Abstract
The comparative study of photosynthetic regulation in the thylakoid membrane of different phylogenetic groups can yield valuable insights into mechanisms, genetic requirements and redundancy of regulatory processes. This review offers a brief summary on the current understanding of light harvesting and photosynthetic electron transport regulation in different photosynthetic eukaryotes, with a special focus on the comparison between higher plants and unicellular algae of secondary endosymbiotic origin. The foundations of thylakoid structure, light harvesting, reversible protein phosphorylation and PSI-mediated cyclic electron transport are traced not only from green algae to vascular plants but also at the branching point between the “green” and the “red” lineage of photosynthetic organisms. This approach was particularly valuable in revealing processes that (1) are highly conserved between phylogenetic groups, (2) serve a common physiological role but nevertheless originate in divergent genetic backgrounds or (3) are missing in one phylogenetic branch despite their unequivocal importance in another, necessitating a search for alternative regulatory mechanisms and interactions.
Similar content being viewed by others
Introduction
The complexity, limitations and possibilities for exploitation of photosynthesis can be fully understood only in the context of extensive comparative studies on photosynthetic organisms as different as unicellular algae are from seed-producing land plants. Direct evolutionary lineages provide sound bases for such studies, but perhaps even more interesting are species that diverged early on in photosynthetic development and arrived at independent solutions for environmental challenges shared by all photosynthetic organisms. The present article aims at a phylogenetic comparison of the photosynthetic machinery in terms of light harvesting and photosynthetic electron transport mechanisms in different eukaryotic photosynthetic organisms. Cyanobacteria, the progenitors of chloroplasts, are excluded here due to profound differences in regulation of light harvesting and electron transfer and will be reviewed elsewhere. Special attention is paid to diatoms not only because their plastids originated from secondary endosymbiosis of a red algae, separate from the green plastid lineage, but also because they possess features typical of both animals and green algae, providing a fascinating example of a “patchwork genome”. Many diatom species are especially suited to rapid changes in light and nutrients, suggesting that the availability of a wider choice of genes has led to unique optimization of photosynthesis in this organism group. Indeed, both functional and structural properties of diatom and higher plant chloroplasts show distinct differences.
The most fundamental difference between plant and diatom chloroplasts is the number of chloroplast envelope membranes. Higher plants have two membranes while chloroplasts originating in a secondary endosymbiosis are engulfed by four (see Figs. 1 and 2 for details on secondary endosymbiosis origin). Apart from this difference, the organization of the thylakoid membrane is strikingly different. In vascular plants the thylakoid membrane is distinctly segregated into stacked regions called grana that are rich in PSII, and unstacked regions called stroma lamellae enriched in PSI (Anderson 1986). Aquatic organisms share a lack of excessive thylakoid stacking and diatoms form no exception (Larkum and Vesk 2003). Diatom thylakoids, as well as those of other Chl a/c-containing organisms, are organized into loosely appressed bands of three lamellae (see Fig. 1, reviewed by Larkum and Vesk 2003) and the number of bands does not change upon acclimation to different light conditions (Lepetit et al. 2012). Unlike land plants, the spatial distribution of photosystems in diatom thylakoids is approximately even between loosely appressed and the stroma-exposed unappressed regions (Pyszniak and Gibbs 1992). Another interesting aspect is that although PSI and PSII of all eukaryotic photosynthetic species share a common cyanobacterial origin, their subunit compositions can still vary between different groups. For instance, the oxygen evolving complex of PSII contains three subunits in higher plants, four subunits in red alga and five in diatoms (Nagao et al. 2010). The role of these additional subunits is still unclear. The difference in PSI subunit composition between plants and diatoms is even more pronounced, with six PSI proteins found in higher plants (Jensen et al. 2007) lacking homologues in diatoms (Grouneva et al. 2011). Conversely diatoms and red algae possess a unique subunit PsaM. Some differences in PSI composition might reflect the lack of state transitions in diatoms (Owens 1986).
Schematic overview of chloroplast and thylakoid structure in diatoms (left) and higher plants (right). Chloroplasts of higher plants have two envelope membranes. They are the product of a primary endosymbiosis in which a prokaryotic photosynthetic organism was engulfed by a heterotrophic eukaryote and reduced to an organelle. Diatom plastids are engulfed by four membranes, a consequence of their origin in a secondary endosymbiosis event, where a heterotrophic eukaryote acquired a photosynthetic eukaryote. The thylakoids of diatoms form triple lamellae. One of those encircles the chloroplast stroma. The pyrenoid is a RubisCO-enriched structure within algal chloroplasts dedicated to CO2-fixation
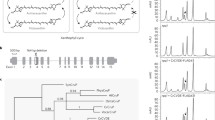
Simplified phylogenetic relationships between main photosynthetic eukaryotic organism groups and the distribution of LHC proteins. The species listed are the ones used in Table 1. Rhodophyta and Heterokonts belong to what is referred to as the “red lineage” in the text. Heterokonts, Cryptophytes and Haptophytes contain Chl a/c and together comprise the group of the Chromophytes. LHC are colour-coded as follows: RedCAP red; LHC green; PsbS pink; LHCSR yellow; ELIP grey; OHP black
Divergent light-harvesting antenna systems
The main characteristic of light-harvesting complex (LHC) proteins is their ability to bind pigments. This is why they are also called CABs (chlorophyll a/b-binding proteins). The three major groups of photosynthetic eukaryotes are distinguished according to their light-harvesting antennae; in Chlorophytes (green algae and land plants) these bind chlorophyll a and b, in Chromophytes (including brown algae and diatoms) chlorophyll a and c, while Rhodophytes (red algae) have only chlorophyll a, but also use extrinsic phycobilin-containing structures called phycobilisomes as light-harvesting antenna (Raven 1970). Diatoms and other Chromophytes originated in a secondary symbiosis event between a non-photosynthetic eukaryote and an organism of red algal origin (Delwiche 1999; Palmer 2003; Moustafa et al. 2009; see Fig. 2 for a simplified view of Chromophyta and Chlorophyta phylogenetic origin), but the LHCs in extant Chromophytes are fucoxanthin-chlorophyll a/c-binding proteins (FCPs) that are more similar to Chlorophytes LHC proteins than to red algal antenna proteins. This peculiarity is in line with the unique genetic footprint of diatoms, in which 16 % of nuclear coding potential can be traced to green origin (Moustafa et al. 2009). There is mounting evidence that the diatom plastid may be the product of multiple endosymbiotic events (Chan et al. 2011), one with an early marine green algae of the Prasinophyceae class (Frommolt et al. 2008).
In addition to chlorophylls, LHCs also bind a number of carotenoids. Carotenoids, mainly β-carotene derivatives such as the pigments of the xanthophyll cycle (Müller et al. 2001), play a central role in photoprotection (see below). Diatoms cannot synthesize pigments of the α-carotene branch (Pennington et al. 1988; Frommolt et al. 2008), so they contain fucoxanthin (β-carotenoid) as their main auxiliary pigment instead of lutein (α-carotenoid), which is the main auxiliary pigment in green algae and plants.
LHC proteins have the task of both optimizing light harvesting and at the same time protecting photosystems from light-induced damage. The extended LHC protein family can be sub-divided into three-helix LHCs (including LHCSR), four-helix PsbS and LHC-like proteins, based on their phylogenetic similarity (see also Fig. 2). For a detailed summary of LHCs, their number of helices and phylogenetic distribution, the reader is specifically referred to Engelken et al. (2010). According to their function, however, LHCSR and PsbS proteins fall into the same category of proteins involved in non-photochemical quenching (NPQ). One theory on the evolution of the LHC family suggests that their primary function might have been entirely photoprotective and that their light-harvesting role was acquired later (Montane and Kloppstech 2000). “Photoprotection” is a collective term for processes that lead to downregulation of light harvesting in favour of excitation energy dissipation (as heat) as well as processes that protect photosystems from over-excitation and detoxify reactive oxygen species.
