Abstract
Plant steroid hormones, brassinosteroids, are essential for growth, development and responses to environmental stresses in plants. Although BR signaling proteins are localized in many organelles, i.e., the plasma membrane, nuclei, endoplasmic reticulum and vacuole, the details regarding the BR signaling pathway from perception at the cellular membrane receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1) to nuclear events include several steps. Brz (Brz220) is a specific inhibitor of BR biosynthesis. In this study, we used Brz-mediated chemical genetics to identify Brz-insensitive-long hypocotyls 2-1D (bil2-1D). The BIL2 gene encodes a mitochondrial-localized DnaJ/Heat shock protein 40 (DnaJ/Hsp40) family, which is involved in protein folding. BIL2-overexpression plants (BIL2-OX) showed cell elongation under Brz treatment, increasing the growth of plant inflorescence and roots, the regulation of BR-responsive gene expression and suppression against the dwarfed BRI1-deficient mutant. BIL2-OX also showed resistance against the mitochondrial ATPase inhibitor oligomycin and higher levels of exogenous ATP compared with wild-type plants. BIL2 participates in resistance against salinity stress and strong light stress. Our results indicate that BIL2 induces cell elongation during BR signaling through the promotion of ATP synthesis in mitochondria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassinosteroids (BRs) are plant steroid hormones that regulate various processes including plant growth and responses to environmental stress, among others. Mutants deficient in BR biosynthesis or signal transduction exhibit phenotypes such as de-etiolated hypocotyls and opened cotyledons in the dark and dwarfism with shortened leaves and stems in the light (Li et al. 1996; Li and Chory 1997). The activation of BR biosynthesis and signal transduction promotes hypocotyl and stem elongation and outward leaf curling. These results showed that BR is necessary for the positive cell elongation and growth of the plant (Belkhadir and Chory 2006).
BR signaling factors are localized in various parts and organelles in the plant cell. BRs are perceived through a plasma membrane localized receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1), a leucine-rich repeat receptor-like serine/threonine kinase that functions in cell elongation (Kinoshita et al. 2005). BRI1 works with the negative regulator BRI1 KINASE INHIBITOR 1 (BKI1) (Wang and Chory 2006) and the positive regulator BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) (Li et al. 2002; Nam and Li 2002), which are anchored to the plasma membrane. BR signals received through these factors on the cell surface are transduced to bri1-5 SUPPRESSOR 1 (BSU1), which is a positive phosphatase (Mora-Garcia et al. 2004) and BRASSINOSTREROID INSENSITIVE 2 (BIN2), which is a negative kinase similar to the glycogen synthase kinase 3-like kinase (Li and Nam 2002) in the cytosol. BIN2 kinase and BSU1 dephosphorylation activities regulate the phosphorylation of BRASSINOZOLE RESISTANT 1/Brz-INSENSITIVE-LONG HYPOCOTYLS 1 (BZR1/BIL1) (Wang et al. 2002; He et al. 2005; Asami et al. 2005) and bri1-EMS-SUPPRESSOR 1 (BES1) (Yin et al. 2002, 2005), which regulate gene expression in the nucleus. Although BR signal transduction from the plasma membrane to the cytosol and nuclei has been described, the BR signaling factors in other organelles have not been clarified.
ATP is well known as an energy source for the growth and life cycle in organisms from plant to animal. Plasma membrane P-ATPase in plants is activated by BRI1 kinase to induce cell wall expansion (Caesar et al. 2011). Reduced glutathione and dithiothreitol inhibited plant hypocotyl elongation; however, hypocotyl elongation could be recovered after treatment with ATP. NADPH oxidase activity and the endogenous production of nitric oxide positively affected hypocotyl elongation with the help of ATP treatment (Tonon et al. 2010). Translocase of the inner membrane 50 (TIM50) and translocase of the inner membrane 21 (TIM21) are mitochondrial proteins, and knock-out mutants of these genes showed a reduction of intracellular ATP levels and short hypocotyls in the dark (Hamasaki et al. 2012; Kumar et al. 2012). These results suggested that ATP plays an important role in plant growth, but a regulatory mechanism between the other plant growth regulator plant hormone and ATP itself is still unknown.
Brz is a specific inhibitor of BR biosynthesis. Brz treatment caused BR-deficient mutant like phenotype onto wild-type plant (Asami et al. 2000). Here, we have isolated and characterized an Arabidopsis mutant, Brz-insensitive-long hypocotyls 2-1D (bil2-1D) using activated tagging mutant lines by Brz. The bil2-1D mutant exhibited positively regulated growth in the hypocotyl, branch and root. bil2-1D responsible gene encodes a novel DnaJ/Hsp40 family protein that is localized in the mitochondria. In this manuscript, we attempted to identify the mitochondrial protein involved in plant growth under BR signal transduction.
Materials and methods
Plant materials, growth conditions and stress treatments
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type plant. Seeds were germinated on medium containing 1/2 Murashige and Skoog (MS) medium (Duchefa, Haarlem, The Netherlands) and 0.8 % phytoagar (Duchefa, Haarlem, The Netherlands) with 1.5 % sucrose and were subsequently transferred to soil. The plants were grown at 22 °C under white light (a 16 h light/8 h dark cycle for long-day conditions). For the genetic analysis of double crossing, the bri1-5 (Wassilewskija[Ws-2]) mutant was used. Seeds of WT and bri1-5 mutant were obtained from ABRC (Arabidopsis Biological Resource Center, Ohio University, Columbus, OH, USA). For the ATP experiment, adenosine 5′-triphosphate disodium salt hydrate (ATP; Sigma-Aldrich, St. Louis, USA) at concentrations of 125 and 250 μM was added to 1/2 MS medium. The ATPase inhibitor oligomycin (Calbiochem, Darmstadt, Germany) was used at concentrations of 25 and 50 μM. The seeds were germinated in darkness for 7 days at 22 °C. To induce salt stress, the seeds were germinated on 1/2 MS plates containing 0 and 125 mM of NaCl for 25 days. For the strong light stress analysis, the seeds were germinated on 1/2 MS medium under strong light (486.2 μmol m−2 s−1) for 25 days. Control plants were germinated in normal light (92.27 μmol m−2 s−1).
