model for C4 grass bioenergy crops. Biofuels Bioprod Biorefin 6(6):640–655. https://doi.org/10.1002/bbb.1357
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev IV, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash T, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Mehboob-ur-Rahman WD, Westhoff P, Mayer KFX, Messing J, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295. https://doi.org/10.1038/nbt.3122
Pičmanová M, Neilson Elizabeth H, Motawia Mohammed S, Olsen Carl E, Agerbirk N, Gray Christopher J, Flitsch S, Meier S, Silvestro D, Jørgensen K, Sánchez-Pérez R, Møller Birger L, Bjarnholt N (2015) A recycling pathway for cyanogenic glycosides evidenced by the comparative metabolic profiling in three cyanogenic plant species. Biochem J 469(3):375–389. https://doi.org/10.1042/bj20150390
Poulton JE (1990) Cyanogenesis in Plants. Plant Physiol 94:401–405
Quinby JR (1974) Sorghum improvement and the genetics of growth. Texas agricultural experiment station
Rooney WL, Blumenthal J, Bean B, Mullet JE (2007) Designing Sorghum as a dedicated bioenergy feedstock. Biofuels Bioprod Biorefin 1(2):147–157. https://doi.org/10.1002/bbb.15
Rosati VC, Blomstedt CK, Møller BL, Garnett T, Gleadow R (2019) The interplay between drought stress, dhurrin and nitrate in the low-cyanogenic Sorghum mutant adult cyanide deficient class 1. Front Plant Sci 10:1458
Sakurada K, Ikegaya H, Ohta H, Fukushima H, Akutsu T, Watanabea K (2009) Effects of oximes on mitochondrial oxidase activity. Toxicol Lett 189:110
Saunders JA, Conn EE (1978) Presence of the cyanogenic glucoside dhurrin in isolated vacuoles from Sorghum. Plant Physiol 61:154–157
Schmidt FB, Cho SK, Olsen CE, Yang SW, Møller BL, Jørgensen K (2018) Diurnal regulation of cyanogenic glucoside biosynthesis and endogenous turnover in cassava. Plant Direct. https://doi.org/10.1002/pld3.38
Simon J, Gleadow RM, Woodrow IE (2010) Allocation of nitrogen to chemical defence and plant functional traits is constrained by soil N. Tree Physiol 30:1111–1117. https://doi.org/10.1093/treephys/tpq049
Sohail MN, Blomstedt CK, Gleadow RM (2020) Allocation of resources to cyanogenic glucosides does not incur a growth sacrifice in Sorghum bicolor (L.) Moench. Plants 9:1791. https://doi.org/10.3390/plants9121791
Sørensen M, Neilson EH, Møller BL (2018) Oximes: unrecognized chameleons in general and specialized plant metabolism. Mol Plant 11:95–117. https://doi.org/10.1016/j.molp.2017.12.014
Takos AM, Rook F (2012) Why biosynthetic genes for chemical defense compounds cluster. Trends Plant Sci 17(7):383–388. https://doi.org/10.1016/j.tplants.2012.04.004
Takos AM, Knudsen C, Lai D, Kannangara R, Mikkelsen L, Motawia MS, Olsen CE, Sato S, Tabata S, Jørgensen K, Møller BL, Rook F (2011) Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J 68(2):273–286. https://doi.org/10.1111/j.1365-313X.2011.04685.x
Tattersall DB, Bak S, Jones PR, Olsen CE, Nielsen JK, Hansen ML, Hoj PB, Moller BL (2001a) Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science 293:1826–1828. https://doi.org/10.1126/science.1062249
Tattersall DB, Bak S, Jones PR, Olsen CE, Nielsen JK, Hansen ML, Hoj PB, Møller BL (2001b) Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science 293(5536):1826–1828. https://doi.org/10.1126/science.1062249
Thayer SS, Conn EE (1981) Subcellular localisation of dhurrin β-glucosidase and hydroxynitrilelayse in the mesophyll cells of Sorghum leaf blades. Plant Physiol 67:617–622
Thorsøe KS, Bak S, Olsen CE, Imberty A, Breton C, Møller BL (2005) Determination of catalytic key amino acids and UDP sugar donor specificity of the cyanohydrin glycosyltransferase UGT85B1 from Sorghum bicolor molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol 139(2):664–673
Woodrow I, Slocum D, Gleadow R (2002) Influence of water stress on cyanogenic capacity in Eucalyptus cladocalyx. Funct Plant Biol 29:103–110
Yu G, Smith D, Zhu H, Guan Y, Lam T (2017) ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36
Acknowledgements
This work was funded in part by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-SC0018409) to JM, by an Australian Research Council Discovery Project DP180101011 to RMG and BLM and by a Novo Nordisk Foundation Distinguished Investigator Grant NNF 0054563 to BLM. We thank Alicia Quinn for assistance with measuring dhurrin concentrations. We thank Ka Man Jasmine Yu for preparing tissue and RNA for analysis of gene expression in TX08001 phytomers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gleadow, R.M., McKinley, B.A., Blomstedt, C.K. et al. Regulation of dhurrin pathway gene expression during Sorghum bicolor development. Planta 254, 119 (2021). https://doi.org/10.1007/s00425-021-03774-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03774-2