Abstract
Main conclusion
Carbonic anhydrases CA1 and CA4 attenuate plant immunity and can contribute to altered disease resistance levels in response to changing atmospheric CO2 conditions.
Abstract
β-Carbonic anhydrases (CAs) play an important role in CO2 metabolism and plant development, but have also been implicated in plant immunity. Here we show that the bacterial pathogen Pseudomonas syringae and application of the microbe-associated molecular pattern (MAMP) flg22 repress CA1 and CA4 gene expression in Arabidopsis thaliana. Using the CA double-mutant ca1ca4, we provide evidence that CA1 and CA4 play an attenuating role in pathogen- and flg22-triggered immune responses. In line with this, ca1ca4 plants exhibited enhanced resistance against P. syringae, which was accompanied by an increased expression of the defense-related genes FRK1 and ICS1. Under low atmospheric CO2 conditions (150 ppm), when CA activity is typically low, the levels of CA1 transcription and resistance to P. syringae in wild-type Col-0 were similar to those observed in ca1ca4. However, under ambient (400 ppm) and elevated (800 ppm) atmospheric CO2 conditions, CA1 transcription was enhanced and resistance to P. syringae reduced. Together, these results suggest that CA1 and CA4 attenuate plant immunity and that differential CA gene expression in response to changing atmospheric CO2 conditions contribute to altered disease resistance levels.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants have evolved a complex immune system to regulate survival from attack by pathogenic microbes and herbivorous insects. Upon perception of microbe-associated molecular patterns (MAMPs) by pattern recognition receptors (PRRs), defense responses are activated, including stomatal closure, production of reactive oxygen species, MAP kinase activation, hormonal signaling, and massive transcriptional reprogramming, which leads to the production of defensive compounds that limit pathogen ingress (Tsuda and Katagiri 2010; Zipfel and Robatzek 2010; Couto and Zipfel 2016). These induced signal outputs collectively lead to pattern-triggered immunity (PTI), which forms the first layer of plant defense to invading microbes. Evidence is accumulating that changing climate conditions can have profound effects on plant resistance pathways (Noctor and Mhamdi 2017; Kazan 2018; Velasquez et al. 2018). Atmospheric CO2 is an important parameter of climate change. Changes in atmospheric CO2 levels can affect disease development in diverse plant-pathogen interactions (Chakraborty et al. 2000; Garrett et al. 2006; Zavala et al. 2013; Mhamdi and Noctor 2016; Zhou et al. 2017; Williams et al. 2018b). Elevated CO2 caused reduced multiplication of potato virus Y in tobacco plants (Matros et al. 2006) and decreased downy mildew severity in soybean plants (Eastburn et al. 2010). In contrast, the susceptibility of wheat plants to Fusarium pseudograminearum was increased by elevated CO2 (Melloy et al. 2014). In tomato, elevated CO2 levels rendered the plants more resistant to Pseudomonas syringae pv. tomato DC3000 (Pst), while the level of resistance against Botrytis cinerea decreased (Zhang et al. 2015). Furthermore, exposure of the model plant species Arabidopsis thaliana (hereafter Arabidopsis) to pre-industrial, current and future levels of atmospheric CO2 uncovered marked effects on plant immunity against diverse (hemi) biotrophic and necrotrophic pathogens (Mhamdi and Noctor 2016; Zhou et al. 2017, 2019; Willams et al. 2018b). Changes in atmospheric CO2 levels not only affect plant-pathogen interactions, but also impact the interaction of plants with mutualistic mycorrhizal fungi and plant growth-promoting rhizobacteria (Werner et al. 2018; Williams et al. 2018a). Hence, to produce climate resilient crops in the future, it is important to understand how changes in atmospheric CO2 levels impact plant–microbe interactions.
Carbonic anhydrases (CAs) are metalloenzymes that were initially purified from red blood cells and mainly function as catalysts in the interconversion of CO2 and bicarbonate (Meldrum and Roughton 1933). There are at least five distinct CA families (α, β, γ, δ, and ε CAs), three of which (α, β, and γ CAs) are ubiquitously distributed among animal, plant, and bacterial species. The widespread distribution and abundance of these CA families underline their evolutionary importance throughout the kingdom of life. βCAs represent the most prominent group of CAs in plants. They are involved in a wide range of biological processes, including CO2 homeostasis, stomatal aperture, respiration, photosynthesis, pH regulation, and anther cell differentiation (Henry 1996; Smith and Ferry 2000; Hu et al. 2010; Engineer et al. 2014; Huang et al. 2017). Arabidopsis contains six βCA genes (AtβCA1-6), which are mostly expressed in aboveground tissues (Wang et al. 2014). Several studies reported on the implication of CAs in plant defense. For instance, the expression of a plastidic CA gene was found to be repressed in potato leaves upon challenge with the potato late blight pathogen Phytophthora infestans (Restrepo et al. 2005). Similarly, expression of the CA gene TC52686 in grapevine was suppressed during infection with the grapevine downy mildew pathogen Plasmopara viticola (Polesani et al. 2008). In contrast, five CA proteins were shown to be more abundant in a proteomic analysis of non-heading Chinese cabbage infected with the downy mildew Hyaloperonospora parasitica (Sun et al. 2014). These reported alterations in CA transcript or protein levels in pathogen-infected plants point to a potential role of CAs in plant defense. Indeed, a positive role in plant defense was demonstrated for a chloroplast-localized CA of tobacco, also known as SALICYLIC ACID (SA)-BINDING PROTEIN 3 (SABP3), as silencing of this CA gene in Nicotiana benthamiana leaves suppressed the hypersensitive response mediated by the Pto:avrPto resistance gene:effector gene pair (Slaymaker et al. 2002). Moreover, CA-silenced N. benthamiana was more susceptible to P. infestans than the wild type, supporting the notion that CAs contribute to disease resistance (Restrepo et al. 2005).
Despite the accumulating evidence for a role of CAs in plant immunity (Wang et al. 2009), little is known about how their regulation or action affects plant-pathogen interactions. CAs are mainly known as responders and actors in atmospheric CO2-mediated signaling. For example, under elevated CO2 conditions, both transcript abundance and enzymatic activity of CAs have been shown to decrease in several plant species (Porter and Grodzinski 1984; Webber et al. 1994; Majeau and Coleman 1996). Moreover, stomatal closure under high CO2 conditions and stomatal opening under low CO2 conditions is hampered in the Arabidopsis CA double-mutant ca1ca4 (Hu et al. 2010). Stomata are entry points of many leaf pathogens. Since activation of PTI triggers the closure of stomata to prevent pathogen entry (Melotto et al. 2008), changes in CA-mediated stomatal aperture may impact disease resistance. Recently, Medina-Puche et al. (2017) reported that several Arabidopsis CAs interact with the transcriptional coregulator NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and NONRECOGNITION OF BTH-4 (NRB4), thereby modulating the perception of the plant defense hormone salicylic acid (SA) in plants. SA is produced during the onset of PTI and plays an important regulatory role in plant immunity (Klessig et al. 2018). Hence, under changing atmospheric CO2 conditions, CA-mediated changes in SA responses may have an effect on the level of disease resistance. Previous observations that SA-dependent defenses in Arabidopsis are modulated under changing atmospheric CO2 conditions (Mhamdi and Noctor 2016; Williams et al. 2018b) support this hypothesis.