Screening for bil2-1D mutants
Approximately 10,000 of the RIKEN GSC Arabidopsis activation tagging lines (Nakazawa et al. 2003) were screened on 1/2 MS medium containing 3 μM Brz (Asami et al. 2000). After growth for 7 days in the dark, seedlings with hypocotyls longer than the controls were identified and transferred to the soil. TAIL-PCR was used to amplify the flanking genomic sequences of the T-DNA of pPCVICE4HPT, as previously described. Total RNA was extracted from the dark-grown 3-day-old seedlings of wild type and bil2-1D plants using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). First-strand cDNA was synthesized with PrimeScript (Takara, Kyoto, Japan), and used in quantitative real-time PCR (qRT-PCR). The qRT-PCR analysis was performed according to the instructions provided for the Thermal Cycler Dice (Takara) using a SYBR Premix ExTaq system (Takara). The following gene-specific primers were used for qRT-PCR analysis: for BIL2, 5′-TGAGTCCCTCAGGTCCCTTA-3′ and 5′-GCCGCCTCCTTGGTAGAG-3′; and for the constitutively expressed control gene ACT2, 5′-CGCCATCCAAGCTGTTCTC-3′ and 5′-TCACGTCCAGCAAGGTCAAG-3′.
Generating transgenic plants
In the recapitulation of the bil2-1D phenotype, BIL2 cDNA was amplified from Arabidopsis Col-0 cDNA with primers for BIL2-Forward 5′-CACCATGAACGCCGCCATTAGAGC-3′, and BIL2-Reverse 5′-TTAGTTCACAGATAGCTTTTCACA-3′ and cloned into pENTR/D-TOPO and subsequently cloned into the binary vector pGWB2 (Invitrogen; Nakagawa et al. 2007) containing a CaMV 35S promoter using a Gateway strategy. To generate RNAi constructs of BIL2, pENTR-BIL2 was subsequently cloned into the binary vector pGWB80 using a Gateway strategy.
For subcellular localization, the BIL2-GFP construct was generated. A 3-kb fragment, including the promoter region and open reading frame (ORF) of BIL2, was amplified from Arabidopsis Col-0 cDNA using primers for the BIL2 promoter: BIL2-GFP-Forward 5′-CACCTTGGAGAGAGATCAAAGAGGAACAATC-3′ and BIL2-GFP-Reverse 5′-GTTCACAGATAGCTTTTCACATAAAGA-3′. The PCR fragment was cloned into pENTR/D-TOPO and subsequently cloned into the binary vector pGWB5 using a Gateway strategy (Invitrogen, Carlsbad, CA, USA).
For the BIL2 promoter and GUS fusion construct, a 1.1-kb fragment, including the first exon and promoter region, was amplified from Arabidopsis Col-0 genomic DNA using primers for BIL2-GUS-Forward 5′-CACCTTGGAGAGAGATCAAAGAGG-3′ and BIL2-GUS-Reverse 5′-CGATTCCCAGGAAGTGCGAC-3′ and cloned into pENTR/D-TOPO and subsequently cloned into the binary vector pGWB3 using a Gateway strategy (Invitrogen).
The resulting p35S-BIL2, BIL2 promoter:BIL2-GFP and BIL2 promoter and GUS fusion constructs were transformed into Col-0 using the floral dipping method. The transgenic plants were screened on 1/2 MS medium containing 25 mg/l of kanamycin.
Quantitative real-time PCR
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen) from light-grown, wild-type 24- (BIL2-OX) and 7-day-old (BIL2-RNAi) seedlings; dark-grown 5-day-old (genes flanking the T-DNA insertion site) seedlings of wild-type and bil2-1D plants; and wild-type and bil2-1D plants transformed with BIL2-OX or BIL2-RNAi. The first-strand cDNA was synthesized using PrimeScript (Takara) and was used in quantitative real-time PCR (qRT-PCR). The qRT-PCR was performed according to the instructions provided for the Thermal Cycler Dice (Takara) using the SYBR Premix ExTaq system (Takara).
The following primers were used: TCH4-Forward 5′-CGAGTCTTGGAACGCTGAT-3′ and TCH4-Reverse 5′-CTTCTTGTTGAAAGCCACGG-3′; CPD-Forward 5′-CACTTCAAAGATGCTCGCACTT and CPD-Reverse 5′-CAGCTCGTAACCGGGACATAG-3′; At2g42020-Forward 5′-GCTTTGGCCGAGTGGTTAAG-3′ and At2g42020-Reverse 5′-AACTCTCGCGGGGAAAC-3′; At2g42030-Forward 5′-AGCCAGCGACGAATTA-3′ and At2g42030-Reverse 5′-CGTCTGAAACGTGACC-3′; At2g42040-Forward 5′-CGTGCATGTGATAAGTAGG-3′ and At2g42040-Reverse 5′-CACACACCCAATACATAGAG-3′; At2g42060-Forward 5′-CTCTGAATCCGAAACC-3′ and At2g42060-Reverse 5′-CAGCGTGAGAAGGATG-3′; At2g42070-Forward 5′-TTCAGATATTCGCGCCTTTA-3′ and At2g42070-Reverse 5′-GGAAGAACAAAGCCAATCCA-3′.
GUS staining
For histochemical detection of GUS expression 2-, 3-, 11- and 28-day-old seedlings of promoter:GUS:BIL2 transgenic plants were used. The samples were stained at 37 °C overnight in GUS staining solution as previously described (Ito and Fukuda 2002). To test the induction of GUS expression, 2- and 3-day-old transgenic seedlings were treated with brassinolide (BL) and Brz for 3 h.
Subcellular localization analysis by fluorescence microscopy
The roots of 5-day-old BIL2-GFP transgenic seedlings were harvested into a freshly prepared staining solution of 500 nM CM-H2XRos (MitoTracker Red; Invitrogen) for 15 min at room temperature. After staining, the seedlings were washed three times in 1/2 MS medium for approximately 10 min (Hedtke et al. 1999).
