Abstract
Main conclusion
The first draft genome for a member of the genus Lavandula is described. This 870 Mbp genome assembly is composed of over 688 Mbp of non-gap sequences comprising 62,141 protein-coding genes.
Lavenders (Lavandula: Lamiaceae) are economically important plants widely grown around the world for their essential oils (EOs), which contribute to the cosmetic, personal hygiene, and pharmaceutical industries. To better understand the genetic mechanisms involved in EO production, identify genes involved in important biological processes, and find genetic markers for plant breeding, we generated the first de novo draft genome assembly for L. angustifolia (Maillette). This high-quality draft reveals a moderately repeated (> 48% repeated elements) 870 Mbp genome, composed of over 688 Mbp of non-gap sequences in 84,291 scaffolds with an N50 value of 96,735 bp. The genome contains 62,141 protein-coding genes and 2003 RNA-coding genes, with a large proportion of genes showing duplications, possibly reflecting past genome polyploidization. The draft genome contains full-length coding sequences for all genes involved in both cytosolic and plastidial pathways of isoprenoid metabolism, and all terpene synthase genes previously described from lavenders. Of particular interest is the observation that the genome contains a high copy number (14 and 7, respectively) of DXS (1-deoxyxylulose-5-phosphate synthase) and HDR (4-hydroxy-3-methylbut-2-enyl diphosphate reductase) genes, encoding the two known regulatory steps in the plastidial isoprenoid biosynthetic pathway. The latter generates precursors for the production of monoterpenes, the most abundant essential oil constituents in lavender. Furthermore, the draft genome contains a variety of monoterpene synthase genes, underlining the production of several monoterpene essential oil constituents in lavender. Taken together, these findings indicate that the genome of L. angustifolia is highly duplicated and optimized for essential oil production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Lavandula (lavenders) is composed of over 32 morphologically distinct species. Among these, a few species including L. angustifolia (English Lavender, or Lavender), L. latifolia (spike lavender), and their hybrid L. x intermedia (Lavandin) are widely grown around the globe for their essential oils (EOs) (Upson et al. 2004), which are frequently used in perfumes, pharmaceutical preparations, cosmetic products, and antiseptics, among others. With an estimated production rate of over 1500 metric tons/annum (mostly extracted from L. angustifolia and L. x intermedia), these oils significantly contribute to the multibillion-dollar flavor and fragrance industry worldwide.
Although lavender has been developed as a model system for studying EO production (Lane et al. 2010) and numerous genes that contribute to EO production in this plant have been reported (Falk et al. 2009; Lane et al. 2010; Demissie et al. 2012), many questions regarding the regulation of EO metabolism and storage in these plants remain unanswered.
The quality of lavender EOs greatly depends on the characteristic scent of the oil, which is determined by certain monoterpenes. For example, the monoterpene camphor contributes an off-odor, and its presence in the oil lowers quality and, hence, market value. Conversely, high levels of linalool and linalyl acetate are desired in lavender oils. Because many high-yield lavender species/cultivars (e.g., L. x intermedia) produce high quantities of undesired constituents (such as camphor), there is great interest in enhancing oil yield and composition in lavenders. These objectives are readily achievable through targeted breeding and/or plant biotechnology. However, an adequate understanding of molecular elements that control the production of EO constituents in lavenders must first be developed.
To better understand the genetic makeup and key pathways that control EO production, secretion, and storage, a first reconstruction of a full-length draft genome assembly has been generated using short-reads from the Illumina platform. This resource can lead to better understanding of the genome architecture, gene clusters, and repeat content and ideally also in the generation of genetic markers for plant breeding programs.