LHC proteins comprising the light-harvesting systems
In higher plants, light-harvesting protein complexes are generally associated with either PSII (LHCII) or PSI (LHCI) (nomenclature by Jansson et al. 1999). The LHCII complex contains the CAB proteins Lhcb1, Lhcb2 and Lhcb3, encoded by the Lhcb1-3 genes, and the minor intrinsic CABs CP29, CP26 and CP24 (encoded by Lhcb4–6). The PSI-specific antenna contains the major CABs Lhca1-4 and the minor subunits Lhca5 and Lhca6 (Jansson et al. 1999). In lower plants such as mosses, ferns and especially algae the situation is less clear-cut. Especially for Chl a/c-containing organisms, the issue is complicated by multiple secondary endosymbiosis events. A recent extensive study of chlorophyll c-containing algae demonstrated that several different LHC subfamilies are common to algal lineages that are only distantly related to each other (Hoffman et al. 2011), suggesting a complex evolutional history of LHCs.
LHC proteins specifically involved in photoprotection
PsbS/LHCSR proteins are involved in the generation of NPQ (Li et al. 2000b) and some representatives are accordingly expressed only under high light conditions. LHCSR (also called LI818 in C. reinhardtii and Lhcx in diatoms) is found in green algae, mosses and diatoms (reviewed in Depauw et al. 2012), but not in vascular plants, red algae or cyanobacteria (Peers et al. 2009). PsbS, on the other hand, occurs in vascular plants. LHCSRs are believed to have been among the first eukaryotic LHCs (Richard et al. 2000; Koziol et al. 2007) and LHC proteins in both chlorophyll a/b and a/c-containing organisms were even suggested to have evolved from a PsbS ancestor (containing four helices) by the loss of one transmembrane helix (Green and Pichersky 1994). This, however, would also suggest a loss of this ancestral protein in the red lineage.
LHCSR proteins have been studied most extensively in C. reinhardtii (Richard et al. 2000). They are not as tightly embedded in the thylakoid membrane as is the case with LHCII/LHCI and are found in stroma-exposed thylakoid regions (Richard et al. 2000). During plastid development, LHCSR proteins are expressed before CABs, further strengthening their protective role, and are suggested to be of ancient origin, preceding the division of green, red and brown algae (members of the Heterokontophyta alongside diatoms) into separate groups (Richard et al. 2000). One example underlining the separate phylogenetic history of PsbS and LHCSR proteins is the fact that the moss Physcomitrella patens not only encodes both proteins but uses both for NPQ formation (Alboresi et al. 2010). In conclusion, NPQ might be one intriguing example of a mechanism that evolved once in aquatic organisms mediated by the LHCSR protein class, which was then succeeded or replaced by a similar mechanism mediated by the PsbS protein.
LHC-like proteins
LHC-like proteins are grouped into ELIPs (early light-induced proteins), SEPs (stress-enhanced proteins), OHPs (one helix proteins) and LHL4 (high light intensity-inducible LHC-like 4) proteins. SEPs and OHPs are found in organisms of both green and red origin, while ELIPs and LHL4 proteins are restricted to the green lineage. Recently, a further subpopulation of LHC-related proteins was characterized in the red lineage and termed RedCAPs (red lineage chlorophyll a/b-binding like proteins, Engelken et al. 2010, see also Fig. 2).
Specific properties of LHCs in diatoms
A recent extensive characterization of LHC proteins from chlorophyll c-containing algae suggested that LHC subfamilies are not lineage-specific but rather the result of multiple gene duplications, gene losses and functional adaptation (Hoffman et al. 2011). Hoffman et al. (2011) also presented a concise likelihood-based phylogenetic tree of LHCs with a special focus on Chl a/c-containing species. This approach resulted in a division into seven major groups. The diversity and complexity of diatom LHCs are underlined by the fact that they have members in all seven LHC subfamilies according to the same classification by Hoffman et al. (2011). Red algae, on the other hand, only have members in group III while green algae LHCs only cluster in group V. This makes the diversity of diatom LHCs surprising given their origin in the red linage (see Fig. 2) and leads to the conclusion that a substantial diversification of LHCs took place in Heterokontophyta, way beyond a simple substitution of phycobilisomes for a LHC-based PSII antenna. There are multiple copies of Lhcx genes (closely related to LHCSR in C. reinhardtii) in diatoms as well (Bailleul et al. 2010; Zhu and Green 2010), which according to the taxonomic lineage of Hoffman et al. (2011) fall into group V of LHCs.
Within the extended LHC family there are a number of proteins specific to PSI only. In chlorophyll c-containing organisms there is the suggestion that PSII-associated LHCs evolved quite separately from the ones specific to PSI (Hoffman et al. 2011) which are of red origin and therefore the proteins are designated as Lhcr, accordingly. In diatoms, PSI specific LHCs are quite numerous and belong mostly to the Lhcr type, while the peripheral antenna is predominantly made up of Lhcf type LHCs (Lepetit et al. 2010; Grouneva et al. 2011).
Evolution of protein phosphorylation as a mechanism for photosynthetic regulation
LHCII phosphorylation has different physiological significance in land plants and green algae
Development of an antenna system capable of adjusting its light-harvesting capacity to changing light intensity/quality, together with mechanisms to regulate its function, has been a vital sequence of events in the evolution of photosynthetic organisms. In addition to acquiring safety valves such as NPQ for efficient dissipation of excess excitation energy, photosynthetic organisms with Chl a/b antenna have evolved reversible light-dependent phosphorylation of LHCII as a mechanism for maintaining redox balance among photosynthetic electron transfer reactions (Bennett 1977; Allen et al. 1981; Grossman et al. 1995). Chl a/b-binding proteins and the regulatory mechanisms of LHCII protein phosphorylation appear to be conserved in evolution, as evidenced by similarities in the light intensity-dependent patterns of LHCII protein phosphorylation in evolutionarily divergent species of liverworts, mosses, ferns, monocots and dicots (Pursiheimo et al. 1998, 2001). In all these plants the strongest level of LHCII protein phosphorylation is detected in low light intensities, whereas exposure to high light leads to down-regulation of LHCII protein phosphorylation (Rintamäki et al. 1997, 2000). A transient down-regulation of LHCII protein phosphorylation has also been reported to occur in green algae in response to photoinhibition (Schuster et al. 1986). In contrast to green plants, thylakoid protein phosphorylation in diatoms (with fucoxanthin-Chl a/c-binding proteins) has not been reported, and no reversible/light-dependent phosphorylation of thylakoid proteins was detected in the Chl a/c-containing algae Ochromonas danica (Gibbs and Biggins 1991). The same could be established for red algae and cyanobacteria that use phycobilisome antenna system (Pursiheimo et al. 1998). Table 1 summarizes a database search of putative phosphorylation sites of the main thylakoid membrane proteins in different species. In addition, it reveals the lack of the phosphoprotein CaS (see below) and the plastid kinase STN7 in two diatom species and one red algae.
The physiological significance and the benefits of low light-induced phosphorylation of thylakoid proteins, LHCII in particular, have long been matters of intensive research (Allen et al. 1981; Tikkanen et al. 2011). Initially, redox-regulated LHCII protein phosphorylation was correlated with state transitions, triggering the migration of phosphorylated (P*) LHCII antenna proteins to granal margins where they bind to the PsaH protein to associate with PSI (Lunde et al. 2000; Kargul et al. 2005). Depege et al. (2003) identified the long-sought LHCII kinase ‘Stt7’ (state transition, thylakoid) from C. reinhardtii, showing that mutation in Stt7 blocked both LHCII phosphorylation and state transitions, while a similar effect was observed in Arabidopsis mutants deficient in the highly similar Stt7 homologue ‘STN7’ (Depege et al. 2003; Bellafiore et al. 2005). In C. reinhardtii thylakoids, Stt7 has been found associated with Cytb6f, LHCII and PSI, but not with PSII (Lemeille et al. 2009). Phosphorylation and migration of the PSII linker protein CP29 have also been reported to be vital for docking of P*LHCII proteins to PSI in C. reinhardtii, but the extent to which CP29 moves between the photosystems seems to depend on the experimental conditions used (Tokutsu et al. 2004; Lemeille et al. 2010). In higher plants, CP29 phosphorylation regulates disassembly of PSII supercomplexes under high light (Fristedt and Vener 2011). The steady state levels of Stt7 and STN7 themselves are also regulated by phosphorylation events upon long-term changes in growth conditions. Phosphorylation of a C-terminal Ser533 of Stt7 in state 2 is not related to the regulation of state transitions (Lemeille et al. 2010), while in Arabidopsis phosphorylation of four sites at the C-terminus controls STN7 turn-over under state 1 conditions (Willig et al. 2011). Further, the activity of STN7 is counterbalanced through the action of the redox-insensitive PP2C-type chloroplast protein phosphatase PPH1/TAP38 required for dephosphorylation of LHCII (Pribil et al. 2010; Shapiguzov et al. 2010). This regulatory action occurs predominantly on stroma-exposed regions of the thylakoid membrane (Shapiguzov et al. 2010). Homologues for PPH1 have also been identified in P. patens, Populus and Brachybodium genomes, suggesting that also the mechanism for dephosphorylation of LHCII may be evolutionarily conserved.