XylT (beta-1,2-xylosyltransferase)::RFP and HDEL (His-Asp-Glu-Leu)::RFP were generated by modifying XylT-GFP and HDEL-GFP and cloning into pGWB (Shoda et al. RIKEN Advanced Science Institute, Saitama, Japan, unpublished data). The BIL2-GFP transformant was generated as previously described. These plant organelle marker-RFP constructs were transformed into BIL2-GFP plants using floral dipping. Images of BIL2-GFP and Mitotracker fluorescence were captured using a LSM 700 laser scanning microscope (Zeiss, Oberkochen, Germany).
ATP measurement
Five-day-old dark mature seedlings were used. The samples were frozen in liquid N2 after harvest and stored at −80 °C. The samples were immersed in sterile water and boiled for 15 min at 98 °C to destroy any ATPases (Yang et al. 2002). Total ATP content in the supernatant was determined using a luciferase-based assay (Kikkoman, Tokyo, Japan), and the luminescence was measured in a Spectra MaX luminometer (Molecular Devices, Sunnyvale, CA, USA). The ATP content was correlated with luminescence by comparison with the ATP standard provided in the kit.
Results
Isolation and characterization of the bil2-1D mutant
Brz is a triazole-type compound that directly binds to the cytochrome P450 steroid C-22 hydroxylase encoded by the DWARF4 (DWF4) gene and specifically inhibits BR biosynthesis. Brz treatment reduces the BR content in plant and causes phenotypes with de-etiolation and dwarfism similar to BR-deficient mutants (Asami et al. 2000, 2001). Mutants that are insensitive to Brz can be activated in BR signaling and biosynthesis; therefore, we screened the brz-insensitive-long hypocotyl (bil) mutant in Arabidopsis. We have isolated the bzr1/bil1 mutant from EMS-mutation lines, and identified BZR1/BIL1 with a bHLH transcription factor that acts as a positive regulator in BR signaling (Wang et al. 2002; Asami et al. 2005; He et al. 2005).
In the dark, wild-type plants are typically etiolated with an elongated hypocotyl and a closed cotyledon. Treatment with Brz inhibits the elongation of hypocotyls and enhances the opening of the cotyledon in the wild-type plant, termed as de-etiolation in the dark. Under these growth conditions, we screened 10,000 Arabidopsis activation tagged lines and isolated a semi-dominant mutant, Brz-insensitive-long hypocotyls 2-1D (bil2-1D). bil2-1D mutant seedlings showed longer hypocotyls and closed cotyledons when grown in the dark on a medium containing Brz (Fig. 1a, b). In the dark without Brz, the hypocotyls of bil2-1D plants were elongated normally and were similar in length to the wild type.
Phenotype and characterization of bil2-1D. bil2-1D mutant showed Brz resistance (a, b). a Hypocotyl elongation of wild-type (WT) and bil2-1D seedlings grown on medium containing 0, 1, and 3 μM Brz in the dark for 7 days. b Hypocotyl length of wild-type (WT) and bil2-1D seedlings grown on medium containing 0, 1 and 3 μM Brz in the dark for 7 days. The results are presented as the mean ± SE (n > 30 seedlings). Triple asterisk indicates significant differences relative to the control at P < 0.001 based on the Student’s t test. c, d Phenotype of wild type and bil2-1D seedlings grown in soil under long-day conditions (16 h light, 8 h dark) for 30 days. Side view (c) and top view (d)
Light-grown bil2-1D mutants showed long petioles and outward-curling leaves similar to the overexpressed plants of the BR receptor, BRI1 (Wang et al. 2001) (Fig. 1c, d). These phenotypes suggested that bil2 mutants enhanced BR signaling.
Identification and characterization of the BIL2 gene
The co-segregation of the Brz-insensitive phenotype with the selection marker and TAIL-PCR indicated a T-DNA insertion in an intergenic region at the end of chromosome II in bil2-1D (Fig. 2a). The At2g42080 gene, located approximately 14.3 kbp downstream of the T-DNA insertion, was overexpressed (Fig. 2b). From T-DNA insertion to the At2g42080 gene, a 7.3-kbp gene was deleted (Fig. 2a). The knock-out mutant for At2g42030 (SALK_061281.28.80.x) and At2g42040 (SALK_007993) demonstrated the expression of each mRNA (Suppl. Fig. S1) and the same phenotype as the wild-type plant (data not shown). The increased expression of the At2g42060 and At2g42070 genes was also identified in bil2-1D compared with the wild type (Suppl. Fig. S1). The transformation and overexpression of genes for At2g42060 and At2g42070 in the wild-type plant showed the same hypocotyl and leaf phenotype as the wild-type plant (data not shown). These results suggested that the bil2-1D mutation is due to the mRNA overexpression of At2g42080 and the gene is named as BIL2.
Novel gene is candidate of bil2-1D mutant. a T-DNA insertion site in bil2-1D is indicated with blue, deletion site with green and the BIL2 gene with a red arrow. b Quantitative real-time PCR analysis of At2g42080 mRNA expression in dark-grown, 3-day-old wild-type (WT) and bil2-1D seedlings. The value was normalized against the expression of the ACTIN2 gene. The error bars indicate standard deviation (n = 3). c Alignment of BIL2 (NP_181738) and its homologs in Arabidopsis thaliana, At3g58020 (NP_191361) and At2g18465 (NP_849977). d Alignment of BIL2 (NP_181738) and its homologs in other plants: Arabidopsis lyrata 1 (XP_002881820), Arabidopsis lyrata 2 (XP_002876451), castor bean Ricinus communis (XP_002530712), soybean Glycine max (XP_003519035), grape Vitis vinifera (XP_002279725), and rice Oryza sativa (NP_001059604). The red box indicates the J domain, and yellow box indicates the HPD motif, which is important for the J domain
BIL2 encodes a novel protein, but is categorized in DnaJ/Heat shock protein 40 (Hsp 40) family, which function as molecular chaperones (Rajan and D’Silva 2009). Ordinary DnaJ/Hsp40 gene family function has chaperone activity to repair the misfolding of nascent polypeptides and protein aggregation during stress (Hartl 1996). DnaJ/Hsp40 contains a J domain, which is essential for interaction with DnaK/Hsp70, and this domain is conserved in BIL2 (Fig. 2c, d). The J domain contains a highly conserved histidine, proline and aspartate (HPD) motif, which is critical for the function of these proteins (Cheetham and Caplan 1998). A BLAST search revealed that two homologous genes of BIL2 were present in Arabidopsis thaliana: At3g58020 and At2g18465 (Fig. 2c) and some homologous genes in Arabidopsis lyrata, castor bean (Ricinus communis), soybean (Glycine max), grape (Vitis vinifera) and rice (Oryza sativa) (Fig. 2d).
