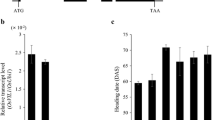
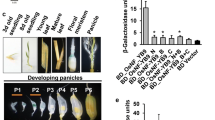
Abstract
Main conclusion
OsNF-YC2 and OsNF-YC4 proteins regulate the photoperiodic flowering response through the modulation of three flowering-time genes ( Ehd1, Hd3a , and RFT1 ) in rice.
Plant NUCLEAR FACTOR Y (NF-Y) transcription factors control numerous developmental processes by forming heterotrimeric complexes, but little is known about their roles in flowering in rice. In this study, it is shown that some subunits of OsNF-YB and OsNF-YC interact with each other, and among them, OsNF-YC2 and OsNF-YC4 proteins regulate the photoperiodic flowering response of rice. Protein interaction studies showed that the physical interactions occurred between the three OsNF-YC proteins (OsNF-YC2, OsNF-YC4 and OsNF-YC6) and three OsNF-YB proteins (OsNF-YB8, OsNF-YB10 and OsNF-YB11). Repression and overexpression of the OsNF-YC2 and OsNF-YC4 genes revealed that they act as inhibitors of flowering only under long-day (LD) conditions. Overexpression of OsNF-YC6, however, promoted flowering only under LD conditions, suggesting it could function as a flowering promoter. These phenotypes correlated with the changes in the expression of three rice flowering-time genes [Early heading date 1 (Ehd1), Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1)]. The diurnal and tissue-specific expression patterns of the subsets of OsNF-YB and OsNF-YC genes were similar to those of CCT domain encoding genes such as OsCO3, Heading date 1 (Hd1) and Ghd7. We propose that OsNF-YC2 and OsNF-YC4 proteins regulate the photoperiodic flowering response by interacting directly with OsNF-YB8, OsNF-YB10 or OsNF-YB11 proteins in rice.






Similar content being viewed by others
Abbreviations
- CO:
-
CONSTANS
- Ehd1:
-
Early heading date 1
- NF-Y:
-
NUCLEAR FACTOR Y transcription factor
- FT:
-
FLOWERING LOCUS T
- GFP:
-
Green fluorescence protein
- GST:
-
Glutathione S-transferase
- HAP:
-
Heme activator protein
- Hd1(3a):
-
Heading date 1(3a)
- LD:
-
Long-day
- ORFs:
-
Open-reading frames
- RFT1:
-
RICE FLOWERING LOCUS T1
- SD:
-
Short-day
- Ubi1:
-
Ubiquitin1
References
Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46:462–476
Cai X, Ballif J, Endo S, Davis E, Liang M, Chen D, DeWald D, Kreps J, Zhu T, Wu Y (2007) A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol 145:98–105
Calvenzani V, Testoni B, Gusmaroli G, Lorenzo M, Gnesutta N, Petroni K, Mantovani R, Tonelli C (2012) Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7:e42902
Cao S, Kumimoto RW, Siriwardana CL, Risinger JR, Holt BF 3rd (2011) Identification and characterization of NF-Y transcription factor families in the monocot model plant Brachypodium distachyon. PLoS One 6:e21805
Chen NZ, Zhang XQ, Wei PC, Chen QJ, Ren F, Chen J, Wang XC (2007) AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J Biochem Mol Biol 40:1083–1089
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18:926–936
Edwards D, Murray JA, Smith AG (1998) Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol 117:1015–1022
Frontini M, Imbriano C, Manni I, Mantovani R (2004) Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 3:217–222
Goda H, Nagase T, Tanoue S, Sugiyama J, Steidl S, Tuncher A, Kobayashi T, Tsukagoshi N, Brakhage AA, Kato M (2005) Nuclear translocation of the heterotrimeric CCAAT binding factor of Aspergillus oryzae is dependent on two redundant localising signals in a single subunit. Arch Microbiol 184:93–100
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131:1855–1867
Hackenberg D, Wu Y, Voigt A, Adams R, Schramm P, Grimm B (2012) Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol Plant 5:876–888
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422:719–722
Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M (2013) Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J 76:36–46
Itoh H, Nonoue Y, Yano M, Izawa T (2010) A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 42:635–638
Jang YH, Park HY, Kim SK, Lee JH, Suh MC, Chung YS, Paek KH, Kim JK (2009) Survey of rice proteins interacting with OsFCA and OsFY proteins which are homologous to the Arabidopsis flowering time proteins, FCA and FY. Plant Cell Physiol 50:1479–1492
Kim SK, Yun CH, Lee JH, Jang YH, Park HY, Kim JK (2008) OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228:355–365
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136:3443–3450
Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ (2008) The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228:709–723
Kumimoto RW, Zhang Y, Siefers N, Holt BF 3rd (2010) NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J 63:379–391
Kwon CT, Koo BH, Kim D, Yoo SC, Paek NC (2015) Casein kinases I and 2α phosphorylate Oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Mol Cells 38:81–88
Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013) CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci 18:157–166
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20:2238–2251
Li C, Distelfeld A, Comis A, Dubcovsky J (2011) Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J 67:763–773
Maity SN, de Crombrugghe B (1998) Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci 23:174–178
Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy Cotyledon mutants of Arabidopsis. Plant Cell 6:1049–1064
Miyoshi K, Ito Y, Serizawa A, Kurata N (2003) OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J 36:532–540
Mu J, Tan H, Hong S, Liang Y, Zuo J (2013) Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant 6:188–201
Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y (2003) Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J 36:82–93
Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL (2007) Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem Biophys Res Commun 360:251–256
Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF 3rd, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24:4777–4792
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, An G (2009) OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant, Cell Environ 32:1412–1427
Siefers N, Dang KK, Kumimoto RW, Bynum WEt, Tayrose G, Holt BF 3rd (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149:625–641
Steidl S, Tuncher A, Goda H, Guder C, Papadopoulou N, Kobayashi T, Tsukagoshi N, Kato M, Brakhage AA (2004) A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J Mol Biol 342:515–524
Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N (2008) Identification, characterization and interaction of HAP family genes in rice. Mol Genet Genom 279:279–289
Tuncher A, Sprote P, Gehrke A, Brakhage AA (2005) The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A.nidulans and human. J Mol Biol 352:517–533
Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J (2010) DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol 153:1747–1758
Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984
Xing Y, Fikes JD, Guarente L (1993) Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J 12:4647–4655
Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40:761–767
Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58:843–856
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, Zhang QF (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol Plant 4:319–330
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484
Acknowledgments
We thank Professors C.D. Han (Gyeongsang National University) and K. Shimamoto (NAIST, Japan) for RNAi vectors. We also thank Prof. C.D. Han for 35S::GFP transgenic rice plants and technical help. We are grateful to Kyung-Sook Lee for preparing transgenic rice lines and the RGRC-NIAS (Japan) and Rural Development Administration (Republic of Korea) for rice cDNA and seeds. This work was supported by grants from the Next-Generation BioGreen 21 Program (PJ007978), Rural Development Administration, Republic of Korea and a Korea University Grant (to J.-K.Kim) and by the BK 21 program (to S.-K.Kim).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, SK., Park, HY., Jang, Y.H. et al. OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 243, 563–576 (2016). https://doi.org/10.1007/s00425-015-2426-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2426-x