Abstract
Main conclusion
The HEUKCHEEM gene plays an important role in spine color formation. A white spine occurs due to two mutations in HEUKCHEEM and is closely related to the regional distribution of these mutants.
Abstract
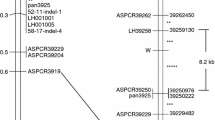
Mapping analysis revealed that the HEUKCHEEM gene is co-segregated with the B locus in the regulation of black spine color development in cucumber fruit. HEUKCHEEM induced the expression of the genes involved in the anthocyanin biosynthetic pathway, leading to the accumulation of anthocyanins in black spines. The transiently over-expressed HEUKCHEEM in cucumber and tobacco plants enhanced the expression of anthocyanin biosynthesis-related genes, leading to anthocyanin accumulation. However, two mutations—insertion of the 6994 bp mutator-like transposable element (MULE) sequence into the second intron and one single-nucleotide polymorphism (SNP) of C to T in the second exon of HEUKCHEEM—were identified in white spines, leading to no accumulation of anthocyanin biosynthesis-related gene transcripts and anthocyanins. Furthermore, association analysis using 104 cucumber accessions with different geographical origins revealed that the types of mutations in HEUKCHEEM are strongly linked to geographical origins. The MULE insertion is found extensively in cucumbers with white spines in East Asia and Australia. However, cucumbers with white spines in other areas could be significantly influenced by a single SNP mutation. Our results provide fundamental information on spine color development in cucumber fruits and spine color-based cucumber breeding programs.







Similar content being viewed by others
Abbreviations
- ANS:
-
Anthocyanin synthase
- (d)CAPS:
-
(Derived) Cleaved amplified polymorphic sequences
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol 4-reductase
- F3H:
-
Flavanone-3-hydroxylase
- INDEL:
-
Insertions and deletions
- MBW:
-
MYB-bHLH-WD40
- MULE:
-
Mutator-like transposable element
- SNP:
-
Single-nucleotide polymorphism
- UFGT:
-
UDP-glucose flavonoid 3-O-glucosyltransferase
- WD-RP:
-
WD-repeat protein
References
Azuma A, Kobayashi S, Goto-Yamamoto N, Shiraishi M, Mitani N, Yakushiji H, Koshita Y (2009) Color recovery in berries of grape (Vitis vinifera L.) ‘Benitaka’, a bud sport of ‘Italia’, is caused by a novel allele at the VvmybA1 locus. Plant Sci 176:470–478
Bendahmane A, Querci M, Kanyuka K, Baulcombe DC (2000) Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J 21:73–81
Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361
Che G, Zhang X (2019) Molecular basis of cucumber fruit domestication. Curr Opin Plant Biol 47:38–46
Chen C, Yin S, Liu X et al (2016) The WD-repeat protein CsTTG1 regulates fruit wart formation through interaction with the homeodomain-leucine zipper I protein Mict. Plant Physiol 171:1156–1168
Chezem WR, Clay NK (2016) Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131:26–43
Cowen NM, Heisel DB (1983) Inheritance of two genes for spine color and linkages in a cucumber cross. J Hered 74:308–309
Czemmel S, Heppel SC, Bogs J (2012) R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249(Suppl 2):S109–S118
Fanourakis NE, Simon PW (1987) Analysis of genetic linkage in the cucumber. J Hered 78:238–242
FAOSTAT (2018) Food and Agriculture Organization of the United Nations. Crop Production Data. http://www.fao.org/faostat/en/#data/QC
He X, Li Y, Lawson D, Xie DY (2017) Metabolic engineering of anthocyanins in dark tobacco varieties. Physiol Plant 159:2–12
Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9987
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071–1083
Jin W, Wang H, Li M, Wang J, Yang Y, Zhang X, Yan G, Zhang H, Liu J, Zhang K (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14:2120–2133
Khoo HE, Azlan A, Tang ST, Lim SM (2017) Anthocyanidins and anthocyanins: colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nur Res 61:1361779
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li Y, Wen C, Weng Y (2013) Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor Appl Genet 126:2187–2196
Li W, Ding Z, Ruan M, Yu X, Peng M, Liu Y (2017) Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci Rep 7:16861–16875
Liu S, Gu X, Miao H, Wang Y, Wng Y, Wehner T, Zhang S (2014) Molecular mapping and candidate gene analysis of black fruit spine gene in cucumber (Cucumis sativus L.). Sci Agric Sin 47:122–132
Liu Y, Tikunov Y, Schouten RE, Marcelis LFM, Visser RGF, Bovy A (2018) Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: a review. Front Chem 6:1–17
Liu M, Zhang C, Duan L, Luan Q, Li J, Yang A, Qi X, Ren Z (2019) CsMYB60 is a key regulator of flavonols and proanthocyanidans that determine the colour of fruit spines in cucumber. J Exp Bot 70:69–84
Lv J, Qi JJ, Shi QX et al (2012) Genetic diversity and population structure of Cucumber (Cucumis sativus L.). PLoS One 7:19
