Abstract
SUMO conjugation affects a broad range of processes in Arabidopsis thaliana, including flower initiation, pathogen defense, and responses to cold, drought and salt stress. We investigated two sequence-related SUMO-specific proteases that are both widely expressed and show that they differ significantly in their properties. The closest homolog of SUMO protease ESD4, ESD4-LIKE SUMO PROTEASE 1 (ELS1, alternatively called AtULP1a) has SUMO-specific proteolytic activity, but is functionally distinct from ESD4, as shown by intracellular localization, mutant phenotype and heterologous expression in yeast mutants. Furthermore, we show that the growth defects caused by loss of ESD4 function are not due to increased synthesis of the stress signal salicylic acid, as was previously shown for a SUMO ligase, indicating that impairment of the SUMO system affects plant growth in different ways. Our results demonstrate that two A. thaliana SUMO proteases showing close sequence similarity have distinct in vivo functions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Conjugation of the small ubiquitin-related modifier (SUMO) to substrate proteins is essential in plants (Saracco et al. 2007; for reviews, see Novatchkova et al. 2004; Geiss-Friedlander and Melchior 2007; Miura et al. 2007a). The process impinges on the response to several forms of stress including drought, heat, cold, phosphate shortage, high salinity, and pathogens (Miura et al. 2005, 2007b; Yoo et al. 2006; Catala et al. 2007; Lee et al. 2007; Conti et al. 2008; van den Burg et al. 2010), and regulates the time of flower initiation (Murtas et al. 2003; Jin et al. 2008). SUMO conjugation utilizes a dedicated set of enzymes (Tang et al. 2008). SUMO activating enzyme SAE activates free SUMO by formation of a thioester with the SUMO carboxyl terminus. SUMO is then transferred to SUMO conjugating enzyme SCE, forming a thioester with the SCE active site Cys. SCE can directly transfer activated SUMO onto an ε-amino group of a substrate Lys residue, if this Lys residue is accessible and in a sequence environment called sumoylation consensus motif, which consists of a hydrophobic residue, followed by Lys, any residue, and Asp or Glu (ΨKxD/E; Anckar and Sistonen 2007). In addition to these SCE-binding motifs, SUMO-binding domains consisting of a short hydrophobic β strand (Hecker et al. 2006) can also increase substrate affinity, or influence the conjugation reaction in other ways. Furthermore, a number of proteins have been identified as SUMO ligases, which enhance modification of substrates lacking any of the above sequence motifs (Johnson 2004; Novatchkova et al. 2004; Geiss-Friedlander and Melchior 2007).
SUMO modification can shield protein surfaces, thereby disrupting protein interactions, or generate new interaction interfaces to promote alternative protein assemblages. Sumoylation can therefore implement changes in protein interactions, and although usually only a small fraction of a protein is in the sumoylated form, the modification can be an essential trigger for proteins to cycle between alternative activity states. Furthermore, it was recently found that attachment of multiple SUMO moieties in the form of poly-SUMO chains can promote proteolytic turnover of substrate proteins (Uzunova et al. 2007; Ulrich 2008).
Most sumoylation events are believed to be reversible, because specific proteases exist that cleave conjugates at the SUMO carboxyl terminus, releasing free SUMO and substrate for further conjugation cycles. SUMO proteases have a second role in the sumoylation cycle. They are also necessary to replenish the SUMO pool with newly synthesized protein, because all SUMO genes encode precursor proteins with carboxyl-terminal extensions, which have to be cleaved for release of mature SUMO. The two roles may, however, be fulfilled preferentially by different proteases that are related in sequence. In the yeast S. cerevisiae, the two SUMO proteases have distinguishable tasks, and only one enzyme is essential for maturation of newly synthesized SUMO (Li and Hochstrasser 1999, 2000; Schwienhorst et al. 2000). Functional differences in SUMO proteases of higher eukaryotes have been extensively studied in mammals, and frequently pertain to differences in subcellular localization (Hay 2007; Yeh 2009). An additional complexity is the occurrence of several different SUMO isoforms in both animals and plants (Novatchkova et al. 2004; Saracco et al. 2007; Budhiraja et al. 2009; van den Burg et al. 2010). Differences in proteolytic activity towards different isoforms have been found for plant SUMO proteases (Chosed et al. 2006; Colby et al. 2006; Budhiraja et al. 2009). In this work, we investigate the SUMO protease from Arabidopsis thaliana that is most similar to ESD4, a previously characterized SUMO protease with a role in plant growth and flower initiation (Reeves et al. 2002; Murtas et al. 2003). The ESD4 homolog, ESD4-LIKE SUMO PROTEASE 1 (ELS1), has a distinct role as shown by mutant phenotype, subcellular localization and complementation of yeast SUMO protease mutants. We also show that the reduced growth of esd4 mutants is not due to increased levels of salicylic acid.