De novo genome assembly of lavender
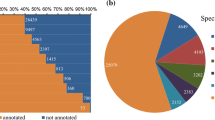
The genome of L. angustifolia (Maillette) was sequenced to a total of ~ 100 × coverage using the Illumina HiSeq 2000 platform combining the use of pair-end and mate pair libraries, all sequenced at 100 bp × 2 setting. The best de novo genome assembly was obtained using FERMI (Li 2012) for contig assembly and OPERA (Gao et al. 2011, 2016) for scaffolding. The initial genome assembly was further improved using in-house transcriptome-based program and GapCloser (Luo et al. 2015). The final draft assembly contains 869,786 077 bp in 84,291 scaffolds with N50 being 96, 735 bp, and the total non-gap sequences (sequences with no gaps) being 688,040,719 bp or 79.1% of the final draft genome. The repeat sequences annotated by RepeatMasker (Smit et al. 2013), RepeatModeler (http://www.repeatmasker.org/RepeatModeler/), and RepeatProteinMasker (http://repeatmasker.org) constitute 42.8% of the lavender draft genome. The genome has a 38.1% GC content. (Table 1 and Fig. 1a).
Summary features of the Lavandula angustifolia (Lavender) draft genome. a Pie chart showing the genome composition. Categories denoted by “*” exclude overlapping repeats and gaps. b Estimation of genome completeness of the lavender draft genome and comparison with other four published plant genomes using Benchmark Universal Single-Copy Orthologues (BUSCO)
Regarding the genome size of lavender, there have been two controversial reports, one suggesting a very large size of 5574 (Zonneveld et al. 2005) and another reporting genome size ranging from 772 Mbp to 880 Mbp (Urwin et al. 2007). The genome size of our draft genome assembly of ~ 870 Mbp is sufficiently close to our own experimentally determined genome size of approximately 850 Mbp using a qPCR-based method (Wilhelm 2003) (data not shown). A similar size prediction was also obtained based on the raw Illumina reads using Kmergenie (Chikhi and Medvedev 2014) (data not shown). Therefore, our data resolve the dispute on lavender genome size by supporting an estimated genome size around 850 Mbp.
The genome completeness determined by Benchmarking Universal Single-Copy Orthologues (BUSCO) (Simão et al. 2015) shows a high-quality draft genome, covering 1292 (89.7%) complete single-copy orthologues (SCO) of the 1440 plant-specific sequences (Embryophyta data set from BUSCO data sets) (Fig. 1b). The presence of 30 fragmented SCOs brings the total SCOs to 1322 and genome completeness to 91.8%. In comparison with the published genomes for several other plants including model plants, the completeness of lavender draft genome was comparable to that of the maize genome (1294 complete SCOs and 31 partial SCOs with completeness at 92%), which was obtained using more resources (Schnable et al. 2009), and is significantly better than the mint genome (848 complete SCOs and 183 partial SCOs; completeness at 71%) (Vining et al. 2017) (Fig. 1b).
An interesting observation among the complete SCOs is the presence of 696 complete duplicated copies. Sequence analysis using BLASTn (Altschul et al. 1990) combined with the BUSCO score analysis revealed that at least 547 (78%) of the 696 complete and duplicated SCOs represent real duplication events in the genome, distinguishing them from possible assembly errors due to genetic heterogeneity between homologous chromosomes. Indeed, possible duplication events resulting in polyploidy ranging from 2n = 36 to 2n = 54 have been reported for L. angustifolia (Upson et al. 2004). In this context, our data indicate that the variety used in this study (Maillette) is most likely a polyploid line.
De novo genome annotation
Using an automated and highly configurable de novo annotation pipeline (MAKER) with transcriptome data (Adal et al. 2018) and NCBI plant reference protein sequences to guide gene prediction for Augustus (Stanke et al. 2006; Campbell et al. 2014; Holt and Yandell 2011), we identified 218,000 initial gene models. This list was subjected to filtering based on minimal gene length and/or homology to known protein sequences to generate the final total 62,141 protein-coding genes. In addition, we have also identified 2003 tRNA and rRNA genes using tRNAScan (Lowe and Eddy 1996) and RNAmmer (Lagesen et al. 2007), respectively, which brings the total number of identified genes to 64,144. The total regions of genes, CDS, and introns cover 26.8, 9.2, and 5.1% of the lavender draft genome, respectively (Fig. 1a and supplementary Table S1). In comparison, lavender genome has a higher (~ 3 times) density of protein-coding sequences (CDS) than the maize genome, but it is more than four times lower than that of Arabidopsis (supplementary Table S1).