The BIL2 gene promotes plant growth
To confirm that the overexpression of At2g42080 caused the bil2-1D phenotype, the At2g42080 coding region was placed immediately downstream of the CaMV 35S promoter and transformed into wild-type Arabidopsis. The obtained BIL2 over-expresser (BIL2-OX) showed long hypocotyls and closed cotyledons when grown in the dark on medium containing Brz (Fig. 3a, b).
BIL2-OX increased the growth of the hypocotyl, inflorescence and roots. a Hypocotyl elongation of wild-type (WT), bil2-1D, BIL2-OX1 and BIL2-OX2 seedlings grown on medium containing 3 μM Brz in the dark for 4 days. b Hypocotyl length of wild type (WT), bil2-1D, BIL2-OX1 and BIL2-OX2 seedlings grown on medium containing 0, 1 and 3 μM Brz in the dark for 4 days. The results are presented as the mean ± SE (n > 30 seedlings). c Real-time PCR analysis of the BIL2 gene expression in the wild-type (WT), BIL2-OX1 and BIL2-OX2 seedlings grown in the light for 24 days. The value was normalized against the expression of the ACTIN2 gene. The error bars indicate standard deviation (n = 3). d Phenotype of wild-type (WT), BIL2-OX1 and BIL2-OX2 seedlings grown in soil under long-day conditions (16 h light, 8 h dark) for 40 days. e–g Measurements indicating primary inflorescence length (e), secondary inflorescence number (f) and branch number (g) of wild-type (WT), BIL2-OX1 and BIL2-OX2 seedlings grown on soil for 40 days. The results are presented as the mean ± SE (n > 15 plants). h Root elongation of wild-type (WT), BIL2-OX1 and BIL2-OX2 seedlings grown on 1/2MS medium light 14 days. Wild-type (WT) is shown in white, BIL2-OX1 in pink and BIL2-OX2 in orange. i Primary root length of wild-type (WT), BIL2-OX1 and BIL2-OX2 grown on 1/2 MS medium in light for 14 and 21 days. j Lateral root number for seedlings grown in light for 14 days. Triple asterisk indicates significant differences relative to the control at P < 0.001 based on the Student’s t test
To analyze the role of BIL2 for plant growth, BIL2-OX and BIL2-RNAi transformants were generated and observed in detail. bil2-1D contains two gene deletions (At2g42030 and At2g42040).
Although the deletion of the two genes did not affect the bil2-1D phenotype (Suppl. Fig. S1), the potentiation effect of the gene deletion must be avoided. Because the BIL2 over-expresser exhibits BIL2 function, we used these transformants for further analysis.
In plants grown in soil under light, the primary inflorescence length of BIL2-OX expressing BIL2 mRNA was longer than that of the wild-type plants (Fig. 3c–e). Although the number of primary inflorescences was similar between BIL2-OX and wild-type plants (data not shown), the number of secondary inflorescences and branches in BIL2-OX were increased compared with the wild-type plant (Fig. 3f, g). BIL2-OX showed longer roots and increased lateral root numbers compared with the wild-type plant (Fig. 3h–j).
In contrast, BIL2-RNA interference vector was constructed downstream of the CaMV 35S promoter and transformed into wild-type Arabidopsis (BIL2-RNAi). Two BIL2-RNAi lines showed that BIL2 mRNA was decreased compared with the wild-type plant (Fig. 4a), showing shorter hypocotyls than wild-type plants when grown in the dark with and without Brz. The shortened hypocotyl of the BIL2-RNAi plants showed a dose-dependent response to the Brz concentration (Fig. 4b, c). Compared with the wild-type plants, the BIL2-RNAi lines did not have decreased homologous genes of BIL2 in Arabidopsis thaliana (Suppl. Fig. S2). Although we could not obtain and analyze the BIL2 gene knock-out mutant, the BIL2-RNAi phenotype suggested an effect of the BIL2 single knockdown. Light-grown BIL2-RNAi transformants showed shorter inflorescence and a tendency of decreased branches compared with wild-type plants (Fig. 4d). These results suggest that BIL2 plays an important role in positive plant growth.
The BIL2-RNAi mutant showed short hypocotyls and short inflorescence. a Real-time PCR analysis of the BIL2 gene expression in the wild-type (WT), BIL2-RNAi1 and BIL2-RNAi2 seedlings grown in the light for 7 days. The value was normalized against the expression of the ACTIN2 gene. The error bars indicate standard deviation (n = 3). b Hypocotyl elongation of wild type (WT) and BIL2-RNAi1 and BIL2-RNAi2 seedlings grown on medium containing 3 μM Brz in the dark for 7 days. c Hypocotyl length of wild-type (WT), BIL2-RNAi1 and BIL2-RNAi2 grown on medium containing 0, 0.3, 1 and 3 μM Brz in the dark for 7 days. The results are presented as the mean ± SE (n > 30 seedlings). Triple asterisk indicates significant differences relative to the control at P < 0.001 based on the Student’s t test. d Phenotype of wild type (WT), BIL2-RNAi1 and BIL2-RNAi2 seedlings grown in soil under long-day conditions (16 h light, 8 h dark) for 40 days
The BR-responsive gene was regulated in BIL2-OX and BIL2-RNAi
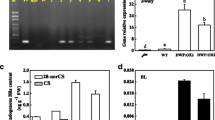
Although the hypocotyl insensitivity of bil2-1D and BIL2-OX against Brz showed that BIL2 protein can be related to BR signaling, the relationship between BIL2 and BR at the molecular level has not been clarified. To reveal the importance of BIL2 for BR signaling, BR-stimulated gene expression in BIL2-OX and BIL2-RNAi transformants was analyzed. Quantitative real-time PCR (qPCR) showed that the BIL2 mRNA expression was higher in BIL2-OX (Fig. 3c) but lower in BIL2-RNAi compared with the wild-type plant (Fig. 4a). The BR-positive regulatory gene THC4 showed higher expression, and the BR biosynthetic gene CPD, which is downregulated with BR stimulation through a feedback mechanism and showed lower expression in BIL2-OX than in the wild-type plant (Fig. 5a, b). In contrast, the BR-positive regulatory gene TCH4 showed lower expression, and CPD showed higher expression in BIL2-RNAi compared with the wild-type plant (Fig. 5c, d). These results suggest that BIL2 plays an important role for BR signaling in the regulation of BR gene expression.