Materials and methods
Plant genotypes and growth conditions
Plant lines used in this work are: SIZ1, At5g60410: siz1-2, siz1-3 (Miura et al. 2005); ESD4, At4g15880: esd4-2 (Murtas et al. 2003); ELS1/AtULP1a, At3g06910: els1-1, 201D11 FLAGdb; els1-2, SAIL_318_C01. SID2, At1g74710: sid2-1 (Wildermuth et al. 2001). sid2-1 mutants and a nahG expressing transgenic line were kindly provided by M. Bartsch and Dr. J. Parker (MPIPZ Cologne, Germany). Genotyping of mutant plants was done by PCR reactions with leaf DNA, using the following primers: siz1-2, GTG GAG GTG GAG ATG ATA ATG CC (459A) together with TGG TTC ACG TAG TGG GCC ATC G (SALK LBa1) to detect the T-DNA insertion allele, and 459A together with CCG AGT CAA TGG AGA GGT ACA TC to detect the WT allele; siz1-3, GGT CTT CAG GCT CTC CCT TAA CT (439A) together with SALK LBa1 to detect the T-DNA insertion allele, and 439A together with GAC TAA TTG CTG GAG TTT AGG TTC to detect the WT allele; esd4-2, CTA ATG GGT GCC GTA GCG ATC ATT C (461A) together with SALK LBa1 to detect the T-DNA insertion allele, and 461A together with TAT CTG CAG AGG GCA CAG ACT AAG TT to detect the WT allele; els1-1, CCA ATC TAG GGT TTT GAA CTC TGA ATT (462A) together with CAG TCA TAG CCG AAT AGC CTC TCC A (Fish2) to detect the T-DNA insertion allele, and 462A together with CAC AAC GAG TGA ACT ACT ATT AGC TT (463A) to detect the WT allele; els1-2, 462A together with GCC TTT TCA GAA ATG GAT AAA TAG CCT TGC TTC C to detect the insertion allele, and 462A together with 463A for WT allele detection; sid2-1, PCR for 40 cycles using primers GCA GTC CGA AAG ACG ACC TCG AG and CTA TCG AAT GAT TCT AGA AGA AGC, followed by Mun I digestion of the ensuing fragment (the mutant allele sid2-1 cannot be digested). The T-DNA insert position in mutant els1-1 was determined by sequencing of the diagnostic PCR fragment indicated above to be …T-DNA–GGG ATT TTG ACC… (the first G abutting the T-DNA sequence being nucleotide 38 after the start codon). The T-DNA insert position of mutant els1-2 was estimated from the size of the diagnostic PCR fragment, in agreement with the information provided at http://signal.salk.edu/cgi-bin/tdnaexpress. Plants used for determination of flowering time were germinated and grown on soil in 16 h light and 8 h dark cycles (long day), or 8 h light 16 h dark cycles (short day) in controlled environment at 25°C.
Vector constructs
ELS1 cDNA was cloned from mRNA by reverse transcription and PCR, and inserted into vector pDONR201 (Invitrogen, Darmstadt, Germany). From there, the ORF was recombined into vector pDEST17 (Invitrogen) for protein expression in E. coli (pDEST17-ELS1). A similar clone with a mutation to convert active site Cys into Ser (TGT to AGT change; pDEST17-ELS1-C461S) was made using standard methods. GFP fused to the carboxyl terminus of ELS1 (ELS1–GFP) was expressed in vector 35S-GW-GFP [a Gateway-compatible plant binary vector with 35S promoter, GFP cassette and conferring kanamycin resistance in plants; a kind gift of Dr. Franziska Turck, MPIPZ, Cologne, Germany) and in vector p3 (a plant binary vector containing a CaMV 35S promoter supplied with two additional enhancer regions (triple 35S promoter), and conferring hygromycin resistance in plants (Yin et al. 2007)]. A construct for expression of the ELS1 promoter fused to GUS used vector pGPTV8-GW-GUS (a binary vector with a Gateway cassette in front of the GUS open reading frame; a kind gift of Dr. Hailong An, MPIPZ, Cologne, Germany). A 2.5-kb promoter fragment of ELS1 ending at the start codon, cloned via PCR into vector pDONR207 (Invitrogen) using oligonucleotides GTC TTC ATT TCC TCC AGC and ACT CTC GGG AAG CAA GCA AG was sequenced and linked to the GUS open reading frame.