Given the importance of CA1 and CA4 in Arabidopsis’ responsiveness to changing CO2 levels (Hu et al. 2010), we chose to investigate the role of these two CAs in plant immunity and determined their effect on CO2-modulated defense using the model plant-pathogen system Arabidopsis P. syringae. We provide evidence that suppression of CA1 and CA4 gene expression is involved in the plant defense response to P. syringae infection and that CA1 and CA4 act as negative regulators of plant immunity, likely through antagonizing SA-mediated signaling. We also found that differential expression of CA1 under different atmospheric CO2 conditions is correlated with an altered level of disease resistance against P. syringae and that CA1 and CA4 are required for the effects of CO2 on disease resistance against P. syringae.
Materials and methods
Cultivation of plants and bacterial strains
For experiments with soil-grown plants, seeds of Arabidopsis thaliana accession Col-0 (Arabidopsis Biological Resource Center (ABRC) stock number CS1092) and mutant ca1ca4 (Hu et al. 2010; kindly provided by Julian Schroeder, UCSD, San Diego, CA, USA) were sown on autoclaved river sand. Two weeks later, seedlings were transferred to 60-ml pots containing a sand/potting soil mixture that was autoclaved twice for 20 min. Plants were grown in a climate chamber with a 10-h day at 20 °C and 14-h night at 18 °C cycle (350 µmol m−2 s−1) with 70% relative humidity. For experiments with different atmospheric CO2 treatments, 2-week-old seedlings in 60-ml pots either stayed in the growth room (ambient; 450 ppm) or were transferred to similar growth rooms with exactly the same conditions, except for CO2 levels, which were high (800 ppm), or low (150 ppm; Zhou et al. 2017). Plants were grown for the remainder of the experiment under different CO2 conditions. The technical specifications of the CO2-controlled growth chambers used in this study were described in detail by Temme et al. (2015).
For experiments with in vitro-grown plants, seeds of Arabidopsis accession Col-0 and mutants aba2-1 (Koornneef et al. 1982; ABRC stock number CS156), coi1-1 (Feys et al. 1994; kindly provided by Jane Glazebrook, University of Minnesota, St. Paul, MN, USA), npr1-1 (Cao et al. 1994; ABRC stock number N3726), ein2-1 (Guzman and Ecker 1990; ABRC stock number N3071), and fls2 (Shan et al. 2008; ABRC stock number SALK_141277) were surface sterilized in gas of a mixture of household chlorine (Glorix original, Unilever, Vlaardingen, the Netherlands) and HCl (37%; 97:3) for 3–4 h. Sterile seeds were subsequently sown on agar plates or in liquid. The agar plates contained Murashige and Skoog (MS) medium (Duchefa Biochemie, Haarlem, the Netherlands), pH 5.9, supplemented with 5 mM MES buffer, 10 g l−1 sucrose and 0.85% (w/v) plant agar (Duchefa Biochemie). When plants were 2-weeks old, they were transferred to liquid medium to be treated with flg22 (see “MAMP treatment”). For experimental conditions in which seeds were sown immediately in liquid MS, see “MAMP treatment”.
Pseudomonas syringae pv. tomato DC3000 (Pst) and its corresponding effector-deficient mutant Pst hrpA− (de Torres et al. 2003; Truman et al. 2006), and Pseudomonas syringae pv. maculicola ES4326 (Psm) and its corresponding coronatine-deficient mutant Psm cor− (Dong et al. 1991) were grown on King’s B medium (King et al. 1954) agar plates supplemented with 50 µg ml−1 rifampicine at 28 °C.
Pseudomonas syringae inoculation and bioassay
Pseudomonas syringae inoculation and the disease resistance assay were performed as described (Van Wees et al. 2013). For dip inoculation, the bacterial inoculum was diluted to a final concentration of 5 × 107 cfu ml−1 of 10 mM MgSO4 containing 0.015% (v/v) Silwet L-77 (Van Meeuwen Chemicals, Weesp, the Netherlands). For pressure infiltration, the bacterial suspension was adjusted to a concentration of 4 × 107 cfu ml−1 unless specified otherwise. Bacterial growth in planta was determined as described (Zhou et al. 2019). Eight biological replicates were included for each time point.
Pseudomonas syringae disease symptoms were scored in the following classes according to their severity: class 1, 0–10% chlorotic or water-soaked area per leaf; class 2, 10–50% chlorotic or water-soaked area per leaf; class 3, > 50% chlorotic or water-soaked area per leaf. Six fully grown and morphologically similar leaves per plant were chosen for scoring and 12 plants were scored per treatment. The average P. syringae disease index per plant was calculated using the formula:
c, the value of the class (1, 2, or 3). The resulting continuous data were tested for normal distribution by the Shapiro–Wilk test and if they passed, they were subsequently subjected to parametric tests for statistical analysis as indicated in the legends.
Botrytis cinerea bioassay
Botrytis cinerea strain B05.10 (Van Kan et al. 1997) was used for the inoculation of 4-week-old plants. Spore inoculation and disease resistance assay were performed as described previously (Van Wees et al. 2013; Zhou et al. 2019). Disease symptoms were scored at 3 days after inoculation. The average disease index was calculated similarly as described above for the P. syringae disease index.
MAMP treatment
For gene expression analysis of plants treated with the MAMPs flg22 or nlp20, 2-week-old in vitro-grown seedlings were transferred from agar plates to 24-well plates containing 1.5 ml of liquid MS with 5 mM MES per well and kept overnight at room temperature. Subsequently, a solution of 0.5 ml of MS + MES supplemented with flg22 (Sigma), or nlp20 (kind gift of Thorsten Nürnberger, Universität Tübingen, Germany (Böhm et al. 2014)) was added to obtain the final concentration (as indicated in the figure legends). The rosette leaves or the whole seedlings were harvested for RNA extraction at indicated time points.
For the growth inhibition assay, seeds of Col-0 and ca1ca4 were surface sterilized and sown in 96-well plates with 200 ml liquid MS + MES per well, supplemented or not with flg22 (1, 10, or 100 nM) from a 100 mM stock solution of flg22. The dry weight was measured when the seedlings were 2-weeks old.
ROS measurement
For the ROS assay, plants were grown at 20 °C in an 8-h light/16-h dark cycle in growth chambers. Leaf discs from 5-week-old plants were floated on water overnight. The water was replaced with 100 µl of a solution containing 20 µM luminol (Sigma), 1 µg horseradish peroxidase (Fluka, Buchs, Switzerland) and 100 nM flg22. ROS production was measured as previously described (Mersmann et al. 2010; Roux et al. 2011). Twelve leaf discs from 5-week-old plants were used for each condition. Luminescence of each sample was measured over 60 min continuously using a high-resolution photon counting system (HRPCS218, Photek, East Sussex, UK) coupled to an aspherical wide lens (Sigma).