We also performed annotation of repetitive elements for the draft genome using RepeatMasker (Smit et al. 2013), which revealed a total of repetitive content ~ 43%, a level which is lower than the most published plant genomes at similar sizes. For examples, the tomato genome (Sato et al. 2012) size ~ 900 Mbp has 68% as repeats (Jouffroy et al. 2016), and the 2104 Mbp maize genome (Schnable et al. 2009) has 82.2% for repeat elements (supplementary Table S1). In comparison, the 119 Mbp model plant Arabidopsis thaliana (Weinig et al. 2002) genome has only 21% for repeat elements (supplementary Table S1). Among the repeats, as expected for most plant genomes, the LTR retrotransposon as the dominant type is contributing to 18% of the genome or 44% of the repeats as expected for most plant genomes. The lower than expected, overall repeat content for lavender could be partly due to selective loss post-genome polyploidization and partly due to biased distribution of gaps in repeat regions. The former is in agreement with the higher density of CDS exons. The latter can be resolved by additional sequencing and using them in closing the gaps.
Genes for EO pathways
Considering that EO production is the main characteristics of the lavender plants, we performed detailed analysis for the genes associated with EO pathways. The draft lavender genome covered all of the known genes encoding the four classes of enzymes required for all the stages of EO biosynthesis (Tables 2, 3 & supplementary Fig. S1). In comparison with the four other published plant genomes, including mint, the only published genome in the mint family as lavender (Vining et al. 2017), we observed that lavender has the largest gene copy numbers for 15 out of the 35 genes involved in EO biosynthesis (Table 2). For methylerythritol pathway (MEP), lavender genome has 13 copies of DXS being one copy higher than that of the mint, while all three other genomes have only a single copy. Interestingly, lavender genome has seven copies of the last gene, HDR, in the MEP pathway, while all other genomes including mint have only a single copy. It is likely that having a larger copy number of the first and last genes for the MEP permits the plant to be more efficient in the production of terpene compounds (essential oils, resins, etc.) as evidenced in other plant like pine (Kim et al. 2009).
In addition, lavender genome has a high copy number for the two precursor enzymes, geranylgeranyl pyrophosphate synthase (GGPPS) and farnesyl pyrophosphate synthase (FPPS). The lavender genome is much more similar to the mint genome by having many EO genes, including 13 of the 15 terpene synthases (TPS) genes critical for EO synthesis, which are absent in the non-mint plants (Table 2), and these genes contribute to the special EO-producing characteristics of the mint family plants.
A surprising finding was that the lavender genome contains only two copies of the R-linalool synthase gene (which contributes most to EO production), while it contains more than two copies of some other terpene synthase genes that contribute minimally to the EO (e.g., limonene synthase and 3-carene synthase) in lavenders (Table 3). Given that linalool synthase is responsible for the production of two of the most abundant EO constitutes (linalool and its acetylated form linalyl acetate, which make up > 80% of the EO in most L. angustifolia and L. intermedia species), and that transcripts for the gene are among the most abundant mRNA species in floral oil glands (Lane et al. 2010), we anticipated a higher copy number in the genome. Taken together, our results indicate that the R-linalool synthase gene is very strongly expressed in floral oil glands.
We also performed orthologous sequences/clusters’ search as a way of validating the genome annotation process using the Orthovenn tool (Wang et al. 2015). There are a total of 8413 clusters which have 76,424 (data not shown) gene sequences shared among all five plant genomes compared, and we were able to find orthologous sequences in Arabidopsis (64), Rice (58), Maize (87), and tomato (776) genomes. We were unable to include mint genome for this analysis due to lack of protein sequences. Tomato shares the largest number of genes with lavender among the plants compared. A search for orthologous sequences containing terpene synthase genes resulted in 20 orthologous clusters divided into characterized and uncharacterized TPS genes. Orthologous sequences for S-linalool synthase gene are presented in all plants and have three copies in tomato genome, one more than lavender. Interestingly, R-linalool synthase is absent in rice and maize, and has a single copy in the tomato genome, one less than lavender (supplementary Table S2). Among the uncharacterized TPS genes, the genes for ent-copalyl diphosphate synthase and solanesyl diphosphate synthase are shared among all plants. The other three genes [cis-abienol synthase, gamma-cadinene synthase, and (+)-epi-alpha-bisabolol synthase] is presented only in lavender (supplementary Table S2).