ELS1 localization
The ELS1p-GUS construct in vector pGPTV8 was transformed into Arabidopsis using floral dip (Clough and Bent 1998), and used for GUS staining by standard methods. ELS1–GFP in vector 35S-GW-GFP was used for floral dip of Arabidopsis, whereas ELS1–GFP in vector p3 was used for Nicotiana benthamiana agroinfection (Bucher et al. 2003). Tissue samples were analyzed using a Leica SP2 AOBS confocal laser scanning microscope.
Experiments in Saccharomyces cerevisiae
The congenic yeast strains used were KU1 (ulp2∆) and YKU8 (ulp1-2). Both mutations cause temperature sensitivity of growth. The ulp1-2 mutation was identified in a screen selecting for spontaneous mutations that suppressed the temperature-sensitive phenotype of the ulp2∆ mutation (K. Uzunova, Univ. of Cologne, Germany and R.J. Dohmen, unpublished results). For complementation studies, ulp2∆ and ulp1-ts cells were transformed with CEN/URA3-based plasmids expressing ESD4 or ELS1 from the galactose-inducible promoter P GAL1 or its weaker derivative P GALS . For expression of ESD4, the cDNA was excised as an XbaI/BamHI fragment from pBLUESCRIPT-ESD4 and cloned into p416GAL1 or p416GALS (Mumberg et al. 1994). The ELS1 cDNA was cloned into the same vector after fusing it to BamHI and EcoRI restriction sites in a PCR reaction that used pDONR201 as a template. To score for complementation, transformants were streaked onto media containing 1% yeast extract, 2% peptone and 2% galactose, followed by incubation at 35°C for 2 days.
Protein purification and in vitro activity assays
ELS1 was purified after expression of a pDEST17 construct using a hexahistidine tag and standard procedures. Incubations for protease activity were carried out as described previously (Murtas et al. 2003).
Protein blotting and antisera
Detection of bands on Western blots was carried out with the ECL detection system (GE Healthcare, Munich, Germany), using anti SUMO1 antibody as described (Murtas et al. 2003) to detect SUMO conjugates, or anti hexahistidine antibody (Qiagen, Hilden, Germany) to detect SUMO fusion and cleavage products. Anti SUMO3 antibody was from Abcam (Cambridge, UK).
RNA gel-blot analysis and RT-PCR
RNA gel blotting was carried out using standard methods. The ELS1 probe was generated using primers ATG AAA AAC CAA TCT AGG G and CAT TAG CTG AGA ATG CAC G. Transcript level of tubulin served as a loading control (probe generated with PCR primers ACA CCA GAC ATA GTA GCA GAA ATC AAG and ACT CGT TGG GAG GAG GAA CT). RT-PCR experiments were carried out as described before (Yin et al. 2007), using primers TTG GAG ACA AGA TGA AGA ACC A and TTG AGA TGG TAG CCC AAC CT to quantitate ELS1 mRNA, and tubulin-specific primers as a control.
Statistical evaluation of leaf number at the time of flowering
A comparison of mean values for the two genotypes (WT vs. els1-1) by a t test gave the following results: LD total leaves, τ = 1.14; LD rosette leaves, τ = 2.16; LD cauline leaves, τ = 3.00; SD total leaves, τ = 1.30; SD rosette leaves, τ = 1.17; SD cauline leaves, τ = 2.62. With a t value of 2.08 for 20 degrees of freedom and P = 95%, and 2.85 for P = 99%, it follows that none of the differences was significant at the 99% level, except for cauline leaves under LD conditions. In the latter case, however, the magnitude of the difference was rather modest (0.6 leaves).