The BR-responsive gene was regulated in BIL2-OX and BIL2-RNAi. a, b Real-time PCR analysis of the regulated BR gene expression in the wild-type (WT), BIL2-OX1 and BIL2-OX2 seedlings grown in the light for 24 days. c, d Real-time PCR analysis of the regulated BR gene expression in the wild-type (WT), BIL2-RNAi1 and BIL2-RNAi2 seedlings grown in the light for 7 days. All values were normalized against the expression of the ACTIN2 gene. The error bars indicate standard deviation (n = 3)
BIL2 plays a role downstream of BRI1 in BR signaling
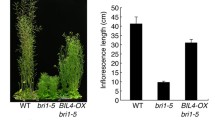
BIL2 functions downstream of BR biosynthesis, as bil2-1D showed resistance against the BR biosynthesis inhibitor Brz (Fig. 1a, b), and functions upstream of BR gene expression (Fig. 5). To determine where BIL2 acts in BR signaling, the genetic interaction between BIL2-OX and the BR receptor mutant bri1-5 was analyzed (Li and Chory 1997). When BIL2-OX1 was crossed with bri1-5, BIL2-OX1 suppressed shorter hypocotyl of bri1-5 in the dark (Fig. 6a) and a shorter inflorescence of bri1-5 in the light (Fig. 6b). These results suggested that the BIL2 plays an important role downstream of BRI1 in BR signaling.
BIL2 plays a role downstream of BRI1 in BR signaling. a, b The BIL2-OX suppresses the dwarf phenotype of BR receptor mutant bri1-5, in the dark (a) and in the light (b). The plants of wild-type (WT), bri1-5 and bri1-5 × BIL2-OX seedlings grown on 3 μM Brz medium in the dark for 7 days (a) or in soil for 40 days (b)
The BIL2 gene is expressed in many plant organs
As BIL2 is a novel gene, the analysis of BIL2-expressed organs can reveal the function of BIL2 in plant growth. To investigate the specific expression of BIL2 in various developmental stages of Arabidopsis, the BIL2 promoter was constructed upstream of glucuronidase (GUS) and transformed into wild-type Arabidopsis. In 2 and 3 days after germination, BIL2 was highly expressed in the hypocotyl after BL treatment (Fig. 7b, e), and low expression was observed in the hypocotyl after Brz treatment (Fig. 7c, f) in comparison with the hypocotyl in 1/2 MS medium (Fig. 7a, d). The qPCR analysis showed that the BIL2 mRNA expression responses to BL and Brz were reproduced in the wild-type plants (Fig. 7g). The BIL2 promoter region does not have a responsive element regulated by BES1 and BZR1/BIL1; thus, an unknown functional element regulated by unknown transacting factors might exist in the BIL2 promoter. At 11 days after germination, BIL2 was highly expressed in the shoot apical meristem (SAM) and lateral root (Fig. 7h, i). At 28 days after germination, BIL2 was expressed in the flowering bud and pollen (Fig. 7j, k). These results suggested that BIL2 plays an important role for plant development in many stages of the plant life cycle.
The BIL2 gene is expressed in many plant organs. BIL2::promoter::GUS expression patterns in transgenic Arabidopsis plants. Dark 2- and 3-day-old seedlings on the 1/2 MS medium (a, d) treated with BL 100 nM (b, e) and 3 μM Brz (c, f). Real-time PCR analysis of BIL2 gene expression in wild-type seedlings grown in the light for 10 days and treated with 100 nM of BL and 3 μM of Brz (g). The values were normalized against expression of the ACTIN2 gene. The error bars indicate the standard deviation (n = 3). Light-grown 11-day-old seedlings of the shoot apical meristem (SAM) (h) and root (i). Light-grown 28-day-old seedlings of flower bud (j) and pollen (l). Scale bars 0.5 mm (a–f, h) and 2 mm (i–k)
The BIL2 protein localized in the mitochondria
To determine the subcellular localization of the BIL2 protein, a BIL2 promoter BIL2 gene-green fluorescent protein (GFP) fusion construct was transformed into wild-type Arabidopsis. Observation by confocal scanning of the BIL2-GFP transgenic seedling roots revealed that the BIL2-GFP was observed in a dot-like structure (Fig. 8a). When co-stained with Mitotracker red, which specifically stains mitochondria (Hedtke et al. 1999) (Fig. 8b), the two staining patterns overlapped (Fig. 8c). The dot-like structure shown by BIL2-GFP did not co-localize with FM4-64 as an endosome marker, XylT-RFP as a Golgi marker and HDEL-RFP as an ER marker (Suppl. Fig. S3). These results indicated that BIL2 was localized to mitochondria.