Medina-Puche L, Molina-Hidalgo FJ, Boersma M, Schuurink RC, Lopez-Vidriero I, Solano R, Franco-Zorrilla JM, Caballero JL, Blanco-Portales R, Munoz-Blanco J (2015) An R2R3-MYB transcription factor regulates eugenol production in ripe strawberry fruit receptacles. Plant Physiol 168:598–614
Migocka M, Papierniak A (2010) Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breeding 28:343–357
Montefiori M, Brendolise C, Dare AP, Lin-Wang K, Davies KM, Hellens RP, Allan AC (2015) In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J Exp Bot 66:1427–1436
Naegele RP, Wehner TC (2016) Genetic resources of cucumber. In: Grumet R, Katzir N, Garcia-Mas J (eds) Genetics and genomics of Cucurbitaceae. Plant genetics and genomics: crops and models, vol 20. Springer, Cham, pp 61–86
Nakata M, Mitsuda N, Herde M, Koo AJK, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M (2013) A bHLH-Type transcription factor, ABA-inducible bHLH-type transcription factor/JA-associated MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25:1641–1656
Nanes BA (2015) Slide Set: reproducible image analysis and batch processing with ImageJ. Biotechniques 59:269–278
Narzisi G, O’Rawe JA, Iossifov I, Fang H, Lee YH, Wang Z, Wu Y, Lyon GJ, Wigler M, Schatz MC (2014) Accurate detection of de novo and transmitted indels within exome-capture data using micro-assembly. Nat Methods 11:1033–1036
Pierce LK, Wehner TC (1990) Review of genes and linkage groups in Cucumber. HortScience 25:605–615
Pitchaimuthu M, Dutta OP, Swamy KRM, Souravi K (2012) Mode of inheritance of bitterness and spine colour in cucumber fruits (Cucumis sativus L.). In: Sari N, Solmaz I, Aras V (eds) Cucurbitaceae 2012. Proceedings of the 10th EUCARPIA meeting on genetics and breeding of Cucurbitaceae, Antalya, Turkey, 15–18 October 2012, pp 70–73
Qi J, Liu X, Shen D, Miao H et al (2013) A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet 45:1510–1515
R Development Core Team (2011) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
Renner SS (2017) A valid name for the Xishuangbanna gourd, a cucumber with carotene-rich fruits. PhytoKeys 85:87–94
Renner SS, Schaefer H, Kocyan A (2007) Phylogenetics of Cucumis (Cucurbitaceae): cucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo). BMC Evol Biol 7:58
Sebastian P, Schaefer H, Telford IR, Renner SS (2010) Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA 107:14269–14273
Shanmugasundarum S, Williams PH, Peterson CE (1971) A cotyledon marker gene in cucumbers. HortScience 6:213–214
Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49:981–994
Strong WJ (1931) Breeding experiments with the cucumber (Cucumis sativus L.). Sci Agr 11:333–346
Sun L, Fan X, Zhang Y, Jiang J, Sun H, Liu C (2016) Transcriptome analysis of genes involved in anthocyanins biosynthesis and transport in berries of black and white spine grapes (Vitis davidii). Hereditas 153:17
Tanaka Y, Brugliera F (2013) Flower colour and cytochromes P450. Phil Trans R Soc B 368:20120432
Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54:733–749
Tkachenko NN (1935) Preliminary results of a genetic investigation of the cucumber, Cucumis sativus L. Appl Plant Breed 9:311–356
Vakalounakis DJ (1992) Heart leaf, a recessive leaf shape marker in cucumber: linkage with disease resistance and other traits. J Hered 83:217–221
Walters SA, Shetty NV, Wehner TC (2001) Segregation and linkage of several genes in cucumber. J Amer Soc Hort Sci 126(4):442–450
Wang G, Qin Z, Zhou X, Zhao C (2007) Genetic analysis and SSR markers of tuberculate trait in Cucumis sativus. Chin Bull Bot 24:168–172
Wang X, Bao K, Reddy UK, Bai Y, Hammar SA, Jiao C, Wehner TC, Ramirez-Madera AO, Weng Y, Grumet R, Fei Z (2018) The USDA cucumber (Cucumis sativus L.) collection: genetic diversity, population structure, genome-wide association studies and core collection development. Hortic Res 5:64
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185
Zhang S, Liu S, Miao H, Wang M, Liu P, Wehner TC, Gu X (2016) Molecular mapping and candidate gene analysis for numerous spines on the fruit of cucumber. J Hered 107:471–477
Acknowledgements
This work was supported by grants from the Bio-industry Technology Development Program (Grant nos. 117043-3, 316087-4) of iPET (Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry) and the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center) (Grant no. PJ01329601) of the Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, C., Win, K.T., Kim, YC. et al. Two types of mutations in the HEUKCHEEM gene functioning in cucumber spine color development can be used as signatures for cucumber domestication. Planta 250, 1491–1504 (2019). https://doi.org/10.1007/s00425-019-03244-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-019-03244-w