Abstract
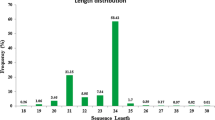
Safflower (Carthamus tinctorius L.) is one of the most important crop plants that has been utilized for production of oleosins. miRNAs (microRNAs) are a class of small and non-coding RNAs that negatively regulate gene expression at post-transcriptional level thus playing a role in plant growth, development, and stress response. In this study, high-throughput Illumina sequencing technology has been used to comprehensively investigate the small RNA transcriptomes of safflower seed, flower, and leaf. It is found that there are at least 236 known miRNAs expressed in safflower, of which 100 miRNAs with relatively high expression abundance exhibited evolutionary conservation across multiple plants. Comparison of their expression abundance among different tissues shows that a total of 116, 133, and 128 miRNAs are significantly differentially expressed with higher abundance or lower abundance between safflower seed/leaf, seed/petal, and leaf/petal. The majority of the most significant differences in miRNA abundance between tissues are tissue-specific miRNAs. In addition, 13 putative novel miRNAs have been identified in safflower. The small RNA transcriptomes obtained in this study provide a basis for further investigation of the physiological roles of identified miRNAs in safflower.


Similar content being viewed by others
Abbreviations
- miRNAs:
-
MicroRNA.
- mRNA:
-
Messenger RNA
- PAGE:
-
Polyacrylamide gel electrophoresis
- DCL:
-
Dicer-like
- nt:
-
Nucleotide
References
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7:986–995
Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13:343–349
Barakat A, Wall K, Leebens-Mack J, Wang YJ, Carlson JE, Depamphilis CW (2007) Large-scale identification of microRNAs from a basal eudicot (Eschscholzia californica) and conservation in flowering plants. Plant J 51:991–1003
Creighton CJ, Reid JG, Gunaratne PH (2009) Expression profiling of microRNAs by deep sequencing. Brief Bioinform 10:490–497
Dsouza M, Larsen N, Overbeek R (1997) Searching for patterns in genomic data. Trends Genet 13:497–498
Fahlgren N, Sullivan CM, Kasschau KD, Chapman EJ, Cumbie JS, Montgomery TA, Gilbert SD, Dasenko M, Backman TW, Givan SA, Carrington JC (2009) Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA 15:992–1002
Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C (2010) Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 16:991–1006
Goodman DH (1964) Safflower: utilization and significance in nutrition and allergy. J Allergy Clin Immunol 35:38–42
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Jia X, Mendu V, Tang G (2010) An array platform for identification of stress-responsive microRNAs in plants. Methods Mol Biol 639:253–269
Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19:2736–2748
Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8:38–44
Klevebring D, Street NR, Fahlgren N, Kasschau KD, Carrington JC, Lundeberg J, Jansson S (2009) Genome-wide profiling of populus small RNAs. BMC Genomics 10:620
Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14:1605–1619
Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ (2008) Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18:571–584
Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106:22534–22539
Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O (2004) In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 18:2237–2242
Poethig RS (2009) Small RNAs and developmental timing in plants. Curr Opin Genet Dev 19:374–378
Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19:421–428
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Song C, Wang C, Zhang C, Korir NK, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11:431
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T (2008) High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9:593
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138:738–749
Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581:1464–1474
Zhang J, Xu Y, Huan Q, Chong K (2009) Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10:449
Zhao CZ, Xia H, Frazier TP, Yao YY, Bi YP, Li AQ, Li MJ, Li CS, Zhang BH, Wang XJ (2009) Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol 10:3
Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C (2008) A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res 18:1456–1465
Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9:149
Acknowledgments
This work was supported by grants from the Special Program for Research of Transgenic Plants (Grant no. 2008ZX08010-002), the National High Technology Research and Development Program of China (863 program, Grant no. 2007AA100503) and the Science and Technology Development Project of Jilin province (Grant no. 20070922).
Author information
Authors and Affiliations
Corresponding authors
Additional information
H. Li and Y. Dong contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2010_1327_MOESM1_ESM.tif
Fig. S1 Evolutionary conserved features of known safflower miRNAs in other plants (Supplementary material 1 TIFF 2717 kb)
425_2010_1327_MOESM2_ESM.xls
Table S1 The detailed expression pattern information of safflower miRNAs between its seed/leaf, seed/petal, and leaf/petal (Supplementary material 2 XLS 95 kb)
Rights and permissions
About this article
Cite this article
Li, H., Dong, Y., Sun, Y. et al. Investigation of the microRNAs in safflower seed, leaf, and petal by high-throughput sequencing. Planta 233, 611–619 (2011). https://doi.org/10.1007/s00425-010-1327-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1327-2