BIL2 may be involved in ATP synthesis in the mitochondria
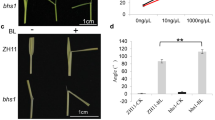
Although the BR signaling factor localized in many plant organelles (Belkhadir et al. 2006), the BR signaling factor in mitochondria had not been determined. To consider the molecular mechanism of plant growth regulation in BR signaling by the mitochondrial protein BIL2, we noted that ATP synthesis that was a major role of mitochondria for plant growth. The relationship between BIL2 and ATP in the hypocotyl elongation was then analyzed. Exogenous ATP promotes hypocotyl elongation and also restores hypocotyl elongation, which was shortened by Brz treatment in the dark (Fig. 9a). The hypocotyl elongation of Arabidopsis is inhibited by oligomycin, an inhibitor of the respiratory chain complex V of the mitochondrial electron transport and a blocker of H+-ATP synthesis. The hypocotyl of BIL2-OX showed partial but significant resistance against oligomycin-induced inhibition of hypocotyl elongation in comparison with the wild-type plant (Fig. 9b). To reveal the direct interaction between ATP synthesis and BIL2, the ATP concentration in the BIL2-OX and wild-type plant was analyzed. In dark-germinated plants for 5 days, the endogenous ATP in two lines of BIL2-OX was higher than in the wild-type plant (Fig. 9c). To analyze the relationship between ATP and BR signaling in Arabidopsis, BR-responsive gene expression was analyzed in the Arabidopsis wild-type plants after a 3-h ATP treatment (Fig. 9). In the plants germinated in the dark for 7 days, TCH4 expression, which is upregulated by BR stimulation, showed increasing dependence on the ATP concentration (Fig. 9d). By contrast, the BR biosynthetic gene CPD, the expression of which is downregulated by BR stimulation through a feedback mechanism, showed decreasing expression that was dependent on the ATP concentration (Fig. 9e). These results suggested that ATP synthesis was promoted through the action of BIL2 in the mitochondria and elongated the hypocotyl against the BR biosynthesis inhibitor Brz. BR signaling-related gene expression was also promoted by ATP synthesis through BIL2.
ATP production by BIL2 promotes hypocotyl elongation. a Hypocotyl elongation by ATP. Hypocotyl lengths of dark 7-day-old seedlings grown in the absence or presence of 1 μM Brz or ATP (125 and 250 μM). Combinatorial treatments are indicated. b Hypocotyl lengths of dark 7-day-old seedlings grown in the absence or presence of oligomycin (OM) (25 and 50 μM). Twenty-five seedlings per treatment were analyzed in each experiment. The data shown are the mean ± SE. c Total ATP concentration in dark-grown 5-day-old seedlings of wild-type (WT), BIL2-OX1 and BIL2-OX2. The data shown are the mean ± SE of three independent experiments. d, e ATP can increase BR signaling. Real-time PCR analysis of regulated BR gene expression in wild-type (WT) seedlings grown in the dark for 7 days and treated with ATP for 3 h
BIL2 increases salt tolerance and strong light tolerance in Arabidopsis
Although the BIL2 gene is a novel gene, it has been classified as a DnaJ gene family member in previous reports (Rajan and D’Silva 2009). The DnaJ gene family is included in heat shock proteins (HSPs) that play roles to protect against proteotoxic stress and to regulate the protein quality control system for assistance (Vos et al. 2008). To reveal the possible molecular function of BIL2, the environmental stress tolerance of the wild-type plant and BIL2-OX was analyzed. For the analysis of salt tolerance, plants grown on medium were divided into groups according to the fresh weight of the above-ground parts in plants grown in 1/2 MS medium and NaCl medium, and the numbers of each group were counted. The number of heavy and surviving plants of BIL2-OX grown on 125 mM NaCl was higher than wild-type plants (Fig. 10a, b). In the analysis of strong light stress tolerance, the number of heavy and surviving plants of BIL2-OX grown in the strong light (486.2 μmol m−2 s−1) was higher than wild-type plants (Fig. 11a, b). The reaction of BRI1-OX was similar to that of wild-type plants under salt stress (Fig. 10) and strong light stress (data not shown). These results suggest that BIL2-OX shows potential resistance against salt and strong light stress that depends on the overexpression of BIL2. Thus, BIL2 maintains its function to protect against proteotoxic stress and can be classified as a DnaJ family protein according to protein function. ATP treatment promoted salt tolerance in the wild-type plants (Suppl. Fig. S4), and the direct treatment of BL caused a weak salt tolerance in the wild-type plants (data not shown). These results showed that although ATP synthesis supported by BIL2 plays important roles, the direct BL signaling effect might be weaker than ATP itself.
BIL2-OX showed resistance against salt stress. a Fresh weight above-ground parts of each plants were divided in four groups (0–5, 5–10, 10–15, and 15–20 mg) and the plant numbers were counted in each group. Individual numbers of the wild-type (WT), BIL2-OX1, BIL2-OX2 and BRI-OX seedlings germinated on half-strength MS medium with (right graphics) or without (left graphics) 125 mM NaCl and under light for 25 days. b BIL2-OX and wild-type (WT) plant phenotypes under salt stress. Photographs of the seedlings were obtained at 25 days after germination. The data shown are the mean ± SE of three independent experiments. Asterisk indicates significant differences relative to the control at P < 0.05 based on Student’s t test
BIL2-OX showed resistance against strong light stress. a The number of plants was counted using the same method as with the salt resistance analysis explained in Fig. 12. The individual numbers of the wild-type (WT), BIL2-OX1 and BIL2-OX2 seedlings germinated on half-strength MS medium under normal light (left graphics) or strong light (left graphics) for 25 days. b BIL2-OX and wild-type (WT) plant phenotype in strong light stress. Photographs of the seedlings were obtained at 25 days after germination. The data shown are the mean values
Discussion
BR regulates many processes in plant growth and development, such as cell division and elongation (Clouse and Sasse 1998). BR biosynthesis-deficient mutants of Arabidopsis, de-etiolated 2 (det2) and dwrf4, (Li et al. 1996; Choe et al. 1998) showed a pleiotropic dwarf phenotype that can be recovered to a wild-type phenotype by feeding of BR (Asami et al. 2005); however, the BR receptor-deficient mutant bri1 displays a pleiotropic dwarf phenotype, including root elongation, which was not rescued by BR (Clouse et al. 1996). BR binds with BRI1, a member of the leucine-rich repeat kinase family (Li and Chory 1997; Kinoshita et al. 2005). The detailed mechanism downstream of BRI1 in BR signaling has been studied, and all these results have been revealed through loss-of-function mutants for the biosynthesis of BR and the BR receptor. BIN2, BZR1/BIL1 and BES1, the other BR signaling components, were identified through gain-of-function mutants.