Results
Characterization of SUMO protease ELS1/AtULP1a
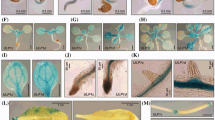
We have previously shown that mutations in the SUMO-specific protease ESD4 (At4g15880) cause early flowering and reduced growth of A. thaliana plants (Reeves et al. 2002; Murtas et al. 2003). The closest homolog of ESD4 in A. thaliana is At3g06910. This ESD4-like gene (called ESD4 LIKE SUMO PROTEASE1, ELS1, in the following; also termed AtULP1a; Kurepa et al. 2003; Novatchkova et al. 2004) codes for a putative SUMO protease. Figure 1a shows a domain diagram of ESD4, ELS1 and the yeast SUMO proteases Ulp1 and Ulp2. A protein sequence alignment (Fig. 1b; Suppl. Fig. S1) indicates that similarity between ESD4 and ELS1 is highest in the carboxyl-terminal protease domain (active site Cys 461 marked by a dot), but also extends towards the amino terminus of the proteins. The ELS1 coding sequence is distributed over a larger 5′ exon and eight smaller downstream exons (Fig. 1c). Figure 2a shows the pattern of GUS activity in a plant expressing an ELS1 promoter–GUS fusion transgene, indicating that the gene is ubiquitously expressed, with high accumulation in the vasculature and in roots. That both of these tissues have a highly active SUMO conjugation system is also suggested by the recent finding that SUMO1 is highly expressed in roots, and SUMO2 in the vasculature (van den Burg et al. 2010). In order to determine expression at later stages of development, tissue of WT plants was harvested for determination of ELS1 mRNA abundance by RT-PCR. The results shown in Fig. 2b indicate that expression of ELS1 mRNA is high in flowers, and persists in siliques, which have an expression level comparable to leaves.
ELS1 protein alignment and gene structure. a ESD4, ELS1 and S. cerevisiae Ulp1 are similar in the carboxyl-terminal protease C48 domain (black box), and contain an amino-terminal extension. b Protein sequence alignment of SUMO proteases EDS4 (top) and ELS1 (bottom). Similarity is most extensive in the carboxyl-terminal catalytic domain, but encompasses the entire length of the proteins. Dot indicates position of the active site Cys residue. Black background was used for identical residues, grey for conserved changes. c Schematic drawing of the intron–exon structure of ELS1. The T-DNA insertion in mutant els1-1 lies in the first exon (thick vertical arrow). d RNA gel blot to demonstrate that T-DNA insertion mutant els1-1 does not express full length ELS1 mRNA. WT, wild type
Expression domains of ELS1. a Expression of ELS1 was assessed with a fusion transgene containing 2.5 kb of ELS1 upstream region, fused to the GUS reporter. Staining for GUS activity in a seedling indicates ubiquitous expression, with high levels in the vasculature and in roots. b RNA from mature plants was used for determination of ELS1 expression in different tissues by reverse transcription-polymerase chain reaction (RT-PCR). Expression is high in flowers, compared to leaves and siliques. TUB tubulin control
To functionally compare ELS1 with ESD4, we determined the intracellular localization of ELS1. Fusion proteins of ELS1 to GFP, carrying GFP either at the carboxyl terminus, or at the amino terminus, were made and cloned into binary expression vectors. Constructs were expressed in Arabidopsis by stable transformation, and in Nicotiana benthamiana leaves by agroinfection (Bucher et al. 2003). Transgenic Arabidopsis seedlings display ELS1–GFP fluorescence in the cytoplasm of root cells (Fig. 3a–d). Because of the generally weak expression, we also investigated distribution in N. benthamiana. In this heterologous host, the majority of the ELS1–GFP fusion protein is also associated with the cytoplasm and/or the endo-membrane system (Fig. 3e–g), and is absent from the nucleus (stained by propidium iodide, red color). In some cases, a faint ELS1–GFP signal was found in the nucleolus (data not shown). While sumoylation is essential for ribosome biogenesis in yeast (Panse et al. 2006), and human SUMO proteases SENP3 and SENP5 are nucleolar proteins (for reviews, see Hay 2007; Yeh 2009), we cannot rule out that the faint nucleolar fluorescence is a side effect of overexpression, or due to the use of a heterologous host. Similarly, a GFP–ELS1 construct had a generally non-nuclear localization, being detectable at membranes and in the cytoplasm (data not shown). In contrast, previous experiments had demonstrated that ESD4 localizes to the nuclear envelope or lamina (Murtas et al. 2003; Xu et al. 2007). We therefore conclude that ELS1 and its closest homolog ESD4 have distinct cellular localizations.