Considering the apparent conservation of redox regulation of LHCII protein phosphorylation, it is intriguing that the physiological significance of LHCII protein phosphorylation seems to differ between green algae and higher plants, as suggested by results obtained with C. reinhardtii and Arabidopsis. In C. reinhardtii, phosphorylation of up to 80 % of the LHCII antenna proteins has been associated with enhanced production of ATP upon a transition from state 1 to state 2 and the shift from linear to cyclic electron flow (Finazzi et al. 1999, 2002), which leads to accumulation of a thylakoid-embedded supercomplex composed of PSI, Cytb6f, FNR, LHCI and LHCII (Iwai et al. 2010). These metabolic actions presumably evolved together with intimate inter-connections between chloroplastic and mitochondrial pathways, contributing to the formation of metabolic flexibility characteristic of unicellular algae (reviewed by Cardol et al. 2011). In higher plants, a considerably smaller fraction of up to 20 % of LHCII proteins are phosphorylated and associate with PSI (Vener 2007). It is noteworthy that in Arabidopsis the low light intensity-induced LHCII phosphorylation and high light-induced LHCII dephosphorylation do not induce changes in the absorption cross section of PSII and PSI as is typical for red/far red light-induced “state transitions” (Tikkanen et al. 2008b, 2010). Results from studies of Arabidopsis stn7 mutants have now led to formulation of a new concept, whereby LHCII protein phosphorylation mediates the balancing of excitation energy of the two photosystems despite fluctuations in white light intensity (Bellafiore et al. 2005; Tikkanen et al. 2006, 2011). Such balancing system apparently involves energy transfer from PSII/LHCII to PSI occurring in thylakoid megacomplex comprised of LHCII, PSII and PSI (Järvi et al. 2011).
PSII core protein phosphorylation as a mechanism to regulate PSII turn-over
Despite the variety of protective and regulatory mechanisms, light-induced damage to photosynthetic membrane protein complexes is an intrinsic property of evolutionarily divergent organisms ranging from cyanobacteria and algal species to higher plants (Powles 1984; Samuelsson et al. 1987; Schuster et al. 1988; Aro et al. 1993; Domingues et al. 2012). The molecular mechanisms of photoinhibition and repair of PSII have been extensively studied during the past decades, and despite some controversy surrounding the primary target of photodamage, the replacement of photodamaged D1 protein through a multi-step process regulated by phosphorylation and assisted by a large catalogue of auxiliary proteins has become relatively well understood (Baena-Gonzalez and Aro 2002; Chi et al. 2012; Mulo et al. 2012).
In higher plants with structurally heterogeneous thylakoid membranes, photochemically active PSII-LHCII supercomplexes become photodamaged in grana thylakoids (Boekema et al. 2000; Tikkanen et al. 2008a). Photodamage to D1 protein triggers monomerization of PSII and dissociation of the LHCII antenna through a yet unknown mechanism, after which the PSII complexes containing damaged D1 protein migrate to stroma-exposed thylakoid regions for repair (Aro et al. 2005; Adam et al. 2006). Reversible phosphorylation of N-terminal threonines in the D1, D2 and CP43 proteins of the PSII core facilitates the PSII repair cycle. These core proteins are mainly phosphorylated by the STN7 paralog STN8, although STN7 kinase also has a role in this process (Bonardi et al. 2005; Vainonen et al. 2005; Tikkanen et al. 2006; Vener 2007). STN8 homologues in other plants including P. patens and the conifer Pinus sitchensis are closely related to the Stl1 kinase in C. reinhardtii (Fig. 3, Depege et al. 2003), suggesting conserved functionality for this kinase across evolution. However, a distinguishable feature of species with less distinct thylakoid organization, such as green algae, mosses, ferns and lycophytes, is a lack of or only minor D1 protein phosphorylation (Pursiheimo et al. 1998; Hamel et al. 2000; Lemeille et al. 2010) despite strong conservation of N-terminal threonine residues in D1 of lower plants, algae and diatoms (see Table 1). The phosphorylation of D2 and CP43 seen in higher plants is conserved in lower land plants and green algae (Pursiheimo et al. 1998; Hamel et al. 2000; Lemeille et al. 2010), but only in ferns the pattern of phosphorylation is clearly responsive to light intensity (Pursiheimo et al. 1998). This observation pinpoints the evolution of a redox-responsive mechanism for modulating CP43 and D2 phosphorylation in ferns and higher plants that does not exist in the more distantly related mosses and liverworts (Pursiheimo et al. 1998) but nevertheless has been observed in C. reinhardtii (Turkina et al. 2006).
Phylogenetic relationship of STN7 and STN8 kinases and their orthologues in plants. A ClustalW alignment of amino acid sequences was imported to MEGA5 to create the bootstrap consensus phylogenetic tree shown, using the Neighbour-Joining method with Poisson correction and bootstrapping at 1,000 replications
PSII core phosphorylation in higher plants has been attributed a role in facilitating the migration of damaged PSII core complexes from grana to stroma thylakoids (Tikkanen et al. 2008a; Goral et al. 2010). It has also been postulated that phosphorylation of the core complex has a role in maintaining the integrity of the damaged PSII complex until re-synthesis of D1 can take place (Baena-Gonzalez and Aro 2002). Sequential dephosphorylation of D1, D2 and CP43 is thought to allow partial disassembly of PSII and degradation of D1 in a coordinated manner (Koivuniemi et al. 1995; Rintamäki et al. 1996; Tikkanen et al. 2008b). Analysis of Arabidopsis stn8 mutants has shown that an inability to phosphorylate PSII core proteins promotes oxidative damage of the photosynthetic protein complexes upon severe high light stress, and suggests that the PSII repair cycle has a vital role in preventing oxidative damage in PSI (Tikkanen et al. in press).
Cyanobacteria and red algae, both with a phycobilisome antenna system for PSII, showed no phosphorylation of the PSII core proteins (Pursiheimo et al. 1998), and the same is most likely true also for prokaryotic prochlorophytes (van der Staay and Staehelin 1994), while mounting evidence suggests that diatoms also lack reversible PSII phosphorylation (Grouneva et al. unpublished results). This correlates with the lack of STN8 homologues in these species, indicating that PSII phosphorylation has evolved as a short-term mechanism to regulate the PSII repair cycle in more developed photosynthetic organisms. Furthermore, it suggests that the protective role of PSII phosphorylation may have evolved together with strict lateral heterogeneity and spatial segregation of the PSII and PSI complexes to the grana and stroma-exposed thylakoid regions in plant chloroplasts, supporting the notion that PSII core protein phosphorylation is more likely connected to migration of PSII complexes in the thylakoid membrane and to the kinetics of protein degradation rather than to the actual process of PSII turnover.
Recent studies show that although diatoms display low susceptibility to photoinactivation, their rates of D1 protein removal from PSII complexes are rather low compared to other phytoplankton (Key et al. 2010) and the small centric diatom T. pseudonana maintains a pool of disassembled PS II lacking the D1 protein (Wu et al. 2011). The elucidation of how these findings correlate with a phosphorylation-independent mechanism of PSII turn-over and energy distribution regulation between the photosystems will be highly intriguing.