Gene expression by qRT-PCR
Total RNA isolation and qRT-PCR were performed as described previously (Oñate-Sánchez and Vicente-Carbajosa 2008; Zhou et al. 2017), using the constitutively expressed reference gene At1g13320 (Czechowski et al. 2005), encoding protein phosphatase PP2AA3, and the \(2^{{ - \Delta \Delta C_{{\text{T}}} }}\) method (Schmittgen and Livak 2008) to calculate relative changes in gene expression. Three biological replicates were taken for each data point. Primers used for qRT-PCR are listed in Supplemental Table S1.
Stomatal aperture measurement
To measure stomatal aperture, a modified protocol of dental resin impressions was used (Geisler et al. 2000; Zhou et al. 2017). Stomata were photographed under an Olympus microscope. Analysis D Olympus Software was used to examine the stomata on the pictures taken. Stomatal aperture was assessed by measuring the width and length of the stomata. At least 20–30 observations per leaf were recorded on at least six leaves per treatment.
Results
Repression of CA1 and CA4 expression upon infection by P. syringae independently of type-III effectors and coronatine
Transcriptional repression of CA genes in response to attack by diverse pathogens has been reported for various plant species. Genevestigator analysis (Zimmermann et al. 2004) of the six β-group members of the Arabidopsis CA genes shows that CA1, CA2, CA4, and CA5 display a predominantly reduced expression pattern in response to infection by diverse plant pathogens, while CA3 and CA6 show a more variable profile (Supplemental Table S2). For this study, we chose to investigate the role of CA1 and CA4 in Arabidopsis immunity to P. syringae under ambient and altered CO2 conditions, because of their previously reported role in CO2-mediated responses (Hu et al. 2010). We first monitored the expression of CA1 and CA4 upon P. syringae infection of Arabidopsis cultivated under ambient CO2 conditions. In mock-infiltrated Col-0 leaves, CA1 and CA4 showed a similar basal expression pattern over time, in which the highest level was reached at 24 h after mock treatment (11:00 am) (Fig. 1a, b). This corroborates with a previously reported finding on diurnal rhythm of CA gene expression in Chlamydononas reinhardtii (Fujiwara et al. 1996), which may be associated with diurnal variations in cellular CO2 levels. At 24 h after pressure infiltration of Arabidopsis leaves with Pst, the expression levels of CA1 and CA4 were significantly suppressed in comparison to the mock treatment (Fig. 1a, b). Together with the Genevestigator results (Supplemental Table S2), these results suggest that repression of CA gene expression is a common plant response to pathogen infection.
Pseudomonas syringae represses the expression of CA1 and CA4 independently of hrpA-dependent effectors and coronatine. aCA1 and bCA4 expression levels relative to the reference gene At1g13320 in leaves of 4-week-old Col-0 plants at 3, 8, and 24 h after pressure infiltration with mock (10 mM MgSO4) or Pst (4 × 106 cfu ml−1). Asterisks indicate statistically significant differences between mock and Pst treatment at specific time points (Student’s t test; *P < 0.05; **P < 0.001; ***P < 0.0001; ns not significant). cCA1 and dCA4 expression levels relative to the reference gene At1g13320 in leaves of 4-week-old Col-0 at 24 h after pressure infiltration with mock (10 mM MgSO4), Psm or Psm cor− (1 × 107 cfu ml−1), or Pst or Pst hrpA− (1 × 108 cfu ml−1). Different letters indicate statistically significant differences between treatments (one-way ANOVA; Fisher’s LSD test; P < 0.05). Error bars represent SD, n = 3 plants
Pseudomonas syringae produces a suite of effector molecules, including the phytotoxin coronatine, which act to suppress plant defenses and promote infection (Mittal and Davis 1995; Brooks et al. 2005; Dou and Zhou 2012). To determine whether these virulence factors have a role in the suppression of CA gene expression, we tested the effect of infection by a Pst hrpA− mutant, which is defective in the type-III secretion system that translocates effectors into the plant host cell, and a Psm cor− mutant, which is defective in coronatine production. We compared the CA1 and CA4 expression levels after infiltration of the leaves with the mutant strains versus their respective wild-type P. syringae strains Pst and Psm. Confirming the findings displayed in Fig. 1a, b, CA1 and CA4 were significantly repressed 24 h after infection with wild-type Pst and Psm (Fig. 1c, d). Infection by the mutant strains Pst hrpA− and Psm cor− repressed CA1 to the same extent as the respective wild-type P. syringae strains (Fig. 1c). Also, CA4 expression was significantly suppressed by the P. syringae mutants, although the effect of Pst hrpA− was less pronounced than that of wild-type Pst (Fig. 1d). Together, this suggests that repression of CA1 and CA4 in Arabidopsis by infection with P. syringae is largely independent of effectors and coronatine.
Suppression of CA gene expression is a MAMP-induced response
Next, we tested whether the suppression of CA gene expression by P. syringae might be a MAMP-induced response. To this end, we examined the expression pattern of CA1 and CA4 in response to treatment with flg22, the 22-amino acid immunogenic epitope of the bacterial MAMP flagellin. As shown in Fig. 2a, the expression of the flg22-induced marker gene FRK1 was significantly enhanced in Col-0 plants from 2 h after flg22 treatment onwards, indicating that the flg22 treatment had been effective. The expression of CA1 and CA4 was examined at 24 h after flg22 application in both wild-type Col-0 and the flg22 receptor mutant fls2 (Gómez-Gómez and Boller 2000; Shan et al. 2008). At 24 h after flg22 application, both CA1 and CA4 were significantly suppressed in Col-0 plants, whereas this repression by flg22 was compromised in the fls2 mutant (Fig. 2b). This indicates that the suppression of CA genes occurs downstream of the recognition of the MAMP flg22. Besides flg22, analysis of available Genevestigator microarray data also show repression of CA1 and/or CA4 by other defense elicitors, such as EF-Tu (elf18), necrosis-inducing Phytophthora protein 1 (NPP1), lipopolysaccharide (LPS), oligosaccharides (OGs), Serratia plymuthiaca HRO-C48 volatiles, and peptide 2 (Pep2; Supplementary Table S3). Also, the Hyaloperonospora arabidopsidis MAMPs necrosis and ethylene-inducing peptide (Nep1)-like proteins (HaNLPs) significantly repress CA1 and CA4 gene expression in Arabidopsis (Oome et al. 2014). Collectively, these results suggest that suppression of CA1 and CA4 gene expression is a general MAMP-induced response in Arabidopsis.