Brz is a specific inhibitor of BR biosynthesis that inhibits the hydroxylation of the C-22 position of the side chain in BR by direct binding to DWF4 enzyme, a cytochrome P450 monooxygenase, through the triazole base of Brz (Asami et al. 2001). To analyze the mechanisms of BR signal transduction, we performed a chemical genetics screening using Brz from gain-of-function mutants. We screened Arabidopsis activation tagging lines and isolated the bil2-1D mutant, which displayed longer hypocotyls characteristic of cell elongation on medium containing Brz in the dark than the wild-type plant. Light-grown bil2-1D exhibited a long petiole phenotype similar to wild-type plants treated with BR or BRI1-OX mutants. The BR marker gene TCH4, the expression of which was upregulated by BR stimulation, was induced in the BIL2-OX but not in the wild-type. Conversely, CPD, the other BR biosynthetic gene, the expression of which is downregulated by BR stimulation, was suppressed in the BIL2-OX but not in the wild-type. BIL2-OX could suppress dwarfing of the BRI1-deficient mutant. BIL2 gene expression was induced by BL treatment and suppressed by Brz, suggesting that BIL2 is directly regulated by BL and is involved in cell elongation through BR signaling.
BIL2 is a novel protein, but analysis of the amino acid sequence revealed that it belongs to the DnaJ/Hsp40 family proteins. DnaJ/Hsp40 proteins are functional partners for DnaK/Heat shock protein 70 (Hsp70s) involved in protein folding, translation, stabilization and protein translocation across cell membrane. All members of DnaJ/Hsp40 contain a “J domain”, which is essential for interaction with DnaK/Hsp70s. The J domain contains a highly conserved histidine, proline and aspartate (HPD) motif, which is critical for their functions. Some members of the DnaJ/Hsp40 protein family contain other conserved regions, such as the glycine/phenylalanine rich region, termed the “G/F region” and a zinc-binding cysteine-rich sequence, termed the “zinc-finger domain”. DnaJ/Hsp40 proteins are classified into three types on the basis of differences in these regions (Szyperski et al. 1994; Cheetham and Caplan 1998). Type I proteins contain all domains/motifs that include the J domain, the G/F region, and the zinc-finger domain. Type II proteins possess the J domain and the G/F region, but lack the zinc-finger domain. Type III proteins possess only the J domain (Kelley 1998; Fan et al. 2004; Walsh et al. 2004). BIL2 is classed as a type III J protein (Rajan and D’Silva 2009). DnaJ/Hsp40 is widely distributed in plants, animals and humans. The DnaJ/Hsp40 protein family is composed of six homologs in Escherichia coli, 22 homologs in Saccharomyces cerevisiae and 41 homologs in humans (Qiu et al. 2006). Arabidopsis has more than 400 DnaJ/Hsp40 protein families (Rajan and D’Silva 2009). These family protein functions have not been elucidated, and the BIL2 functions were not known. The J domain amino acid sequence of BIL2 is similar to the human DnaJ protein that possesses a tetratricopeptide repeat 2 (TPR2). The TPR proteins function in various cellular processes, including cell-cycle control, mitochondrial and peroxisomal protein transport, stress response, and protein kinase inhibition (Goebl and Yanagida 1991). DnaJ/Hsp40 proteins regulate DnaK/Hsp70 and other proteins as a function of chaperones. The DnaJ/Hsp40 protein TPR2 (DnaJC7) is involved in the folding of many proteins, and TPR2 mediates the retrograde transfer of substrates from Hsp90 to Hsp70 (Brychzy et al. 2003).
BIL2-GUS expression was observed in hypocotyl during early stage and strongly expressed in pollen during in the flower developmental stage. BR biosynthetic and signaling deficient mutants showed reduced pollen number, viability, and release efficiency (Ye et al. 2010). MALE GAMETOPHYTE DEFECTIVE 1 (MGP1) encodes the FAd subunit of mitochondrial F1F0-ATP synthase in Arabidopsis which was highly expressed in pollen and plays important roles in pollen formation (Li et al. 2010).
BIL2 localization was detected in the mitochondria. Although BR signaling proteins are localized in many organelles, i.e., cellular membrane, nuclei, endoplasmic reticulum (ER) and vacuole (Belkhadir et al. 2006), this study is the first to report BR signaling protein localization in the mitochondria. Our most interesting but difficult discovery during the BIL2 analysis was the localization of BIL2 in mitochondria. We would like to discuss regulation mechanism of BR signaling and plant growth by the mitochondrial protein.
ATP is a vital factor in plant growth, essentially representing a major energy source of the cell. Most of the plant ATP is primarily produced in the mitochondria and secondarily in the chloroplast (Haferkamp et al. 2011). Therefore, understanding the mechanisms involved in ATP synthesis in the mitochondria is important. We hypothesized that ATP synthesis in the mitochondria by ATPase promotes hypocotyl elongation, and ATPase folding or stabilization might be facilitated through BIL2 as a DnaJ/Hsp40 function in the mitochondria. Although wild-type seedlings grown in medium containing Brz had shorter hypocotyls, BIL2-OX had a longer hypocotyl due to its resistance to Brz. The wild-type hypocotyl phenotype was recovered after treatment with ATP in medium containing Brz. These results showed that ATP plays an important role in hypocotyl elongation against Brz. The mitochondrial ATPase inhibitor oligomycin, which blocks the respiratory chain complex, inhibited hypocotyl elongation, but hypocotyl elongation of BIL2-OX was resistant to oligomycin compared to the wild-type plant. BIL2-OX produced higher exogenous ATP than the wild-type plant. These results support our hypothesis that BIL2 facilitates the folding or stability of ATPase in the mitochondria during plant growth (Fig. 12).
Environmental stress causes misfolding, aggregation and degradation for each organelle protein in the plant. In plant mitochondria, these proteotoxic damages have been observed, and ATP generation plays an important role in stress resistance (Jacoby et al. 2011). BIL2-OX showed resistance against salinity stress and strong light stress. ATP treatment promoted salinity resistance in wild-type Arabidopsis grown in medium containing NaCl (Suppl. Fig. S4). BIL2 is classified as a member of the DnaJ/Hsp40 family (Rajan and D’Silva 2009), but the actual function of the BIL2 protein has not been elucidated. The effects of BIL2 resistance against environmental stress might support the function of BIL2 as a DnaJ/Hsp40 molecular chaperone in the plant mitochondria. Further analysis will reveal the function of BIL2 in detail.