Subcellular localization of ELS1 protein. a–d ELS1–GFP fusion protein was expressed in Arabidopsis and assessed by microscopy. Images show the hypocotyl–root transition of a young seedling. a Red channel detects chloroplasts of the hypocotyl. b Green channel detects ELS1–GFP throughout the cytoplasm. c Phase contrast image. d Overlay image. e–g Section of an N. benthamiana leaf cell after transient expression of ELS1–GFP. ELS1–GFP localizes to endo-membranes and to the cytoplasm, but not to the nucleus (panel e). Consequently, staining of nuclear DNA by propidium iodide (f) does not overlap with ELS1–GFP expression domains (e). Note that there is a gap between the membrane decorated by ELS1-GFP (green), and the stained DNA (red), indicating that the nuclear periphery does not contain ELS1–GFP (overlay image g)
We next tested whether ELS1 is a SUMO-specific protease. The ORF was expressed in E. coli with a hexahistidine tag, purified and used in an in vitro proteolysis assay as previously described (Murtas et al. 2003). Figure 4 shows that ELS1/AtULP1a can cleave sequences from the carboxyl terminus of SUMO1 (At4g26840). The protein is sensitive to N-ethyl maleimide (NEM), a broad specificity inhibitor of cysteine proteases. Furthermore, substitution of the predicted catalytic center Cys at position 461 by Ser resulted in an inactive protein (Fig. 4, third lane). These results demonstrate that ELS1/AtULP1a acts as a SUMO protease in vitro and confirm the experiments of Chosed et al. (2006).
Desumoylation activity of ELS1. ELS1, or its C461S derivative, in which the predicted active site cysteine was mutated, and SUMO1 with carboxyl-terminal extension were purified from E. coli via amino-terminal His tag and combined for incubation. Aliquots from these reactions were subjected to Western blotting with anti hexahistidine antibody. ELS1, but not ELS1C461S, was capable of cleaving the extension (ext.) off the carboxyl terminus of SUMO1
An insertion mutant in ELS1 was obtained from the FLAGdb collection (line 201D11, Ws genetic background; Samson et al. 2002). Sequencing of a PCR-generated genomic fragment revealed the position of the T-DNA in the first exon, which interrupts the Gly codon of position 13 (Fig. 1b, c; “Materials and methods”). The mutant was called els1-1. The putative loss of gene function in the els1-1 mutant was indicated by the absence of mRNA in an RNA gel blot experiment (Fig. 1d). The appearance of els1-1 mutants is generally similar to WT plants, except for slightly reduced growth (Fig. 5) and thinner stems. Staining of tissue sections suggested that the vasculature of the els1-1 mutant is less developed (data not shown). However, the relationship of this trait to ELS1 remains to be determined, because mutant allele els1-2, which carries a T-DNA insertion in ELS1 in the Col-0 background, does not lead to thin stems. Interestingly, it was recently reported that the phenotype of a mutation in a ubiquitin-specific protease depended strongly on the genetic background (Schmitz et al. 2009). Protein gel blot experiments indicated that, unlike esd4 mutants, the T-DNA insertion in els1-1 did not cause a gross change in intracellular abundance of SUMO conjugates (Fig. 6, lanes 1–3), suggesting a more limited role in SUMO regulation.