Expression of defense-related marker gene FRK1 and CA1 and CA4 in response to flg22 treatment. a Expression of FRK1 relative to the reference gene At1g13320 in response to treatment with flg22 (500 nM) or water (mock) in 2-week-old Col-0 seedlings at specified time points after treatment (30 min and 2, 4, and 6 h). Asterisks indicate statistically significant differences between mock and flg22 treatment at specific time points (Student’s t test; *P < 0.05; **P < 0.01; ns not significant). bCA1 and CA4 expression levels relative to the reference gene At1g13320 in 2-week-old seedlings of Col-0 and fls2, 24 h after flg22 (125 nM) or mock treatment. cCA1 expression levels relative to the reference gene At1g13320 in 2-week-old seedlings of Col-0, aba2-1, coi1-1, npr1-1, and ein2-1 at 8 h after flg22 (500 nM) or mock treatment. Asterisks indicate statistically significant differences between mock and flg22 treatment within each genotype (two-way ANOVA; Fisher’s LSD test; ***P < 0.001; **P < 0.01; *P < 0.05; ns not significant). Error bars represent SD, n = 3 plants
Plant hormones such as SA, ethylene (ET), jasmonic acid (JA), and abscisic acid (ABA) have all been implicated in the regulation of PTI (Tsuda and Katagiri 2010; Cao et al. 2011). To investigate whether these hormones play a role in MAMP-induced suppression of CA genes, we determined the expression of CA1 in response to flg22 treatment in mutants impaired in synthesis of ABA (aba2-1), or responsiveness to JA (coi1-1), SA (npr1-1), or ET (ein2-1). We observed that the suppression of CA1 by flg22 occurred to the same extent in the mutants aba2-1, coi1-1, and npr1-1 as in wild-type Col-0 (Fig. 2c). In contrast, the ein2-1 mutant did not display suppression of CA1 gene expression in response to flg22 treatment; however, it has been demonstrated that ET signaling is required for the steady-state expression of the Arabidopsis flg22 receptor gene FLS2 (Boutrot et al. 2010; Mersmann et al. 2010). The lack of flg22-mediated suppression of CA1 in ein2 may, therefore, be explained by a diminished recognition of flg22 rather than diminished signaling downstream of recognition by FLS2. The results with the ABA-, JA-, and SA-related mutants suggest that ABA-, JA-, and SA-dependent signaling are not likely to be important for in flg22-mediated suppression of CA1 gene expression.
Enhanced MAMP responsiveness in mutant ca1ca4
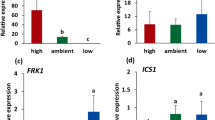
After MAMP perception, multiple responses are activated (e.g. oxidative burst, stomatal closure, and SA accumulation), which are often accompanied by a substantial transcriptional reprogramming (Yu et al. 2017). To gain insight in the function of CAs in plant immunity, we examined several flg22-induced responses in the ca1ca4 double mutant, which carries T-DNA insertions in the CA1 and CA4 genes (Hu et al. 2010). Figure 3a shows that flg22 induced FRK1 to a significantly higher level in ca1ca4 than in Col-0. Likewise, significantly augmented transcript levels of the SA biosynthesis gene ICS1 were induced in the ca1ca4 mutant after flg22 application (Fig. 3b). Transcript levels of the flg22 receptor gene FLS2 remained unaltered in ca1ca4 compared to Col-0 (Fig. 3c), indicating that the effects of CA1 and CA4 on defense-related gene expression are not due to differences in FLS2 expression. Similar to flg22, the oomycete MAMP nlp20, which is the active 20-amino acid immunogenic epitope of HaNLPs (Böhm et al. 2014), also induced enhanced transcript levels of the SA-responsive genes FRK1 (Fig. 3a) and ICS1 in ca1ca4 (Fig. 3b), suggesting that CA1 and CA4 broadly affect MAMP-induced transcription of the defense-related marker genes.
Augmented defense responses in the ca1ca4 mutant upon flg22 and nlp20 treatment. aFRK1, bICS1, and cFLS2 expression levels relative to the reference gene At1g13320 in 2-week-old seedlings of wild-type Col-0 and mutant ca1ca4 plants at 24 h after flg22 (500 nM), nlp20 (100 nM) or mock treatment. Asterisks indicate statistically significant differences between Col-0 and ca1ca4 within the same treatment (Student’s t test; *P < 0.05; **P < 0.01). Error bars represent SD, n = 3 seedlings. d Dry weight of 2-week-old seedlings of Col-0 and ca1ca4 cultivated in the presence of 0, 1, 10, or 100 nM flg22. Depicted are the averages of dry weight per seedling. Asterisks indicate statistically significant differences between Col-0 and ca1ca4 within the same treatment (Student’s t test; *P < 0.05). Error bars represent SD, n = 8 seedlings. e ROS burst induced by flg22 (100 nM) or mock treatment in leaf discs of Col-0, ca1ca4, and fls2. Depicted in the left panel are photon counts in each genotype after mock or flg22 treatment at indicated time points after flg22 treatment. The right panel depicts cumulative ROS production (photon counts) within 1 h after flg22 treatment. Asterisks indicate statistically significant differences between mutants and wild-type Col-0 (one-way ANOVA, Fisher’s LSD test; *P < 0.05; ***P < 0.001). Error bars represent SE, n = 4/12 (mock/flg22) leaf discs
Flg22 treatment causes strong growth inhibition in Arabidopsis seedlings (Gómez-Gómez et al. 1999). To assay for flg22-mediated growth inhibition, Col-0 and ca1ca4 seedlings were grown for 2 weeks in the presence of flg22 after which their dry weight was determined. Col-0 displayed more than 50% growth reduction after treatment of 1 nM flg22 (Fig. 3d). Interestingly, growth of ca1ca4 was reduced to a significantly greater extent than Col-0 after treatment with 1 nM and 10 nM flg22. Another feature of the flg22-induced defense response is the generation of reactive oxygen species (ROS; Nühse et al. 2007). The flg22-triggered ROS burst was significantly enhanced in the ca1ca4 mutant compared to that in Col-0 (Fig. 3e). Together, these results show that ca1ca4 plants display an augmented response to flg22 treatment, resulting in enhanced defense-related gene expression and a greater MAMP-mediated inhibition of seedling growth. From this, we conclude that CA1 and CA4 play a role in repressing MAMP-mediated defense responses.
CA1 and CA4 reduce resistance to Pst
To further investigate the function of CAs in plant disease resistance, we tested the responsiveness of Col-0 and ca1ca4 to infection with Pst. Pathogen-induced stomatal closure to inhibit pathogen entry has been established as an important defense response in plant resistance against P. syringae pathogens (Melotto et al. 2006). Therefore, we first determined whether CA1 and CA4, which are highly abundant in guard cells and control stomatal aperture (Hu et al. 2010), play a role in Pst-induced stomatal closure and opening. We tested stomatal responsiveness in Col-0 and ca1ca4 after treatment with Pst by dip inoculation, upon which the bacteria enter the leaf interior through stomatal openings. Consistent with previous results (Melotto et al. 2006), Col-0 plants reacted by closing their stomata between 1 and 2.5 h after Pst inoculation, and subsequent reopening at 4 h (Fig. 4a, b). As demonstrated previously (Hu et al. 2010), we observed that the stomatal aperture of ca1ca4 is significantly higher than that of Col-0 (Fig. 4a). Moreover, Pst-induced stomatal closure was delayed in ca1ca4 and became only apparent at 7 h after inoculation (Fig. 4a, b). The delayed stomatal closure triggered by Pst infection in ca1ca4 supports the notion that CA1 and CA4 are involved in Pst-induced stomatal movements.