Conclusion
In conclusion, the lavender genome assembly reported here represents the first relatively complete high-quality draft genome. The initial analysis of the genome sequences reveals a genome optimized for EO production by having large copy numbers of many EO pathway genes and genes unique to EO-producing plants. Future work may involve the estimation of the level of genome heterozygosity, identification of any genome duplication events during the course of the plant’s evolution, closing of the gaps in the draft assembly using long sequencing platforms, and optical mapping technologies to map the genome sequences to their chromosomal locations. Given the economic status of lavender and its applications of EOs in many industries, the lavender draft genome sequences can serve as a significant genomics resource for the lavender research communities.
Author contribution statement
SSM and PL designed and funded the research. SSM, AMA and LSS prepared gDNA for sequencing and provided ESTs and transcriptome sequences used in annotating the draft assembly. RPNM and PL conducted bioinformatics work to generate the draft genome assembly and annotate it. RPNM, PL and SSM prepared the manuscript. All authors read and approved the manuscript.
Abbreviations
- EO:
-
Essential oils
- qPCR:
-
Quantitative real-time PCR
- BUSCO:
-
Benchmarking universal single-copy orthologues
- MEP:
-
Methylerythritol pathway
- MVA:
-
Mevalonate pathway
- GGPPS:
-
Geranylgeranyl pyrophosphate synthase
- FPPS:
-
Farnesyl pyrophosphate synthase
- DXS:
-
1-Deoxyxylulose-5-phosphate synthase
- HDR:
-
4-Hydroxy-3-methylbut-2-enyl diphosphate reductase
- TPS:
-
Terpene synthases
References
Adal AM, Sarker LS, Malli RPN et al (2018) RNA-Seq in the discovery of a sparsely expressed scent-determining monoterpene synthase in lavender (Lavandula). Planta. https://doi.org/10.1007/s00425-018-2935-5
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Campbell MS, Law M, Holt C et al (2014) MAKER-P: a Tool Kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol 164:513–524. https://doi.org/10.1104/pp.113.230144
Chikhi R, Medvedev P (2014) Informed and automated k-mer size selection for genome assembly. Bioinformatics 30:31–37. https://doi.org/10.1093/bioinformatics/btt310
Demissie ZA, Cella MA, Sarker LS et al (2012) Cloning, functional characterization and genomic organization of 1,8-cineole synthases from Lavandula. Plant Mol Biol 79:393–411. https://doi.org/10.1007/s11103-012-9920-3
Falk L, Biswas K, Boeckelmann A et al (2009) An effcient method for the micropropagation of lavenders: regeneration of a unique mutant. J Essent Oil Res 21:225–228. https://doi.org/10.1080/10412905.2009.9700154
Gao S, Nagarajan N, Sung WK (2011) Opera: reconstructing optimal genomic scaffolds with high-throughput paired-end sequences. Lect Notes Comput Sci 6577 LNBI:437–451. https://doi.org/10.1007/978-3-642-20036-6_40 (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics)
Gao S, Bertrand D, Chia BKH, Nagarajan N (2016) OPERA-LG: efficient and exact scaffolding of large, repeat-rich eukaryotic genomes with performance guarantees. Genome Biol 17:102. https://doi.org/10.1186/s13059-016-0951-y
Holt C, Yandell M (2011) MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform 12:491. https://doi.org/10.1186/1471-2105-12-491
Jouffroy O, Saha S, Mueller L et al (2016) Comprehensive repeatome annotation reveals strong potential impact of repetitive elements on tomato ripening. BMC Genom 17:624. https://doi.org/10.1186/s12864-016-2980-z
Kim YB, Kim SM, Kang MK et al (2009) Regulation of resin acid synthesis in Pinus densiflora by differential transcription of genes encoding multiple 1-deoxy-d-xylulose 5-phosphate synthase and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase genes. Tree Physiol 29:737–749. https://doi.org/10.1093/treephys/tpp002