Growth habit of mutants impaired in activity of components of SUMO conjugation and de-conjugation. a, b WT plant (a accession Ws) next to els1-1 plant (b) after growth in short day conditions. c–e A mutation in the ELS1 gene does not change the growth characteristics of esd4-2 mutants. Two esd4-2/esd4-2 ELS1/els1-1 plants (c, e), and one esd4-2 els1-1 double mutant (d) are siblings of the same esd4-2/esd4-2 ELS1/els1-1 mother plant. Differences in growth between plants homozygous or heterozygous for the els1-1 mutation are small, compared to effects of genetic background or environment (plant age ca. 16 weeks). f–j A block in salicylic acid biosynthesis by mutation sid2-1 does not alleviate dwarfism of esd4 mutants. f WT plant, g sid2-1 mutant, h plant homozygous for sid2-1, and heterozygous for the esd4-2 mutation; i sid2-1 esd4-2 double mutant, j esd4-2 single mutant. Plants were grown in parallel under the same conditions for 5 weeks, insets in panels i and j are close-ups. Scale bars represent 5 cm
Determination of in vivo SUMO conjugate levels. Lanes 1–3 extracts of WT (accession Ws), of esd4, and of els1-1 mutant leaves were used for a protein gel blot probed with anti SUMO1 antibody. Whereas SUMO conjugate levels are significantly elevated in esd4 cells, no significant change compared to the WT was detected in els1-1. A section containing the Rubisco large subunit band of a filter stained with Ponceau S is shown below the gel as a loading control. Lanes 4–6 extracts from WT (accession Col-0), from esd4-2 els1-1 double mutants, and from plants homozygous for esd4-2 and heterozygous for the els1-1 mutation were compared by blotting and hybridization with anti SUMO1 antibody (top panel), and anti SUMO3 antibody (bottom panel). The middle panel shows a control gel stained with Coomassie blue to document protein concentration in the extracts. Molecular weight marker positions are shown to the left of the blots. A dot indicates position of free SUMO1
We determined the time of flowering of WT and els1-1 plants by measuring leaf numbers (Table 1). Statistical analysis (cf. “Materials and methods”) indicated no significant difference in the number of rosette leaves, or in total leaf number. The number of cauline leaves formed before flowering under long-day conditions differs, but the magnitude of the difference is small (0.6 leaves; Table 1; “Materials and methods”). The same results were obtained by comparing mutants carrying another allele, els1-2, with its WT accession Col-0 (data not shown). In contrast, a strong, statistically significant difference in flowering time between WT and esd4 plants was previously reported (Reeves et al. 2002). We conclude that, unlike ESD4, ELS1 is not a determinant of flowering time.
Complementation of yeast (S. cerevisiae) mutants by expression of Arabidopsis SUMO proteases
Saccharomyces cerevisiae has two SUMO-specific proteases, Ulp1 and Ulp2 (Fig. 1a). They have the same type of protease domain, but different amino-terminal domains. Ulp2 has, in addition, a carboxyl-terminal extension following the catalytic domain. Both ELS1 and ESD4 have an amino-terminal (but no carboxyl-terminal) extension and based on their sequence may therefore be grouped with yeast protease Ulp1, rather than with Ulp2 (Novatchkova et al. 2004). We cloned the ELS1 and ESD4 ORFs into plasmids for expression in yeast, using two types of vectors. One vector, carrying the galactose-inducible GAL1 promoter, allows a relatively high expression level. The other vector expresses the Arabidopsis SUMO proteases under control of a truncated variant of the GAL1 promoter, which results in attenuated galactose-inducibility, while maintaining tight repression by glucose (Mumberg et al. 1994; GALS in Fig. 7). Complementation of mutations in either ULP1 or ULP2 was tested using both vectors. ULP1 is essential, and the complementation assay used the temperature sensitivity of growth of an ulp1-ts mutant with similar phenotypes as reported previously for another ulp1 allele (Li and Hochstrasser 1999). In contrast, mutants with a deletion of the ULP2 gene are viable, but sensitive to a variety of stresses including elevated temperature (Li and Hochstrasser 2000; Schwienhorst et al. 2000). The temperature-sensitive phenotype of a ULP2 deletion allele (ulp2Δ) was used to score complementation by an Arabidopsis cDNA. Figure 7 shows that ELS1 can complement the ulp1-ts mutant, whereas the ESD4 cDNA cannot. In striking contrast, the ulp2Δ temperature sensitivity is not complemented by ELS1, but by ESD4 cDNA, when expressed from a weak promoter. We therefore conclude that ELS1 and ESD4 differ in their ability to complement yeast mutants. While it is the essential function of ULP1 to generate mature SUMO, ULP2 predominately acts on polymeric SUMO chains (Li and Hochstrasser 1999; Schwienhorst et al. 2000; Bylebyl et al. 2003). These results suggest that in vivo ELS1 has a similar role in the SUMO cycle to ULP1 while ESD4 has that of ULP2.
