Abstract
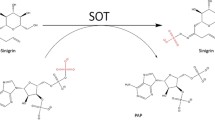
The Arabidopsis thaliana gene AtSgpp (locus tag At2g38740), encodes a protein whose sequence motifs and expected structure reveal that it belongs to the HAD hydrolases subfamily I, with the C1-type cap domain (Caparrós-Martín et al. in Planta 237:943–954, 2013). In the presence of Mg2+ ions, the enzyme has a phosphatase activity over a wide range of phosphosugar substrates. AtSgpp promiscuity is preferentially detectable on d-ribose-5-phosphate, 2-deoxy-d-ribose-5-phosphate, 2-deoxy-d-glucose-6-phosphate, d-mannose-6-phosphate, d-fructose-1-phosphate, d-glucose-6-phosphate, dl-glycerol-3-phosphate, and d-fructose-6-phosphate. Site-directed mutagenesis analysis of the putative signature sequence motif-5 (IAGKH), which defines its specific chemistry, brings to light the active-site residues Ala-69 and His-72. Mutation A69M, changes the pH dependence of AtSgpp catalysis, and mutant protein AtSgpp-H72K was inactive in phosphomonoester dephosphorylation. It was also observed that substitutions I68M and K71R slightly affect the substrate specificity, while the replacement of the entire motif for that of homologous dl-glycerol-3-phosphatase AtGpp (MMGRK) does not switch AtSgpp activity to the specific targeting for dl-glycerol-3-phosphate.



Similar content being viewed by others
Abbreviations
- HAD:
-
Haloacid dehalogenase-like hydrolase proteins
- MBP:
-
Maltose-binding protein
References
Allen KN, Dunaway-Mariano D (2004) Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem Sci 29:495–503
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ames BN (1966) Assay of inorganic phosphate, total phosphate, and phosphatases. Methods Enzymol 8:115–118
Bradford MM (1976) A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L (2006) Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol 361:1003–1034
Caparrós-Martín JA, Reiland S, Köchert K, Cutanda MC, Culiáñez-Macia FA (2007) Arabidopsis thaliana AtGpp1 and AtGpp2: two novel low molecular weight phosphatases involved in plant glycerol metabolism. Plant Mol Biol 63:505–517
Caparrós-Martín JA, McCarthy-Suárez I, Culiáñez-Macià FA (2013) HAD hydrolase function unveiled by substrate screening: enzymatic characterization of Arabidopsis thaliana subclass I phosphosugar phosphatase AtSgpp. Planta 237:943–954
Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E (1998) A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem 273:14107–14112
Cormack B (1991) Directed mutagenesis using the polymerase chain reaction. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology, vol 1. Wiley, New York, pp 8.5.1–8.5.9
Corpet F, Servantm F, Gouzy J, Kahn D (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res 28:267–269
Daughtry KD, Huang H, Malashkevich V, Patskovsky Y, Liu W, Ramagopal U, Sauder JM, Burley SK, Almo SC, Dunaway-Mariano D, Allen KN (2013) Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide. Biochemistry 52:5372–5386
Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Kim SH, Pokhrel S, Yoo YJ (2008) Mutation of non-conserved amino acids surrounding catalytic site to shift pH optimum of Bacillus circulans xylanase. J Mol Catal B Enzym 55:130–136
Koonin EV, Tatusov RL (1994) Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J Mol Biol 244:125–132
Kuznetsova E, Proudfoo M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF (2006) Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem 281:36149–36161
Lahiri SD, Zhang G, Dai J, Dunaway-Mariano D, Allen KN (2004) Analysis of the substrate specificity loop of the HAD superfamily cap domain. Biochemistry 43:2812–2820
Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN (2006) Diversification of function in the haloacid dehalogenase enzyme superfamily: the role of the cap domain in hydrolytic phosphorus-carbon bond cleavage. Bioorg Chem 34:394–409
Lambert C, Leonard N, De Bolle X, Depiereux E (2002) ESyPred3D: prediction of proteins 3D structures. Bioinformatics 18:1250–1256
Lu Z, Dunaway-Mariano D, Allen KN (2005) HAD superfamily phosphotransferase substrate diversification: structure and function analysis of HAD subclass IIB sugar phosphatase BT4131. Biochemistry 44:8684–8696