CA1 and CA4 influence stomatal aperture and resistance to P. syringae. a Stomatal aperture in leaves of 4-week-old wild-type Col-0 and mutant ca1ca4 plants at 1, 2.5, 4, and 7 h after dip inoculation with Pst (5 × 107 cfu ml−1). Indicated are the averages of stomatal aperture (± SD) of six leaves. Different letters indicate statically significant differences between mock and Pst treatment within the same genotype (two-way ANOVA; Fisher’s LSD test; P < 0.01; ns not significant). Indications above the brackets specify whether there is an overall statistically significant difference between Col-0 and ca1ca4 at specific time points (***P < 0.001; *P < 0.05). Error bars represent SD, n = 6 leaves. b Stomatal apertures in Pst-treated leaves relative to mock (10 mM MgSO4)-treated leaves at 0, 1, 2.5, 4, 7 h after treatment. Asterisks indicate statistically significant differences between Col-0 and ca1ca4 at specific time points (Student’s t test; **P < 0.01; *P < 0.05; ns not significant). Error bars represent SD, n = 6 leaves. c Bacterial growth in 4-week-old Col-0 and ca1ca4 plants at 2 and 4 days after dip inoculation with Pst (5 × 107 cfu ml−1). Indicated are the averages of log10-transformed bacterial titers per leaf area. Asterisks indicate statistically significant differences between Col-0 and ca1ca4 at specific time points (Student’s t test; *P < 0.05; ns not significant). Error bars represent SD, n = 8 plants. d Disease symptom severity on 4-week-old Col-0 and ca1ca4 plants at 2, 3, and 4 days after pressure infiltration with Pst (6 × 105 cfu ml−1). Indicated is the average of the disease index calculated from the percentage of leaves in three different disease severity classes. Class 1, 0–10% chlorotic or water-soaked area per leaf; class 2, 10–50% chlorotic or waters-oaked area per leaf; class 3, > 50% chlorotic or waters-oaked area per leaf. Asterisks indicate statistically significant differences between Col-0 and ca1ca4 at specific time points (Student’s t test; **P < 0.01; ns not significant). Error bars represent SD, n = 12 plants
Next, we performed disease resistance assays with Col-0 and ca1ca4 in which growth of Pst and disease symptoms were monitored. Double-mutant ca1ca4 exhibited a bacterial titer that was significantly lower than that of Col-0 plants at 4 d after dip inoculation with Pst (Fig. 4c). Plants were also inoculated with Pst by pressure infiltration, which bypasses stomatal defense. Figure 4d shows that the ca1ca4 mutant developed significantly fewer disease symptoms than Col-0 plants at 3 and 4 days after infiltration. Together, these results show that CA1 and CA4 negatively impact disease resistance to Pst, with no clear role for stomatal defense, suggesting that post-invasion defenses are antagonized.
CA1 and CA4 antagonize SA-responsive gene expression upon Pst infection
In Arabidopsis, SA plays an important role in activating defense against P. syringae (Pieterse et al. 2012). To investigate whether CA1 and CA4 interfere with SA-dependent defenses, we infiltrated leaves of Col-0 and ca1ca4 plants with Pst and subsequently monitored expression levels of the SA-responsive genes PR1, PR2, FRK1, ICS1, WRKY22, and WRKY29. Figure 5 shows that all tested SA-responsive genes were induced by Pst to a significantly higher level in ca1ca4 than in Col-0 and most of the genes showed a slightly enhanced basal expression level in the ca1ca4 mutant. Conversely, the JA-responsive marker gene VSP2 was significantly suppressed in Pst-infected ca1ca4 plants (Fig. 5). These data suggest that CA1 and CA4 may modulate plant immunity by affecting SA- and JA-dependent plant responses.
SA- and JA-responsive gene expression in the mutant ca1ca4 upon infection by P. syringae. PR1, PR2, FRK1, ICS1, WRKY22, WRKY29, and VSP2 expression levels relative to the reference gene At1g13320 in 4-week-old Col-0 and ca1ca4 plants 24 h after infiltration with Pst (4 × 107 cfu ml−1) or 10 mM MgSO4 (mock). Error bars represent SD, n = 3 plants. Different letters indicate statically significant differences between Col-0 and ca1ca4 within the same treatment (two-way ANOVA; Fisher’s LSD test; P < 0.05). Indications above the brackets specify whether there is an overall statistically significant difference between mock and Pst treatment (**P < 0.01)
CA1 and CA4 are involved in atmospheric CO2-affected disease resistance against Pst
Atmospheric CO2 levels have been shown to influence plant development and defense (Velasquez et al. 2018). In Arabidopsis, plant growth and the level of resistance against P. syringae is also impacted by changes in atmospheric CO2 levels (Zhou et al. 2017, 2019). In the present study, we tested the role of CA1 and CA4 in the effect of changes in atmospheric CO2 levels on Arabidopsis disease resistance against Pst. We found that the disease resistance of Arabidopsis Col-0 plants against Pst was decreased under high CO2 (800 ppm) compared with that under ambient CO2 (450 ppm), whereas it was enhanced under low CO2 conditions (150 ppm; Fig. 6a), confirming previous findings (Zhou et al. 2017). In the ca1ca4 double mutant, the level of Pst resistance was at all three atmospheric CO2 levels as high as that observed in Col-0 under low CO2 (Fig. 6a), suggesting that in wild-type plants, the CAs play a role in the modulation of atmospheric CO2-affected disease resistance to Pst. This was confirmed by the observation that the Pst-mediated suppression of CA1 gene expression becomes stronger with decreasing CO2 concentrations in the atmosphere (Fig. 6b).
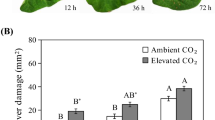
The role of CAs in atmospheric CO2-modulated disease resistance to Pst and B. cinerea. Arabidopsis wild-type Col-0 and mutant ca1ca4 plants were grown under high (800 ppm), ambient (450 ppm) and low (150 ppm) levels of atmospheric CO2 until 4-week old and dip inoculated with Pst (a, b) or drop inoculated with B. cinerea (c). a Disease severity in Col-0 and ca1ca4 at 4 d after dip inoculation with Pst (4 × 107 cfu ml−1). Shown is the average Pst disease index calculated from the percentage of six leaves per plant belonging to different disease severity classes. Error bars represent SD, n = 12 plants. bCA1 expression levels relative to the reference gene At1g13320 in Col-0 grown under high, ambient and low levels of atmospheric CO2 at 24 h after dip inoculation with Pst (1 × 108 cfu ml−1) or 10 mM MgSO4 (mock). Error bars represent SD, n = 3 plants. c Disease severity in Col-0 and ca1ca4 plants inoculated with B. cinerea (1 × 106 spores ml−1). Disease symptoms were scored 4 days after inoculation. Shown is the average of the disease index calculated from the percentage of leaves in four different disease severity classes. Error bars represent SD, n = 12 plants. Different letters indicate statistically significant differences between CO2 treatments within the same genotype. Indications above the brackets specify the interaction (Arabidopsis genotype × CO2 conditions) between Col-0 and ca1ca4 and the three CO2 conditions (two-way ANOVA; Fisher’s LSD test; *P < 0.05; ***P < 0.001; ns not significant).
In Arabidopsis, changes in atmospheric CO2 levels can also affect disease resistance against necrotrophic fungi (Williams et al. 2018b; Zhou et al. 2019). To test whether CAs play a role in this process, we tested the resistance of Arabidopsis plants to the necrotrophic pathogen B. cinerea at three different atmospheric CO2 levels. We found that high CO2-grown Col-0 plants developed less disease symptoms compared with plants grown under ambient and low CO2 conditions (Fig. 6c), confirming that Arabidopsis disease resistance against B. cinerea increases as atmospheric CO2 levels increase (Zhou et al. 2019). Mutant ca1ca4 also displayed increasing levels of disease severity with increasing CO2 levels, which was similar to that observed in Col-0 (Fig. 6c), suggesting that CA1 and CA4 do not influence atmospheric CO2-altered disease resistance to B. cinerea.