Components and regulation of photosynthetic electron flow through cyclic electron transport
Along with other acclimation mechanisms, the cyclic electron transfer (CET) around PSI also participates in ensuring efficient photosynthesis and maintenance of metabolic homeostasis in chloroplasts (Rumeau et al. 2007; Shikanai 2007; Johnson 2011). CET, where electrons are shunted from ferredoxin (Fd) back to PSI, has been considered to occur via two routes: (1) the putative Fd-plastoquinone reductase (FQR) complex or (2) the NAD(P)H dehydrogenase (NDH)-like complex (Ifuku et al. 2011; Peng et al. 2011).
The Arabidopsis crr2 pgr5 double mutant defective in both putative routes of CET shows a clear phenotype of stunted growth and pale leaves, providing strong evidence that CET is essential for normal photosynthetic growth in higher plants (Munekage et al. 2004); however, the physiological importance of CET remains somewhat puzzling. It has been suggested that CET is needed to balance the ratio of ATP to NADPH and to protect PSI from photoinhibition (Shikanai 2007), but its significance, particularly in C3 plants, continues to be discussed extensively and even controversially (Bendall and Manasse 1995; Allen 2003; Joliot and Joliot 2002, 2006; Johnson 2005, 2011; Nogales et al. 2012). In contrast to higher plants, the importance of CET in green algae is more evident, where it is tightly coupled with state transitions (Finazzi et al. 1999, 2002). During state 1, when LHCII is associated with PSII, linear electron transport (LET) predominates, while upon transition to state 2 the proportion of CET markedly increases (Finazzi et al. 2002). Such a connection between state transitions and CET has not been demonstrated to exist in higher plants (Havaux 1992).
Despite extensive research, the exact molecular components of CET are still largely unknown. For example, as the FQR complex has not yet been characterized, it has been speculated that a supercomplex comprising PSI and Cytb6f, together with additional proteins like ferredoxin NADP+ oxidoreductase (FNR), Fd, and PGRL1 (see below), forms the hypothetical FQR complex in eukaryotes (DalCorso et al. 2008). Indeed, such a supercomplex was recently characterized from the green algae C. reinhardtii, and was proved to be involved in CET (Iwai et al. 2010).
NDH complexes in electron transfer
Plant thylakoid membranes contain a NDH complex homologous to the mitochondrial NADH dehydrogenase complex and the E. coli NDH-1 complex. In land plants, the NDH complex is composed of more than 30 subunits, which are both nuclear- and plastid-encoded (for recent reviews, see Ifuku et al. 2011; Peng et al. 2011). Based on genetic and biochemical studies, these subunits presumably form four subcomplexes: (1) the membrane subcomplex, (2) subcomplexes A and (3) B, and (4) the lumenal subcomplex. Of these, the subcomplex B as well as the lumenal subcomplex are specific to higher plants, and composed entirely of nuclear-encoded subunits (Peng et al. 2011). Furthermore, in higher plants NDH forms a supercomplex with PSI (Peng et al. 2008; Peng and Shikanai 2011), and the minor LHCI proteins Lhca5 and Lhca6 are involved in binding the NDH complex to PSI (Peng et al. 2009).
Despite extensive research and partial purification of the NDH complex from several species, the electron donor(s) for NDH has long remained elusive. While the thylakoid NDH complex has homology to the E. coli NDH-1 complex, the nuoE, nuoF and nuoG genes encoding the subunits that form the electron input/activity domain in E. coli have not been found in chloroplast or cyanobacterial genomes. However, three novel components of NDH (CRR31, CRRJ, and CRRL) were recently characterized from Arabidopsis, together with compelling evidence that NDH accepts electrons directly from Fd (Yamamoto et al. 2011). The C-terminal region of the CRR31 protein is thought to form a binding site for Fd, analogous to the structurally similar PsaE subunit on the stromal side of PSI (Yamamoto et al. 2011). Accordingly, the correct definition of the NDH should now be “NDH-like complex” instead of “NAD(P)H dehydrogenase” (Ifuku et al. 2011; Yamamoto et al. 2011).
Although the cyanobacterial NDH-1 complex bears strong structural homology to the chloroplast NDH-like complex in plants, there are important functional differences. Cyanobacterial NDH-1 participates not only in CET, but also in respiration and in carbon acquisition (Battchikova et al. 2011). These additional functions are reflected in the subunit composition of cyanobacterial NDH-1 complexes, with subunits NdhD3, NdhF3, CupA, and CupS of the NDH-1S subcomplex specialized for carbon acquisition (Zhang et al. 2004; Battchikova et al. 2011). These subunits are missing from the NDH-like complex of higher plants, while several nuclear-encoded NDH-proteins such as PnsL1–5 and PnsB1–5 are specific to plants (Suorsa et al. 2009; Ifuku et al. 2011; Peng et al. 2011, 2012; Yamamoto et al. 2011).
The functional significance of NDH-like complexes in C3 plants has been difficult to assess. Arabidopsis and tobacco ndh mutants do not show any visible phenotype under controlled growth conditions (reviewed in Suorsa et al. 2009; Ifuku et al. 2011), suggesting that the alternative FQR-mediated pathway (see below) is largely responsible for CET in C3 plants (Munekage et al. 2004; Okegawa et al. 2008). In C4 plants, on the other hand, the NDH complex is considered to have an important role in directing CET for ATP production in bundle sheath chloroplasts (Majeran et al. 2008; Majeran and van Wijk 2009). Another suspected role for the NDH-like complex is in chlororespiration, where the plastoquinone (PQ) pool becomes oxidized via transfer of electrons to molecular oxygen, serving as a safety valve during stress to protect the PQ pool from over-reduction (Niyogi 2000; Peltier and Cournac 2002). The functional role of the NDH-like complex in C3 plants becomes even more puzzling due to its presence in etioplasts prior to greening and thylakoid biogenesis with the assembly of the major photosynthetic pigment-protein complexes (Kanervo et al. 2008; Peng et al. 2008). During de-etiolation, the NDH-like complex rapidly starts to interact with PSI, and after just 48 h in light, the PSI-NDH supercomplex is assembled (Peng et al. 2008). This result may support the speculated co-operation between NDH and the plastid terminal oxidase PTOX, which is known to be crucial for the early leaf development and for the proper chloroplast biogenesis (Rosso et al. 2006, 2009). Alternatively, the etioplastic NDH-like complex might be physiologically nonfunctional until the appearance of its assembly partner, PSI.
The NDH-like complex is one of the best examples of the versatility in regulation of photosynthetic electron transfer that exists among phylogenetically divergent species. The NDH-like complex is conserved in cyanobacteria, the chloroplast ancestor, at one end of the spectrum, and in higher plants at the other, but there are “gaps” in the distribution of the NDH-like complex among evolutionary divergent groups (reviewed in Martin and Sabater 2010; Peltier et al. 2010). For instance, the plastid genome of the conifer Pinus thunbergii lacks the ndh genes (Wakasugi et al. 1994), as does that of Welwitschia (McCoy et al. 2008), a member of the Gnetales order that is phylogenetically classified as a sister group of the Pinaceae family (Braukmann et al. 2009). Nonetheless, NDH is encoded in the plastomes of the conifer Cryptomeria japonica (Hirao et al. 2008) and the cycad Cycas taitungensis (Wu et al. 2007), demonstrating an absence of uniformity in the occurrence of the NDH-like complex among gymnosperms. Descending further down the evolutionary line, the plastomes of the common stonewort Chara vulgaris (Charales; Turmel et al. 2006) and the moss Physcomitrella (Sugiura et al. 2003) contain ndh genes, while they are absent from red algae, diatoms (Oudot-Le Secq et al. 2007) and many species of green algae including C. reinhardtii and Chlorella (reviewed in Peltier et al. 2010). Some members of the Prasinophyceae group of green algae are exceptional, for example Ostreococcus does not encode NDH, whereas some other species of the group do (Turmel et al. 2009).
An alternative ‘type II’ NDH (Ndh-2) is a single subunit enzyme responsible for PQ pool reduction in C. reinhardtii and is thought to compensate for the lack of the NDH-like complex in that algae (Jans et al. 2008; Desplats et al. 2009), although Ndh-2 also exists in species that possess NDH-like complexes, including cyanobacteria and Arabidopsis, where its role is not understood. Conversely, heterogeneity in the occurrence of Ndh-2 in diatoms, which lack NDH-like complexes, has been observed (Grouneva et al. 2011).