Abbreviations
- BIL2:
-
Brz-insensitive-long hypocotyls 2
- BIL2-OX:
-
BIL2 overexpression
- BRI1:
-
BRASSINOSTEROID INSENSITIVE 1
- BKI1:
-
BRI1 KINASE INHIBITOR 1
- BAK1:
-
BRI1-ASSOCIATED RECEPTOR KINASE 1
- BSU1:
-
bri1-5 suppressor 1
- BIN2:
-
BRASSINOSTREROID INSENSITIVE 2
- BZR1:
-
BRASSINOZOLE RESISTANT 1
- BES1:
-
bri1-EMS-SUPPRESSOR 1
- BR:
-
Brassinosteroid
- BL:
-
Brassinolide
- Brz:
-
Brz220
- CaMV:
-
Cauliflower mosaic virus
- DnaJ:
-
J-protein, Hsp40
- Hsp40:
-
40 kDa heat shock protein
- HPD motif:
-
Histidine, proline, aspartate motif
- MS:
-
Murashige and Skoog
- RNAi:
-
RNA interference
- TAIL-PCR:
-
Thermal asymmetric interlaced PCR
References
Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123:93–100
Asami T, Mizutani M, Fujioka S, Goda H, Min YK, Shimada Y, Nakano T, Takatsuto S, Matsuyama T, Nagata N, Sakata K, Yoshida S (2001) Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J Biol Chem 276:25687–25691
Asami T, Nakano T, Fujioka S (2005) Plant brassinosteroid hormones. Vitam Horm 72:479–504
Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314:1410–1411
Belkhadir Y, Wang X, Chory J (2006) Arabidopsis brassinosteroid signaling pathway. Sci STKE 2006(364):cm5
Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WM (2003) Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J 22:3613–3623
Caesar K, Elgass K, Chen Z, Huppenberger P, Witthoft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K (2011) A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J 66:528–540
Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3:28–36
Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10:231–243
Clouse SD, Sasse JM (1998) BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49:427–451
Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111:671–678
Fan CY, Lee S, Ren HY, Cyr DM (2004) Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell 15:761–773
Goebl M, Yanagida M (1991) The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci 16:173–177
Haferkamp I, Fernie AR, Neuhaus HE (2011) Adenine nucleotide transport in plants: much more than a mitochondrial issue. Trends Plant Sci 16:507–515
Hamasaki H, Yoshizumi T, Takahashi N, Higuchi M, Kuromori T, Imura Y, Shimada H, Matsui M (2012) SD3, an Arabidopsis thaliana homolog of TIM21, affects intracellular ATP levels and seedling development. Mol Plant 5:461–471
Hartl FU (1996) Molecular chaperones in cellular protein folding. Nature 381:571–579
He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307:1634–1638
Hedtke B, Meixner M, Gillandt S, Richter E, Borner T, Weihe A (1999) Green fluorescent protein as a marker to investigate targeting of organellar RNA polymerases of higher plants in vivo. Plant J 17:557–561
Ito J, Fukuda H (2002) ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14:3201–3211
Jacoby RP, Taylor NL, Millar AH (2011) The role of mitochondrial respiration in salinity tolerance. Trends Plant Sci 16:614–623
Kelley WL (1998) The J-domain family and the recruitment of chaperone power. Trends Biochem Sci 23:222–227
Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433:167–171
Kumar S, Yoshizumi T, Hongo H, Yoneda A, Hara H, Hamasaki H, Takahashi N, Nagata N, Shimada H, Matsui M (2012) Arabidopsis mitochondrial protein TIM50 affects hypocotyl cell elongation through intracellular ATP level. Plant Sci 183:212–217
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295:1299–1301
Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272:398–401
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110:213–222
Li WQ, Zhang XQ, Xia C, Deng Y, Ye D (2010) MALE gametophyte defective 1, encoding the FAd subunit of mitochondrial F1F0-ATP synthase, is essential for pollen formation in Arabidopsis thaliana. Plant Cell Physiol 51:923–935
Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18:448–460
Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, Suzuki K, Muto S, Matsui M (2003) Activation tagging, a novel tool to dissect the functions of a gene family. Plant J 34:741–750
Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110:203–212
Qiu XB, Shao YM, Miao S, Wang L (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63:2560–2570
Rajan VB, D’Silva P (2009) Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Integr Genomics 9:433–446
Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wuthrich K (1994) NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc Natl Acad Sci USA 91:11343–11347
Tonon C, Cecilia Terrile M, Jose Iglesias M, Lamattina L, Casalongue C (2010) Extracellular ATP, nitric oxide and superoxide act coordinately to regulate hypocotyl growth in etiolated Arabidopsis seedlings. J Plant Physiol 167:540–546
Vos MJ, Hageman J, Carra S, Kampinga HH (2008) Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry 47:7001–7011
Walsh P, Bursac D, Law YC, Cyr D, Lithgow T (2004) The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep 5:567–571
Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313:1118–1122
Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410:380–383
Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2:505–513
Yang NC, Ho WM, Chen YH, Hu ML (2002) A convenient one-step extraction of cellular ATP using boiling water for the luciferin-luciferase assay of ATP. Anal Biochem 306:323–327
Ye Q, Zhu W, Li L, Zhang S, Yin Y, Ma H, Wang X (2010) Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc Natl Acad Sci USA 107:6100–6105
Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191
Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120:249–259
Acknowledgments
We thank Dr. Tsuyoshi Nakagawa (Shimane University) for the gift of the gateway vectors, pGWB2, pGWB80, pGWB5, and pGWB3. This work was supported in part by funding from the Program for Promotion of Basic Research Activities for Innovation Bioscience (PROBRAIN) to T.N. and T.A., and CREST, Japan Science and Technology Agency to T.N. and T.A.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Bekh-Ochir, D., Shimada, S., Yamagami, A. et al. A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta 237, 1509–1525 (2013). https://doi.org/10.1007/s00425-013-1859-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1859-3