Discussion
During the last decade, our understanding of the mechanisms involved in plant immune signaling greatly increased (Couto and Zipfel 2016; Cheng et al. 2019; Nobori and Tsuda 2019). Evidence is accumulating that climate change parameters can have profound effects on plant immunity (Noctor and Mhamdi 2017; Kazan 2018; Velasquez et al. 2018). As one of the core characteristics of global climate change, the increasing atmospheric CO2 level has been shown to affect various plant-pathogen systems (Chakraborty et al. 2000; Garrett et al. 2006; Yáñez-López et al. 2014). In the pressent study, we revealed that the β-carbonic anhydrases CA1 and CA4 of Arabidopsis modulate plant immune responses and that they are likely involved in CO2-modulated plant defense against Pst.
Upon P. syringae infection, expression of the CA1 and CA4 genes in Arabidopsis was strongly repressed (Fig. 1). This occurred largely independently of hrpA-dependent effectors and coronatine (Fig. 1c, d). We further demonstrated that repression of CA1 and CA4 is triggered by the MAMPs flg22 and nlp20 (Fig. 2). This suggests that repression of CA1 and CA4 is part of the Arabidopsis defense response when under attack by P. syringae. The inability of the ein2-1 mutant to repress CA1 expression (Fig. 2c) most likely results from the strongly reduced expression of FLS2 in ein2-1 (Boutrot et al. 2010; Mersmann et al. 2010). However, a role for ET signaling in repression of CA1 expression downstream of flg22 recognition cannot be ruled out. Nonetheless, the repression of CA1 by flg22 occurred independently of ABA, JA, or SA signaling (Fig. 2c).
In Arabidopsis, perception of flg22 triggers multiple responses, such as activation of defense-related genes and growth inhibition (Yu et al. 2017). Our results with the double-mutant ca1ca4 show significantly enhanced expression levels of two defense-related marker genes, FRK1 and ICS1, as well as stronger growth inhibition compared to wild-type plants upon treatment with flg22 (Fig. 3). The FLS2 expression levels were unaffected by the ca1ca4 mutation (Fig. 3c), suggesting the enhanced activation of plant immune responses by flg22 is not likely due to an enhanced capacity of flg22 recognition. This is supported by the observation that another MAMP, nlp20, also triggered enhanced expression of the defense-related marker genes FRK1 and ICS1 in ca1ca4 (Fig. 3a, b). Collectively, these data indicate that suppression of CAs is part of the basal plant immune response, thereby positively contributing to the activation of defenses against the pathogen encountered.
CAs have been reported to control CO2 homeostasis and stomatal aperture. Consistent with previous findings (Henry 1996; Smith and Ferry 2000; Hu et al. 2010), our results showed that the ca1ca4 mutant displayed greater stomatal aperture than wild-type Col-0 plants (Fig. 4a). Moreover, stomatal closure, which is part of the defense response induced upon inoculation with Pst, is delayed in ca1ca4 as it is detected starting at 4 h while in Col-0, closure is already evident at 1 h after inoculation (Fig. 4a, b). This points to a positive role of the CAs in stomatal defenses. However, despite the larger opening of the stomata throughout the first 7 h and the delay in the stomatal closure response, the ca1ca4 mutant exhibited enhanced resistance to Pst compared to wild-type Col-0 in both the Pst dipping and infiltration assays (Fig. 4c, d). Although CA1 and CA4 positively regulate stomatal defenses, these results suggest that they negatively influence other post-invasion plant defense responses, likely those mediated by SA signaling, as the ca1ca4 mutant showed significantly augmented SA-responsive gene expression upon infection with Pst (Fig. 5).
Previous studies have shown a positive involvement of CAs in plant defense against avirulent Pst strains. For instance, in tobacco, silencing of the CA known as SA-BINDING PROTEIN 3 (SABP3), led to suppression of the Pto:avrPto-mediated hypersensitive defense response (Slaymaker et al. 2002). Also in Arabidopsis, the orthologue AtSABP3, which is also named CA1 (used in this study), is required for expression of full defense against the avirulent bacterial pathogen Pst carrying avrB (Wang et al. 2009). Our results showed that CA1 gene expression was similarly repressed upon infection by wild-type Pst and its correspondent effector mutant Pst hrpA− (Fig. 1c). Still, the CA1 protein could be a potential target of type-III effectors of Pst as the abundance of CA1 was shown to be reduced to a lesser extent upon infection by wild-type Pst than by its correspondent effector mutant Pst hrpA− (Jones et al. 2006). The dual role of CAs in defense against virulent and avirulent Pst strains suggests differential actions of CAs during compatible and incompatible interactions between a host and its pathogens.
One important aspect of prevailing climate changes is the elevation of atmospheric CO2 and this has boosted research on plant defenses under elevated CO2 conditions (Restrepo et al. 2005; Polesani et al. 2008; Noctor and Mhamdi 2017; Kazan 2018; Velasquez et al. 2018; Williams et al. 2018b). CAs are important enzymes in CO2 metabolism and we addressed whether they play a role in atmospheric CO2—affected plant disease resistance. Previous reports have shown that elevated CO2 increased the disease resistance of tomato plants against Pst (Li et al. 2014; Zhang et al. 2015). In contrast, we found enhanced disease susceptibility to P. syringae and increased resistance against B. cinerea in Arabidopsis plants grown at increasing CO2 levels (Fig. 6; Zhou et al. 2017, 2019). This suggests that the response of plants to changes in the level of atmospheric CO2 is plant species specific.
Arabidopsis defense against P. syringae and B. cinerea is largely regulated by SA and JA signaling, respectively (Glazebrook 2005). Previously, we showed that increasing atmospheric CO2 levels lowered the level of resistance against Pst, while the level of resistance against B. cinerea increased (Zhou et al. 2019). This opposite effect on resistance against the (hemi)biotroph Pst and the necrotroph B. cinerea is likely due to the antagonistic interaction between the SA and the JA defense pathways (Pieterse et al. 2012). In this study, we confirmed our previous observation that with increasing CO2 levels, Arabidopsis resistance against Pst decreases, while resistance against B. cinerea increases (Fig. 6). Moreover, we show that in mutant ca1ca4, the effect of the atmospheric CO2 level on Pst resistance is lost, while for B. cinerea, the CO2 effect on resistance remains unaltered in comparison to Col-0 (Fig. 6). These results suggest that CAs predominantly have an impact on SA-mediated resistance.