PGR5, PGRL1 and CaS in regulation of electron transport. Importance in LET or CET?
The CET pathway occurring via a hypothetical FQR complex was initially suggested to involve the PROTON GRADIENT REGULATION 5 protein (PGR5). More recent research has introduced new components for the FQR complex as well as suggested a novel function for the PGR5 protein. The phylogeny and function of PGR5 as well as the putative interaction with the PGRL protein are shortly discussed below.
The PGR5 protein was first characterized from Arabidopsis by Toshiharu Shikanai and co-workers (Munekage et al. 2002). The pgr5 mutant was shown to lack a proton gradient across the thylakoid membrane, and thus also lacked NPQ induction (Munekage et al. 2002). Furthermore, PGR5 functions in photoprotection of PSI (Munekage et al. 2002) and is indispensable for the maintenance of PSI under fluctuating light conditions (Suorsa et al. 2012). PGR5 is proposed to mediate electron transfer via the Fd-mediated CET pathway (Munekage et al. 2002; Shikanai 2007); however, our recent results suggest that the primary role of PGR5 lies in the regulation of LET instead, because inhibition of PSI in pgr5 by high light pulses was found to be largely prevented by blocking LET (Suorsa et al. 2012). The relative activity of particularly the FQR-mediated CET as compared to LET seems to increase upon environmental stress conditions. Indeed, drought stress induces accumulation of the PGR5 proteins in Arabidopsis (Lehtimäki et al. 2010). In maize mesophyll cells, cold stress switches the preferable route of CET from NDH-dependent into FQR-dependent (Savitch et al. 2011). In either cases, the components of the NDH-dependent CET route were shown not to respond to the stress conditions (Lehtimäki et al. 2010; Savitch et al. 2011).
Genes encoding PGR5 are present in all plant and algae genomes sequenced so far (Peltier et al. 2010) while the open reading frame ssr2016 encodes a PGR5 homologue in cyanobacteria (Yeremenko et al. 2005). Like PGR5, expression of ssr2016 is upregulated by stress, while both the pgr5 and ssr2016 mutants of Arabidopsis and cyanobacteria, respectively, exhibit notably similar defects in CET (Yeremenko et al. 2005; Suorsa et al. 2012). PGR5 and ssr2016 gene products are not functionally identical, however, as the prominent fluctuating light phenotype of Arabidopsis (Suorsa et al. 2012) is not seen in the cyanobacterial ssr2016 mutant (Aro et al. unpublished results).
Interestingly, the C. reinhardtii CET-related supercomplex that contained PSI, Cytb6f, and several additional proteins (Iwai et al. 2010) lacked PGR5, but instead contained a Volvocales-specific Cytb6f subunit PETO (Iwai et al. 2010). PETO is known to be reversibly phosphorylated under state 2 (Hamel et al. 2000, Table 1). Although the loss of the soluble PGR5 protein during thylakoid preparation cannot be ruled out, PGR5 may be replaced with phosphorylated PETO in C. reinhardtii, thus explaining the tight connection between state transitions and CET (Iwai et al. 2010).
PGRL1 (PGR5-LIKE PHOTOSYNTHETIC PHENOTYPE 1) is a thylakoid-associated protein existing in two isoforms; PGRL1A and PGRL1B (DalCorso et al. 2008). The PGRL1A protein interacts with PGR5, FNR, PSI and Cytb6f (DalCorso et al. 2008), suggesting a role in CET. The recent discovery that PGRL1A is phosphorylated by the STN8 kinase (Reiland et al. 2011) may provide an additional physiological link between CET and chloroplast redox regulation, which affects the activity of the STN8 kinase (Bonardi et al. 2005; Vainonen et al. 2005).
PGRL1, unlike PGR5, seems to be specific to eukaryotes, as it is absent from cyanobacteria (DalCorso et al. 2008; Peltier et al. 2010). Green algae and diatoms each possess one PGRL (Petroutsos et al. 2009; Grouneva et al. 2011) that appears to play a broader physiological role than the higher plant homologues. Upon iron starvation, which is particularly harmful for PSI (Guikema and Sherman 1984), the amount of the PGRL1 protein increases in C. reinhardtii (Petroutsos et al. 2009). Furthermore, the corresponding RNAi mutant cells with lower level of the PGRL1 protein showed altered response to iron both at photosynthetic and at the cellular level (Petroutsos et al. 2009). Therefore, it was proposed that the PGRL1 protein plays a dual role in C. reinhardtii by functioning both in CET and in iron metabolism (Petroutsos et al. 2009). In line with this theory, iron starvation up-regulates CET in cyanobacteria (Sandstrom et al. 2002; Ivanov et al. 2012).
The calcium-sensing receptor CaS resides in the thylakoid membrane of higher plants and is phosphorylated by STN8 in response to high light intensity (Vainonen et al. 2008). The C-terminal phosphorylation site in Arabidopsis CaS occurs within a region containing characteristic motifs for interaction with 14-3-3 proteins and FHA domains, both generally functioning in phosphorylation-dependent signal transduction (Li et al. 2000a; Fulgosi et al. 2002; Hammet et al. 2003; Vainonen et al. 2008). The CaS C-terminus also carries a rhodanese-like domain, yet another feature known to be involved in signal transduction, although the catalytic residues required for rhodanese activity do not occur in CaS. A CaS homologue in C. reinhardtii was also identified as a phosphoprotein, but its phosphorylation status was shown to be independent of light-induced redox signals (Lemeille et al. 2010). Even so, CaS was recently shown to be vital for NPQ in C. reinhardtii, linking photoprotection of PSII with chloroplast calcium signalling (Petroutsos et al. 2011). The characteristics of CaS suggest its involvement in signal transduction across the thylakoid membrane, triggered by calcium, light and/or redox changes and likely effecting phosphorylation-dependent cascades via protein–protein interactions in the chloroplast.
CaS homologues have not been found in the red lineage (Table 1). However, the death-specific protein (DSP), occurring exclusively in diatoms, displays two calcium-binding domains. In addition, DSP contains a region with a high degree of amino acid sequence homology to PGR5 (Chung et al. 2008). DSP expression is upregulated by disruption of photosynthetic electron flow (Chung et al. 2008), suggesting redox sensitivity that may link calcium signalling with photosynthetic regulation similar to the role of CaS in the green lineage. This intriguing possibility of functional counterparts carrying out calcium sensing and photoprotection in divergent species points an important role for calcium signalling in photosynthesis regulation.
Conclusions
Some photosynthetic regulatory mechanisms discussed here are conserved throughout organism groups and are based on a similar genetic make-up. One example is the PGR5-mediated CET. On the other hand, there are mechanisms that serve a similar overall purpose in phylogenetically divergent lines but nevertheless rely on unrelated/yet unknown gene products to do so. The involvement of reversible thylakoid protein phosphorylation in the PSII repair cycle and light-harvesting regulation provides an intriguing instance of such divergence between photosynthetic groups. The following conclusions are drawn concerning the phylogeny of the most studied regulatory mechanisms of thylakoid light harvesting and electron transfer processes in chloroplasts:
1. The LHCSR-based mechanism for NPQ in aquatic species has been superseded by PsbS, another member of the extended LHC family in vascular plants, but both the LHCSR- and the PsbS-mediated mechanisms can be highly effective or even coexist, as is the case in mosses.
2. Reversible phosphorylation of thylakoid proteins displays a clear development and accumulation in the course of evolution towards higher plants. It might therefore be connected to an increasing complexity of thylakoid organization (stacking) and facilitate the movement of both LHCs and photosystem core complexes by introducing positive charges/shifting the ratio of positive and negative charges between protein complexes and membrane lipids. There is mounting evidence of reversible thylakoid protein phosphorylation missing in the red lineage. This fact provides a clear example of two genetically alternative mechanisms for photosynthesis regulation. In the case of light harvesting in diatoms, a high PSII/PSI ratio might counterbalance the lack of spatial segregation between a “slow” PSII and a “fast” PSI. Back in 2004, nearly half of diatom putative proteins had no function assigned to them based on sequence homology (Armbrust et al. 2004), being either diatom-specific or species-unique genes (Bowler et al. 2008). It can be speculated, therefore, that among them are proteins of unknown structure that nevertheless display a similar function to known proteins in the green lineage.