Lagesen K, Hallin P, Rødland EA et al (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res 35:3100–3108. https://doi.org/10.1093/nar/gkm160
Lane A, Boecklemann A, Woronuk GN et al (2010) A genomics resource for investigating regulation of essential oil production in Lavandula angustifolia. Planta 231:835–845. https://doi.org/10.1007/s00425-009-1090-4
Li H (2012) Exploring single-sample snp and indel calling with whole-genome de novo assembly. Bioinformatics 28:1838–1844. https://doi.org/10.1093/bioinformatics/bts280
Lowe TM, Eddy SR (1996) TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res 25:955–964. https://doi.org/10.1093/nar/25.5.0955
Luo R, Liu B, Xie Y et al (2015) Erratum to “SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler” [GigaScience, (2012), 1, 18]. Gigascience 4:1. https://doi.org/10.1186/s13742-015-0069-2
Sato S, Tabata S, Hirakawa H et al (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641. https://doi.org/10.1038/nature11119
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science (80-) 326:1112–1115. https://doi.org/10.1126/science.1178534
Simão FA, Waterhouse RM, Ioannidis P et al (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Smit A, Hubley R, Green P (2013) RepeatMasker Open-4.0. 2013-2015. http://www.repeatmasker.org
Stanke M, Keller O, Gunduz I et al (2006) AUGUSTUS: a b initio prediction of alternative transcripts. Nucl Acids Res 34:W435–W439. https://doi.org/10.1093/nar/gkl200
Upson T, Andrews S, Royal Botanic Gardens K (2004) The genus Lavandula. SciTech book news, vol 28. Book News Inc., Portland, p 442
Urwin NAR, Horsnell J, Moon T (2007) Generation and characterisation of colchicine-induced autotetraploid Lavandula angustifolia. Euphytica 156:257–266. https://doi.org/10.1007/s10681-007-9373-y
Vining KJ, Johnson SR, Ahkami A et al (2017) Draft genome sequence of Mentha longifolia and development of resources for mint cultivar improvement. Mol Plant 10:323–339. https://doi.org/10.1016/j.molp.2016.10.018
Wang Y, Coleman-Derr D, Chen G, Gu YQ (2015) OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucl Acids Res 43:W78–W84. https://doi.org/10.1093/nar/gkv487
Weinig C, Ungerer MC, Dorn LA et al (2002) Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics 162:1875–1884. https://doi.org/10.1038/35048692
Wilhelm J (2003) Real-time PCR-based method for the estimation of genome sizes. Nucl Acids Res 31:56. https://doi.org/10.1093/nar/gng056
Zonneveld BJM, Leitch IJ, Bennett MD (2005) First nuclear DNA amounts in more than 300 angiosperms. Ann Bot 96:229–244. https://doi.org/10.1093/aob/mci170
Acknowledgements
This work was supported through grants to SSM and PL from the Natural Sciences and Engineering Research Council of Canada (Discovery Grant), Canada Research Chair Program (PL), and the Investment Agriculture Foundation of B.C. through programs it delivers on behalf of Agriculture and Agri-Food Canada and the B.C. Ministry of Agriculture and was made possible by Compute Canada/SHARCNET high-performance computing facilities.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Accession numbers
The raw sequencing data generated from this research have been deposited into NCBI sequence read archive (SRA) database under accession SRP161522 ( bioproject accession PRJN490378).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Malli, R.P.N., Adal, A.M., Sarker, L.S. et al. De novo sequencing of the Lavandula angustifolia genome reveals highly duplicated and optimized features for essential oil production. Planta 249, 251–256 (2019). https://doi.org/10.1007/s00425-018-3012-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-018-3012-9