Williams et al. (2018b) also tested the effect of both elevated and sub-ambient levels of atmospheric CO2 on disease caused by necrotrophic (Plectosphaerella cucumerina) and biotrophic (H. arabidopsidis) pathogens. They observed enhanced resistance against the necrotroph under elevated CO2 conditions, corroborating our findings with B. cinerea (Fig. 6c). However, in contrast to our observations, resistance against the biotroph was also enhanced under elevated CO2 conditions. Similar observations were done by Mhamdi and Noctor (2016) who found enhanced resistance against both Pst and B. cinerea under conditions of elevated CO2. However, in the latter, study plants were grown at long day conditions (16 h light/8 h dark versus 10 h light/14 h dark in our study) and at very high CO2 levels (3000 ppm versus 800 ppm in our study), suggesting that effects of CO2 on pathogen resistance are conditionally determined by prevailing environmental factors. Interestingly, both Williams et al. (2018b) and Mhamdi and Noctor (2016) provided evidence that the effect of changed atmospheric CO2 levels on plant immunity is associated with cellular redox status. We found that the mutant ca1ca4 developed a stronger oxidative burst in response to flg22 treatment than did wild-type plants (Fig. 3e), confirming the notion that CAs may modulate plant immunity via changes in cellular redox processes. Different atmospheric CO2 levels may also directly affect the growth rate or pathogenicity of the microbial pathogens, but we did not test to what extend this contributed to the disease outcome in our experiments.
In conclusion, our results show that induction of defense responses in P. syringae-infected Arabidopsis plants results in the repression of CA1 and CA4 gene expression. This leads to the alleviation of CA-mediated suppression of SA-dependent defenses and consequently increased disease resistance against Pst (Fig. 7). Changes in atmospheric CO2 influence CA activity, which as a result impact SA-dependent defenses against Pst, possibly via changes in the cellular redox status (Mhamdi and Noctor 2016). Collectively, our results provide new leads for future investigations on plant adaptation to global environmental changes. A more comprehensive analysis of the exact function of CAs in plant defense, including the βCAs that were not investigated here, will be subject of future study.
A model of CO2-modulated, MAMP-induced suppression of CA1 and CA4 that alleviate attenuation of SA-dependent defenses during the plant immune response to infection by P. syringae. Upon attack by P. syringae, plants recognize the flg22 epitope of the MAMP flagellin, resulting in repression of CA1 and CA4 gene expression. In uninduced plants, CAs have an antagonizing effect on PTI-mediated responses. Recognition of flg22 results in suppression of CAs and increased defense-related gene expression, ultimately leading to enhancement of SA-dependent resistance to P. syringae and inhibition of plant growth. At a low atmospheric CO2 level, repression of CA1 by Pst is enhanced, while at a high CO2 level, CA1 repression is reduced. This contributes to an increase versus a decrease in resistance levels against Pst at low and high CO2 levels, respectively. Arrows, induction; blocked lines, repression
Author contribution statement
YZ and SCMVW designed the experiments. YZ performed most of experiments and analyzed the data. Other authors assisted in experiments and discussed the results. YZ, IAV-V, CMJP, and SCMVW wrote the manuscript.
Abbreviations
- CA:
-
Carbonic anhydrase
- ET:
-
Ethylene
- JA:
-
Jasmonic acid
- MAMP:
-
Microbe-associated molecular pattern
- Pst :
-
Pseudomonas syringae pv. tomato DC3000
- Psm :
-
Pseudomonas syringae pv. maculicola 4326
- PTI:
-
Pattern-triggered immunity
- SA:
-
Salicylic acid
References
Böhm H, Albert I, Oome S, Raaymakers TM, Van den Ackerveken G, Nürnberger T (2014) A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Path 10:e1004491
Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP (2010) Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc Natl Acad Sci USA 107:14502–14507
Brooks DM, Bender CL, Kunkel BN (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol Plant Pathol 6:629–639
Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6:1583–1592
Cao F, Yoshioka K, Desveaux D (2011) The roles of ABA in plant–pathogen interactions. J Plant Res 124:489–499
Chakraborty S, Tiedemann A, Teng P (2000) Climate change: potential impact on plant diseases. Environ Pollut 108:317–326
Cheng YT, Zhang L, He SY (2019) Plant-microbe interactions facing environmental challenge. Cell Host Microbe 26:183–192
Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16:537–552
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dong X, Mindrinos M, Davis KR, Ausubel FM (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3(1):61–72. https://doi.org/10.1105/tpc.3.1.61
Dou D, Zhou J-M (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12:484–495
de Torres M, Sanchez P, Fernandez-Delmond I, Grant M (2003) Expression profiling of the host response to bacterial infection the transition from basal to induced defense responses. Plant J 33:665–676
Eastburn DM, Degennaro MM, Delucia EH, Dermody O, Mcelrone AJ (2010) Elevated atmospheric carbon dioxide and ozone alter soybean diseases at SoyFACE. Glob Change Biol 16:320–330
Engineer C, Ghassemian M, Anderson J, Peck S, Hu H, Schroeder J (2014) Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513:246–250
Feys BJ, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6:751–759
Fujiwara S, Ishida N, Tsuzuki M (1996) Circadian expression of the carbonic anhydrase gene, Cah1, in Chlamydomonas reinhardtii. Plant Mol Biol 32:745–749
Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE (2006) Climate change effects on plant disease: genomes to ecosystems. Annu Rev Phytopahol 44:489–509
Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12:2075–2086
Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227
Gómez-Gómez L, Boller T (2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1012
Gómez-Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18:277–284
Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2:513–523
Henry RP (1996) Multiple roles of carbonic anhydrase in cellular transport and metabolism. Annu Rev Physiol 58:523–538
Hu H, Boisson Dernier A, Israelsson Nordström M, Böhmer M, Xue S, Ries A, Godoski J, Kuhn JM, Schroeder JI (2010) Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat Cell Biol 12:87–93
Huang J, Li Z, Biener G, Xiong E, Malik S, Eaton N, Zhao CZ, Raicu V, Kong H, Zhao D (2017) Carbonic anhydrases function in anther cell differentiation downstream of the receptor-like kinase EMS1. Plant Cell 29:1335–1356
Jones AM, Thomas V, Bennett MH, Mansfield J, Grant M (2006) Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol 142:1603–1620
Kazan K (2018) Plant-biotic interactions under elevated CO2: a molecular perspective. Environ Exp Bot 153:249–261
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307
Klessig DF, Choi HW, Dempsey DA (2018) Systemic acquired resistance and salicylic acid: past, present, and future. Mol Plant Microbe Interact 31:871–888
Koornneef M, Jorna M, Brinkhorst-Van der Swan D, Karssen C (1982) The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 61:385–393
Li X, Sun Z, Shao S, Zhang S, Ahammed GJ, Zhang G, Jiang Y, Zhou J, Xia X, Zhou Y, Yu J, Shi K (2014) Tomato-Pseudomonas syringae interactions under elevated CO2 concentration: the role of stomata. J Exp Bot 66:307–316