3. There appears to be no clear, “unbroken” phylogenetic line of the plastid NDH-like complex. In conclusion, it can be speculated that the selection pressure for plastid NDH is either relatively low (species that do not possess this protein complex also lack the entire set of encoding genes) or that its function in eukaryotes is redundant and in consequence a secondary loss took place in certain species (pine) or organism groups (diatoms).
4. PGR5, in contrast, is present throughout eukaryotic photosynthetic organism groups and Arabidopsis mutants show a severe phenotype under fluctuating light conditions.
Abbreviations
- CET:
-
Cyclic electron transport
- FQR:
-
Fd-plastoquinone reductase
- LET:
-
Linear electron transport
- LHC:
-
Light-harvesting complex
- NDH:
-
NAD(P)H dehydrogenase
- NPQ:
-
Non-photochemical quenching
- PS:
-
Photosystem
References
Adam Z, Rudella A, van Wijk K (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9:234–240
Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T (2010) Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA 107:11128–11133
Allen J (2003) Cyclic, pseudocyclic and noncyclic photophosphorylation: new links in the chain. Trends Plant Sci 8:15–19
Allen J, Bennett J, Steinback K, Arntzen C (1981) Chloroplast protein-phosphorylation couples plastoquinone redox state to distribution of excitation-energy between photosystems. Nature 291:25–29
Anderson JM (1986) Photoregulation of the composition, function, and structure of thylakoid membranes. Annu Rev Plant Physiol 37:93–136
Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kroger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Aro E, Virgin I, Andersson B (1993) Photoinhibition of photosystem-2—inactivation, protein damage and turnover. Biochim Biophys Acta 1143:113–134
Aro E, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamäki E (2005) Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56:347–356
Baena-Gonzalez E, Aro E (2002) Biogenesis, assembly and turnover of photosystem II units. Philos Trans R Soc Lond Ser B-Biol Sci 357:1451–1459
Bailleul B, Rogato A, de Martino A, Coesel S, Cardol P, Bowler C, Falciatore A, Finazzi G (2010) An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc Natl Acad Sci USA 107:18214–18219
Battchikova N, Eisenhut M, Aro E (2011) Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim Biophys Acta 1807:935–944
Bellafiore S, Bameche F, Peltier G, Rochaix J (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433:892–895
Bendall D, Manasse R (1995) Cyclic photophosphorylation and electron-transport. Biochim Biophys Acta 1229:23–38
Bennett J (1977) Phosphorylation of chloroplast membrane polypeptides. Nature 269:344–346
Boekema E, van Breemen J, van Roon H, Dekker J (2000) Arrangement of photosystem II supercomplexes in crystalline macrodomains within the thylakoid membrane of green plant chloroplasts. J Mol Biol 301:1123–1133
Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437:1179–1182
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kroger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jezequel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Braukmann TWA, Kuzmina M, Stefanovic S (2009) Loss of all plastid ndh genes in Gnetales and conifers: extent and evolutionary significance for the seed plant phylogeny. Curr Genet 55:323–337
Cardol P, Forti G, Finazzi G (2011) Regulation of electron transport in microalgae. Biochim Biophys Acta 1807:912–918
Chan CX, Yang EC, Banerjee T, Yoon HS, Martone PT, Estevez JM, Bhattacharya D (2011) Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr Biol 21:328–333
Chi W, Sun X, Zhang L (2012) The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochim Biophys Acta 1817:239–246
Chung C, Hwang SL, Chang J (2008) Nitric oxide as a signaling factor to upregulate the death-specific protein in a marine diatom, Skeletonema costatum, during blockage of electron flow in photosynthesis. Appl Environ Microbiol 74:6521–6527
DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schuenemann D, Finazzi G, Joliot P, Barbato R, Leister D (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132:273–285
Delwiche CF (1999) Tracing the thread of plastid diversity through the tapestry of life. Am Nat 154:S164–S177
Depauw FA, Rogato A, Ribera d’Alcala M, Falciatore A (2012) Exploring the molecular basis of responses to light in marine diatoms. J Exp Bot 63:1575–1591
Depege N, Bellafiore S, Rochaix J (2003) Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299:1572–1575
Desplats C, Mus F, Cuine S, Billon E, Cournac L, Peltier G (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J Biol Chem 284:4148–4157
Domingues N, Matos AR, Marques da Silva J, Cartaxana P (2012) Response of the diatom Phaeodactylum tricornutum to photooxidative stress resulting from high light exposure. PLoS ONE 7:e38162
Engelken J, Brinkmann H, Adamska I (2010) Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol Biol 10:233
Finazzi G, Furia A, Barbagallo R, Forti G (1999) State transitions, cyclic and linear electron transport and photophosphorylation in Chlamydomonas reinhardtii. Biochim Biophys Acta 1413:117–129
Finazzi G, Rappaport F, Furia A, Fleischmann M, Rochaix J, Zito F, Forti G (2002) Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii. EMBO Rep 3:280–285
Fristedt R, Vener AV (2011) High light induced disassembly of photosystem II supercomplexes in Arabidopsis requires STN7-dependent phosphorylation of CP29. PLoS ONE 6:e24565
Frommolt R, Werner S, Paulsen H, Goss R, Wilhelm C, Zauner S, Maier UG, Grossman AR, Bhattacharya D, Lohr M (2008) Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol Biol Evol 25:2653–2667
Fulgosi H, Soll J, Maraschin S, Korthout H, Wang M, Testerink C (2002) 14-3-3 proteins and plant development. Plant Mol Biol 50:1019–1029
Gibbs P, Biggins J (1991) Invivo and invitro protein-phosphorylation studies on Ochromonas danica, an alga with a chlorophyll-a/c fucoxanthin binding-protein. Plant Physiol 97:388–395
Goral TK, Johnson MP, Brain APR, Kirchhoff H, Ruban AV, Mullineaux CW (2010) Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: effects of photoinhibition and protein phosphorylation. Plant J 62:948–959
Green B, Pichersky E (1994) Hypothesis for the evolution of 3-Helix Chl a/b and Chl a/c light-harvesting antenna proteins from 2-helix and 4-helix ancestors. Photosynth Res 39:149–162
Grossman A, Bhaya D, Apt K, Kehoe D (1995) Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu Rev Genet 29:231–288
Grouneva I, Rokka A, Aro E (2011) The thylakoid membrane proteome of two marine diatoms outlines both diatom-specific and species-specific features of the photosynthetic machinery. J Proteome Res 10:5338–5353
Guikema J, Sherman L (1984) Influence of iron deprivation on the membrane-composition of Anacystis nidulans. Plant Physiol 74:90–95
Hamel P, Olive J, Pierre Y, Wollman F, de Vitry C (2000) A new subunit of cytochrome b(6)f complex undergoes reversible phosphorylation upon state transition. J Biol Chem 275:17072–17079
Hammet A, Pike B, McNees C, Conlan L, Tenis N, Heierhorst J (2003) FHA domains as phospho-threonine binding modules in cell signaling. IUBMB Life 55:23–27
Havaux M (1992) Photoacoustic measurements of cyclic electron flow around photosystem-I in leaves adapted to light-states-1 and light-states-2. Plant Cell Physiol 33:799–803
Hirao T, Watanabe A, Kurita M, Kondo T, Takata K (2008) Complete nucleotide sequence of the Cryptomeria japonicia D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species. BMC Plant Biol 8:70
Hoffman GE, Sanchez Puerta MV, Delwiche CF (2011) Evolution of light-harvesting complex proteins from Chl c-containing algae. BMC Evol Biol 11:101
Ifuku K, Endo T, Shikanai T, Aro E (2011) Structure of the chloroplast NADH dehydrogenase-like complex: Nomenclature for nuclear-encoded subunits. Plant Cell Physiol 52:1560–1568
Ivanov AG, Sane PV, Simidjiev I, Park Y-, Huner NPA, Öquist G (2012) Restricted capacity for PSI-dependent cyclic electron flow in ΔpetE mutant compromises the ability for acclimation to iron stress in Synechococcus sp. PCC 7942 cells. Biochim Biophys Acta 1817:1277–1284
Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464:1210–1213
Jans F, Mignolet E, Houyoux P, Cardol P, Ghysels B, Cuine S, Cournac L, Peltier G, Remacle C, Franck F (2008) A type II NAD(P) H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci USA 105:20546–20551
Jansson S, Green B, Grossman A, Hiller R (1999) A proposal for extending the nomenclature of light-harvesting proteins of the three transmembrane helix type. Plant Mol Biol Rep 17:221–222
Järvi S, Suorsa M, Paakkarinen V, Aro E (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J 439:207–214
Jensen PE, Bassi R, Boekema EJ, Dekker JP, Jansson S, Leister D, Robinson C, Scheller HV (2007) Structure, function and regulation of plant photosystem I. Biochim Biophys Acta 1767:335–352
Johnson G (2005) Cyclic electron transport in C-3 plants: fact or artefact? J Exp Bot 56:407–416
Johnson GN (2011) Physiology of PSI cyclic electron transport in higher plants. Biochim Biophys Acta 1807:384–389
Joliot P, Joliot A (2002) Cyclic electron transfer in plant leaf. Proc Natl Acad Sci USA 99:10209–10214
Joliot P, Joliot A (2006) Cyclic electron flow in C3 plants. Biochim Biophys Acta 1757:362–368
Kanervo E, Singh M, Suorsa M, Paakkarinen V, Aro E, Battchikova N, Aro E (2008) Expression of protein complexes and individual proteins upon transition of etioplasts to chloroplasts in pea (Pisum sativum). Plant Cell Physiol 49:396–410
Kargul J, Turkina M, Nield J, Benson S, Vener A, Barber J (2005) Light-harvesting complex II protein CP29 binds to photosystem I of Chlamydomonas reinhardtii under state 2 conditions. FEBS J 272:4797–4806
Key T, McCarthy A, Campbell DA, Six C, Roy S, Finkel ZV (2010) Cell size trade-offs govern light exploitation strategies in marine phytoplankton. Environ Microbiol 12:95–104
Koivuniemi A, Aro E, Andersson B (1995) Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry (NY) 34:16022–16029
Koziol AG, Borza T, Ishida K, Keeling P, Lee RW, Durnford DG (2007) Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol 143:1802–1816
Larkum AW, Vesk M (2003) Algal plastids: their fine structure and properties. In: Larkum AW, Douglas SE, Raven JA (eds) Photosynthesis in algae. Kluwer Academic Publishers, Dordrecht, pp 11–28
Lemeille S, Willig A, Depege-Fargeix N, Delessert C, Bassi R, Rochaix J (2009) Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol 7:664–675
Lemeille S, Turkina MV, Vener AV, Rochaix J (2010) Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. MolCell Proteomics 9:1281–1295
Lepetit B, Volke D, Gilbert M, Wilhelm C, Goss R (2010) Evidence for the existence of one antenna-associated, lipid-dissolved and two protein-bound pools of diadinoxanthin cycle pigments in diatoms. Plant Physiol 154:1905–1920
Lepetit B, Goss R, Jakob T, Wilhelm C (2012) Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth Res 111:245–257
Li J, Lee GI, Van Doren SR, Walker JC (2000a) The FHA domain mediates phosphoprotein interactions. J Cell Sci 113:4143–4149
Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000b) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403:391–395
Lunde C, Jensen P, Haldrup A, Knoetzel J, Scheller H (2000) The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408:613–615
Majeran W, van Wijk KJ (2009) Cell-type-specific differentiation of chloroplasts in C4 plants. Trends Plant Sci 14:100–109
Majeran W, Zybailov B, Ytterberg AJ, Dunsmore J, Sun Q, van Wijk KJ (2008) Consequences of C-4 differentiation for chloroplast membrane Proteomes in maize mesophyll and bundle sheath cells. Mol Cell Proteomics 7:1609–1638
Martin M, Sabater B (2010) Plastid ndh genes in plant evolution. Plant Physiol Biochem 48:636–645
McCoy SR, Kuehl JV, Boore JL, Raubeson LA (2008) The complete plastid genome sequence of Welwitschia mirabilis: an unusually compact plastome with accelerated divergence rates. BMC Evol Biol 8:130
Montane M, Kloppstech K (2000) The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene 258:1–8
Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D (2009) Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324:1724–1726
Müller P, Li X, Niyogi K (2001) Non-photochemical quenching. A response to excess light energy. Plant Physiol 125:1558–1566
Mulo P, Sakurai I, Aro E (2012) Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta 1817:247–257
Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110:361–371
Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429:579–582
Nagao R, Moriguchi A, Tomo T, Niikura A, Nakajima S, Suzuki T, Okumura A, Iwai M, Shen J, Ikeuchi M, Enami I (2010) Binding and functional properties of five extrinsic proteins in oxygen-evolving photosystem II from a marine centric diatom, Chaetoceros gracilis. J Biol Chem 285:29191–29199
Niyogi K (2000) Safety valves for photosynthesis. Curr Opin Plant Biol 3:455–460
Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I (2012) Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci USA 109:2678–2683
Okegawa Y, Kagawa Y, Kobayashi Y, Shikanai T (2008) Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol 49:825–834
Oudot-Le Secq M, Grimwood J, Shapiro H, Armbrust EV, Bowler C, Green BR (2007) Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol Genet Genomics 277:427–439
Owens TG (1986) Light-harvesting function in the diatom Phaeodactylum tricornutum: II. Distribution of excitation energy between the photosystems. Plant Physiol 80:739–746
Palmer J (2003) The symbiotic birth and spread of plastids: how many times and whodunit? J Phycol 39:4–11
Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462:518–521
Peltier G, Cournac L (2002) Chlororespiration. Annu Rev Plant Biol 53:523–550
Peltier G, Tolleter D, Billon E, Cournac L (2010) Auxiliary electron transport pathways in chloroplasts of microalgae. Photosynth Res 106:19–31
Peng L, Shikanai T (2011) Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol 155:1629–1639
Peng L, Shimizu H, Shikanai T (2008) The Chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J Biol Chem 283:34873–34879
Peng L, Fukao Y, Fujiwara M, Takami T, Shikanai T (2009) Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21:3623–3640
Peng L, Yamamoto H, Shikanai T (2011) Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim Biophys Acta 1807:945–953
Peng L, Fukao Y, Fujiwara M, Shikanai T (2012) Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 24:202–214
Pennington F, Guillard R, Liaaenjensen S (1988) Algal carotenoids. 40. Carotenoid distribution patterns in Bacillariophyceae (Diatoms). Biochem Syst Ecol 16:589–592
Pesaresi P, Pribil M, Wunder T, Leister D (2011) Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: the roles of STN7, STN8 and TAP38. Biochim Biophys Acta 1807:887–896
Petroutsos D, Terauchi AM, Busch A, Hirschmann I, Merchant SS, Finazzi G, Hippler M (2009) PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii. J Biol Chem 284:32770–32781
Petroutsos D, Busch A, Janssen I, Trompelt K, Bergner SV, Weinl S, Holtkamp M, Karst U, Kudla J, Hippler M (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23:2950–2963
Powles S (1984) Photoinhibition of photosynthesis induced by visible-light. Annu Rev Plant Physiol Plant Mol Biol 35:15–44
Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D (2010) Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol 8:e1000288
Pursiheimo S, Rintamäki E, Baena-Gonzalez E, Aro E (1998) Thylakoid protein phosphorylation in evolutionally divergent species with oxygenic photosynthesis. FEBS Lett 423:178–182
Pursiheimo S, Mulo P, Rintamäki E, Aro E (2001) Coregulation of light-harvesting complex II phosphorylation and lhcb mRNA accumulation in winter rye. Plant J 26:317–327
Pyszniak A, Gibbs S (1992) Immunocytochemical localization of photosystem-I and the fucoxanthin-Chlorophyll-a/c light-harvesting complex in the diatom Phaeodactylum tricornutum. Protoplasma 166:208–217
Raven P (1970) A multiple origin for plastids and mitochondria. Science 169:641