Lu Z, Dunaway-Mariano D, Allen KN (2008) The catalytic scaffold of the haloalkanoic acid dehalogenase enzyme superfamily acts as a mold for the trigonal bipyramidal transition state. Proc Natl Acad Sci USA 105:5687–5692
Morais MC, Zhang W, Baker AS, Zhang G, Dunaway-Mariano D, Allen KN (2000) The crystal structure of Bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry 39:10385–10396
Morais MC, Zhang G, Zhang W, Olsen DB, Dunaway-Mariano D, Allen KN (2004) X-ray crystallographic and site-directed mutagenesis analysis of the mechanism of Schiff-base formation in phosphonoacetaldehyde hydrolase catalysis. J Biol Chem 279:9353–9361
Nielsen JE (2007) Analysing the pH-dependent properties of proteins using pKa calculations. J Mol Graph Model 25:691–699
Norbeck J, Pahlman AK, Akhtar N, Blomberg A, Adler L (1996) Purification and characterization of two isoenzymes of dl-glycerol-3-phosphatase from Saccharomyces cerevisiae. Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J Biol Chem 271:13875–13881
Olsen DB, Hepburn TW, Moos M, Mariano PS, Dunaway-Mariano D (1988) Investigation of the Bacillus cereus phosphonoacetaldehyde hydrolase. Evidence for a Schiff base mechanism and sequence analysis of an active-site peptide containing the catalytic lysine residue. Biochemistry 27:2229–2234
Rández-Gil F, Blasco A, Prieto JA, Sanz P (1995) DOGR1 and DOGR2: two genes from Saccharomyces cerevisiae that confer 2-deoxyglucose resistance when overexpressed. Yeast 11:1233–1240
Rao KN, Kumaran D, Seetharaman J, Bonanno JB, Burley SK, Swaminathan S (2006) Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications. Protein Sci 15:1735–1744
Schagger H, von Jagow G (1987) Tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379
Schenk PM, Baumann S, Mattes R, Steinbiss HH (1995) Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques 19:196–200
Selengut JD (2001) MDP-1 is a new and distinct member of the haloacid dehalogenase family of aspartate-dependent phosphohydrolases. Biochemistry 40:12704–12711
Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH (2011) Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theory Comput 7:2284–2295
Sussman I, Avron M (1981) Characterization and partial purification of dl-glycerol-1-phosphatase from Dunaliella salina. Biochim Biophys Acta 661:199–204
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Wang W, Cho HS, Kim R, Jancarik J, Yokota H, Nguyen HH, Grigoriev IV, Wemmer DE, Kim S-H (2002) Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J Mol Biol 319:421–431
Zhang G, Mazurkie AS, Dunaway-Mariano D, Allen KN (2002) Kinetic evidence for a substrate-induced fit in phosphonoacetaldehyde hydrolase catalysis. Biochemistry 41:13370–13377
Zhang G, Morais MC, Dai J, Zhang W, Dunaway-Mariano D, Allen KN (2004) Investigation of metal Ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily. Biochemistry 43:4990–4997
Acknowledgments
We acknowledge Professors Montserrat Pagès (CSIC Barcelona, Spain), Thomas Kupke (University of Heidelberg, Germany) and Manuel Hernández (University Polytechnic of Valencia, Spain) for their warm support. We also thank the advice and provision of plasmid pSBETa by Dr Florence Vignols and Yves Meyer (University of Perpignan, France); the computer software helps by Ramón Nogales-Rangel and Alexis González-Policarpo; the Eugenio Grau-Ferrando kind advice and help for sequencing; the Dr Angela Batcheller support in the English edition of the manuscript. This work was funded by the 10-month research contract MEC-FEDER to J. A. C.-M., 10-month research contract JAE-DOC to I. M.-S., and by the research project BIO2006-10138 from the MEC-FEDER of Spain to F. A. C.-M.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Caparrós-Martín, J.A., McCarthy-Suárez, I. & Culiáñez-Macià, F.A. The kinetic analysis of the substrate specificity of motif 5 in a HAD hydrolase-type phosphosugar phosphatase of Arabidopsis thaliana . Planta 240, 479–487 (2014). https://doi.org/10.1007/s00425-014-2102-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2102-6