Majeau N, Coleman JR (1996) Effect of CO2 concentration on carbonic anhydrase and ribulose-1, 5-bisphosphate carboxylase/oxygenase expression in pea. Plant Physiol 112:569–574
Matros A, Amme S, Kettig B, Buck-Sorlin GH, Sonnewald U, Mock H-P (2006) Growth at elevated CO2 concentrations leads to modified profiles of secondary metabolites in tobacco cv. Samsun NN and to increased resistance against infection with potato virus Y. Plant Cell Environ 29:126–137
Medina-Puche L, Castello M, Canet J, Lamilla J, Colombo M, Tornero P (2017) β-carbonic anhydrases play a role in salicylic acid perception in Arabidopsis. PLoS ONE 12:e0181820
Meldrum NU, Roughton FJW (1933) Carbonic anhydrase. Its preparation and properties. J Physiol 80:113–142
Melloy P, Aitken E, Luck J, Chakraborty S, Obanor F (2014) The influence of increasing temperature and CO2 on Fusarium crown rot susceptibility of wheat genotypes at key growth stages. Eur J Plant Pathol 140:19–37
Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126:969–980
Melotto M, Underwood W, He S-Y (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol 46:101–122
Mersmann S, Bourdais G, Rietz S, Robatzek S (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154:391–400
Mhamdi A, Noctor G (2016) High CO2 primes plant biotic stress defences through redox-linked pathways. Plant Physiol 172:929–942
Mittal S, Davis KR (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 8:165–171
Nobori T, Tsuda K (2019) The plant immune system in heterogeneous environments. Curr Opin Plant Biol 50:58–66
Noctor G, Mhamdi A (2017) Climate change, CO2, and defense: the metabolic, redox, and signaling perspectives. Trends Plant Sci 22:857–870
Nühse TS, Bottrill AR, Jones AME, Peck SC (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J 51:931–940
Oñate-Sánchez L, Vicente-Carbajosa J (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1:93
Oome S, Raaymakers TM, Cabral A, Samwel S, Böhm H, Albert I, Nürnberger T, Van den Ackerveken G (2014) Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc Natl Acad Sci USA 111:16955–16960
Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Polesani M, Desario F, Ferrarini A, Zamboni A, Pezzotti M, Kortekamp A, Polverari A (2008) cDNA-AFLP analysis of plant and pathogen genes expressed in grapevine infected with Plasmopara viticola. BMC Genomics 9:142
Porter MA, Grodzinski B (1984) Acclimation to high CO2 in bean carbonic anhydrase and ribulose bisphosphate carboxylase. Plant Physiol 74:413–416
Restrepo S, Myers K, Del Pozo O, Martin G, Hart A, Buell C, Fry W, Smart C (2005) Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Mol Plant Microbe Interact 18:913–922
Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, De Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23:2440–2455
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Shan LB, He P, Li JM, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4:17–27
Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF (2002) The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA 99:11640–11645
Smith KS, Ferry JG (2000) Prokaryotic carbonic anhydrases. FEMS Microbiol Rev 24:335–366
Sun C, Wang L, Hu D, Riquicho ARM, Liu T, Hou X, Li Y (2014) Proteomic analysis of non-heading Chinese cabbage infected with Hyaloperonospora parasitica. J Proteomics 98:15–30
Temme AA, Liu JC, Cornwell WK, Cornelissen JHC, Aerts R (2015) Winners always win: growth of a wide range of plant species from low to future high CO2. Ecol Evol 5:4949–4961
Truman W, Zabala MT, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46:14–33
Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13:459–465
Van Kan J, Van't Klooster J, Wagemakers C, Dees D, Van der Vlugt-Bergmans C (1997) Cutinase A of Botrytis cinerea is expressed, but not essential, during penetration of gerbera and tomato. Mol Plant Microbe Interact 10:30–38
Van Wees SCM, Van Pelt JA, Bakker PAHM, Pieterse CMJ (2013) Bioassays for assessing jasmonate-dependent defenses triggered by pathogens, herbivorous insects, or beneficial rhizobacteria. Meth Mol Biol 1011:35–49
Velasquez A, Castroverde C, He S (2018) Plant-pathogen warfare under changing climate conditions. Curr Biol 28:619–634
Wang YQ, Feechan A, Yun BW, Shafiei R, Hofmann A, Taylor P, Xue P, Yang FQ, Xie ZS, Pallas JA, Chu CC, Loake GJ (2009) S-Nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J Biol Chem 284:2131–2137
Wang M, Zhang Q, Liu FC, Xie WF, Wang GD, Wang J, Gao QH, Duan K (2014) Family-wide expression characterization of Arabidopsis beta-carbonic anhydrase genes using qRT-PCR and Promoter: GUS fusions. Biochimie 97:219–227
Webber AN, Nie GY, Long SP (1994) Acclimation of photosynthetic proteins to rising atmospheric CO2. Photosynth Res 39:413–425
Werner GDA, Zhou YL, Pieterse CMJ, Kiers ET (2018) Tracking plant preference for higher-quality mycorrhizal symbionts under varying CO2 conditions over multiple generations. Ecol Evol 8:78–87
Williams A, Petriacq P, Beetling D, Cotton T, Ton J (2018a) Impacts of atmospheric CO2 and soil nutritional value on plant responses to rhizosphere colonization by soil bacteria. Front Plant Sci 9:1493
Williams A, Petriacq P, Schwarzenbacher R, Beerling D, Ton J (2018b) Mechanisms of glacial-to-future atmospheric CO2 effects on plant immunity. New Phytol 218:752–761
Yáñez-López R, Torres-Pacheco I, Guevara-González RG, Hernández-Zul MI, Quijano-Carranza JA, Rico-García E (2014) The effect of climate change on plant diseases. Afr J Biotechnol 11:2417–2428
Yu X, Feng B, He P, Shan L (2017) From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55:109–137
Zavala JA, Nabity PD, DeLucia EH (2013) An emerging understanding of mechanisms governing insect herbivory under elevated CO2. Annu Rev Entomol 58:79–97
Zhang S, Li X, Sun Z, Shao S, Hu L, Ye M, Zhou Y, Xia X, Yu J, Shi K (2015) Antagonism between phytohormone signalling underlies the variation in disease susceptibility of tomato plants under elevated CO2. J Exp Bot 66:1951–1963
Zhou Y, Vroegop-Vos I, Schuurink RC, Pieterse CMJ, Van Wees SCM (2017) Atmospheric CO2 alters resistance of Arabidopsis to Pseudomonas syringae by affecting abscisic acid accumulation and stomatal responsiveness to coronatine. Front Plant Sci 8:700
Zhou Y, Van Leeuwen SK, Pieterse CMJ, Bakker PAHM, Van Wees SCM (2019) Effect of atmospheric CO2 on plant defense against leaf and root pathogens of Arabidopsis. Eur J Plant Pathol 154:31–42
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Zipfel C, Robatzek S (2010) Pathogen-associated molecular pattern-triggered immunity: veni, vidi…? Plant Physiol 154:551–554
Acknowledgements
We thank Dr Jane Glazebrook for the Psm and Psm cor− bacteria. This work was supported by a China Scholarship Council (CSC) PhD scholarship (to Y.Z.), VIDI Grant no. 11281 of the Netherlands Organization for Scientific Research (NWO/STW to S.C.M.V.W), a grant from the Gatsby Charitable Foundation (to C.Z.), a long-term post-doctoral fellowship from the European Molecular Biology Organization (ALTF 657-2013; to D.V.d.D), and ERC Advanced Investigator Grant no. 269072 of the European Research Council (to C.M.J.P).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, Y., Vroegop-Vos, I.A., Van Dijken, A.J.H. et al. Carbonic anhydrases CA1 and CA4 function in atmospheric CO2-modulated disease resistance. Planta 251, 75 (2020). https://doi.org/10.1007/s00425-020-03370-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-020-03370-w