Reiland S, Finazzi G, Endler A, Willig A, Baerenfaller K, Grossmann J, Gerrits B, Rutishauser D, Gruissem W, Rochaix J, Baginsky S (2011) Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc Natl Acad Sci USA 108:12955–12960
Richard C, Ouellet H, Guertin M (2000) Characterization of the LI818 polypeptide from the green unicellular alga Chlamydomonas reinhardtii. Plant Mol Biol 42:303–316
Rintamäki E, Kettunen R, Aro E (1996) Differential D1 dephosphorylation in functional and photodamaged photosystem II centers—dephosphorylation is a prerequisite for degradation of damaged D1. J Biol Chem 271:14870–14875
Rintamäki E, Salonen M, Suoranta U, Carlberg I, Andersson B, Aro E (1997) Phosphorylation of light-harvesting complex II and photosystem II core proteins shows different irradiance-dependent regulation in vivo—application of phosphothreonine antibodies to analysis of thylakoid phosphoproteins. J Biol Chem 272:30476–30482
Rintamäki E, Martinsuo P, Pursiheimo S, Aro E (2000) Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc Natl Acad Sci USA 97:11644–11649
Rosso D, Ivanov AG, Fu A, Geisler-Lee J, Hendrickson L, Geisler M, Stewart G, Krol M, Hurry V, Rodermel SR, Maxwell DP, Huner NPA (2006) IMMUTANS does not act as a stress-induced safety valve in the protection of the photosynthetic apparatus of arabidopsis during steady-state photosynthesis. Plant Physiol 142:574–585
Rosso D, Bode R, Li W, Krol M, Saccon D, Wang S, Schillaci LA, Rodermel SR, Maxwell DP, Huner NPA (2009) Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21:3473–3492
Rumeau D, Peltier G, Cournac L (2007) Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Envir 30:1041–1051
Samuelsson G, Lonneborg A, Gustafsson P, Oquist G (1987) The susceptibility of photosynthesis to photoinhibition and the capacity of recovery in high and low light grown cyanobacteria, Anacystis nidulans. Plant Physiol 83:438–441
Sandstrom S, Ivanov A, Park Y, Oquist G, Gustafsson P (2002) Iron stress responses in the cyanobacterium Synechococcus sp PCC7942. Physiol Plant 116:255–263
Savitch LV, Ivanov AG, Gudynaite-Savitch L, Huner NPA, Simmonds J (2011) Cold stress effects on PSI photochemistry in Zea mays: differential increase of FQR-dependent cyclic electron flow and functional implications. Plant Cell Physiol 52:1042–1054
Schuster G, Dewit M, Staehelin L, Ohad I (1986) Transient inactivation of the thylakoid photosystem-II light-harvesting protein-kinase system and concomitant changes in intramembrane particle-size during photoinhibition of Chlamydomonas reinhardtii. J Cell Biol 103:71–80
Schuster G, Timberg R, Ohad I (1988) Turnover of thylakoid photosystem-II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J Biochem 177:403–410
Shapiguzov A, Ingelsson B, Samol I, Andres C, Kessler F, Rochaix J, Vener AV, Goldschmidt-Clermont M (2010) The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc Natl Acad Sci USA 107:4782–4787
Shikanai T (2007) Cyclic electron transport around photosystem I: genetic approaches. Annu Rev Plant Biol 58:199–217
Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M (2003) Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res 31:5324–5331
Suorsa M, Sirpiö S, Aro E (2009) Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol Plant 2:1127–1140
Suorsa M, Jarvi S, Grieco M, Nurmi M, Pietrzykowska M, Rantala M, Kangasjarvi S, Paakkarinen V, Tikkanen M, Jansson S, Aro EM (2012) PROTON GRADIENT REGULATION5 is essential for proper acclimation of Arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell. doi:10.1105/tpc.112.097162
Tikkanen M, Piippo M, Suorsa M, Sirpiö S, Mulo P, Vainonen J, Vener AV, Allahverdiyeva Y, Aro E (2006) State transitions revisited—a buffering system for dynamic low light acclimation of Arabidopsis (vol 62, pg 795, 2006). Plant Mol Biol 62:795
Tikkanen M, Nurmi M, Kangasjärvi S, Aro E (2008a) Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim Biophys Acta 1777:1432–1437
Tikkanen M, Nurmi M, Suorsa M, Danielsson R, Mamedov F, Styring S, Aro EM (2008b) Phosphorylation-dependent regulation of excitation energy distribution between the two photosystems in higher plants. Biochim Biophys Acta 1777:425–432
Tikkanen M, Grieco M, Kangasjarvi S, Aro EM (2010) Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol 152:723–735
Tikkanen M, Grieco M, Aro E (2011) Novel insights into plant light-harvesting complex II phosphorylation and ‘state transitions’. Trends Plant Sci 16:126–131
Tokutsu R, Teramoto H, Takahashi Y, Ono TA, Minagawa J (2004) The light-harvesting complex of photosystem I in Chlamydomonas reinhardtii: protein composition, gene structures and phylogenic implications. Plant Cell Physiol 45:138–145
Turkina MV, Kargul J, Blanco-Rivero A, Villarejo A, Barber J, Vener AV (2006) Environmentally modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol Cell Proteomics 5:1412–1425
Turmel M, Otis C, Lemieux C (2006) The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol 23:1324–1338
Turmel M, Gagnon M, O’Kelly CJ, Otis C, Lemieux C (2009) The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of Prasinophytes and the origin of the secondary chloroplasts of Euglenids. Mol Biol Evol 26:631–648
Vainonen J, Hansson M, Vener A (2005) STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J Biol Chem 280:33679–33686
Vainonen JP, Sakuragi Y, Stael S, Tikkanen M, Allahverdiyeva Y, Paakkarinen V, Aro E, Suorsa M, Scheller HV, Vener AV, Aro E (2008) Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J 275:1767–1777
van der Staay G, Staehelin L (1994) Biochemical-characterization of protein-composition and protein-phosphorylation patterns in stacked and unstacked thylakoid membranes of the prochlorophyte Prochlorothrix hollandica. J Biol Chem 269:24834–24844
Vener AV (2007) Environmentally modulated phosphorylation and dynamics of proteins in photosynthetic membranes. Biochim Biophys Acta 1767:449–457
Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, Sugiura M (1994) Loss of all Ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinusthunbergii. Proc Natl Acad Sci USA 91:9794–9798
Willig A, Shapiguzov A, Goldschmidt-Clermont M, Rochaix J (2011) The phosphorylation status of the chloroplast protein kinase STN7 of Arabidopsis affects its turnover. Plant Physiol 157:2102–2107
Wu C, Wang Y, Liu S, Chaw S (2007) Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol 24:1366–1379
Wu H, Cockshutt AM, McCarthy A, Campbell DA (2011) Distinctive photosystem II photoinactivation and protein dynamics in marine diatoms. Plant Physiol 156:2184–2195
Yamamoto H, Peng L, Fukao Y, Shikanai T (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23:1480–1493
Yeremenko N, Jeanjean R, Prommeenate P, Krasikov V, Nixon P, Vermaas W, Havaux M, Matthijs H (2005) Open reading frame ssr2016 is required for antimycin A-sensitive photosystem I-driven cyclic electron flow in the cyanobacterium Synechocystis sp PCC 6803. Plant Cell Physiol 46:1433–1436
Zhang P, Battchikova N, Jansen T, Appel J, Ogawa T, Aro E (2004) Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803. Plant Cell 16:3326–3340
Zhu S, Green BR (2010) Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochim Biophys Acta 1797:1449–1457
Acknowledgments
The authors gratefully acknowledge funding by the Academy of Finland, project numbers 138703 (MS) and 250216 (IG), CoE projects 118637 (EMA), 218157 (SK) and 130595 (SK) as well as support by the Marie Curie Initial Training Network COSI (GA-215174).
Author information
Authors and Affiliations
Corresponding author
Additional information
A contribution to the Special Issue on Evolution and Biogenesis of Chloroplasts and Mitochondria.
Rights and permissions
About this article
Cite this article
Grouneva, I., Gollan, P.J., Kangasjärvi, S. et al. Phylogenetic viewpoints on regulation of light harvesting and electron transport in eukaryotic photosynthetic organisms. Planta 237, 399–412 (2013). https://doi.org/10.1007/s00425-012-1744-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-012-1744-5