Complementation of yeast mutants in SUMO proteases by Arabidopsis proteins. a Schematic drawing of the constructs used. An inducible yeast promoter (either strong or weak) is followed by Arabidopsis SUMO protease cDNA of either ESD4, or ELS1. The vectors also contain sequences for selection (URA3) and single copy propagation (CEN ARS) in yeast. b Growth of yeast mutants carrying either a ts allele of ULP1 or lacking the ULP2 gene is normally impaired at elevated temperature. In contrast, ELS1 (but not ESD4) supports growth of ulp1-ts mutant cells, and low levels of ESD4 (but not ELS1) support growth of ulp2Δ mutant cells at elevated temperature. Strains were grown for 2 days at the semi-permissive temperature of 35°C
Double mutant analysis of Arabidopsis SUMO proteases ESD4 and ELS1
Lack of co-localization and distinct mutant phenotypes of els1 and esd4 mutants makes direct functional redundancy or overlap in function unlikely, but does not exclude all forms of mutual influence. In order to reveal interactions between ELS1 and ESD4, we constructed the double mutant esd4-2 els1-1. esd4-2 is a loss of function mutation in the Col-0 background (Murtas et al. 2003) and apparently affects growth and fertility of plants more severely than the esd4-1 allele, which is in the Ler background. esd4-2 plants are almost completely sterile under our standard growth conditions. A double heterozygous esd4-2 els1-1 plant was backcrossed to Col-0, and F2 plants were genotyped. Both esd4-2 els1-1 double homozygous, and esd4-2/esd4-2ELS1/els1-1 plants could be identified. esd4-2/esd4-2 ELS1/els1-1 plants carried more seeds than esd4-2 single mutants. Progeny plants of esd4-2/esd4-2 ELS1/els1-1 parents were grown and genotyped. Interestingly, 36 of 38 plants were again ELS1/els1-1, indicating that the els1-1 mutation, if present in one of the two gametes (we suspect the male gametes), acts as a fertility restorer. However, apart from fertility, there is little difference in greenhouse growth habit between esd4-2, esd4-2 els1-1 and esd4-2/esd4-2 ELS1/els1-1 plants. Figure 5c–e shows a comparison of two esd4-2/esd4-2 ELS1/els1-1 plants (plants c, e), and one esd4-2 els1-1 double homozygote (plant d). Growth differences due to the mixed (Ws/Col-0) genetic background and/or to environmental influences exceed those caused by loss of ELS1 function.
We also determined the level of SUMO conjugates in esd4-2 els1-1 double mutants and in sibling plants with genotype esd4-2/esd4-2 ELS1/els1-1 (Fig. 6, lanes 5, 6). Consistent with the results shown in Fig. 6, lanes 1–3, which indicate only a minor influence of the els1-1 mutation on SUMO conjugate levels, the double mutant does not have more SUMO conjugates than the single esd4 mutant.
Growth defect of esd4 mutant plants is SA-independent
In contrast to ELS1, SUMO protease ESD4 is a determinant of flowering time (Reeves et al. 2002). As recently reported, mutants in SUMO ligase SIZ1 (At5g60410) are also early flowering (Jin et al. 2008). Whereas siz1 mutants are impaired in SUMO conjugation, esd4 mutants accumulate SUMO conjugates, presumably as a consequence of decreased SUMO removal from substrates. Correct flower initiation may therefore require both efficient sumoylation and efficient de-sumoylation. Yet another similarity between siz1 and esd4 mutants is that both have a small stature, indicating significant growth retardation. It was recently shown that siz1 plants have elevated levels of salicylic acid, and that their stunted growth is largely abolished if the nahG reading frame, encoding a salicylate hydroxylase from Pseudomonas bacteria, is expressed in these mutants (Lee et al. 2007; Jin et al. 2008). We were therefore interested in whether the reduced growth of esd4 mutants is also suppressed by a decrease in intracellular SA levels. To that end, the esd4-2 mutation was combined with a nahG transgene, and, in separate crosses, with the sid2-1 mutation, which affects SA biosynthesis by abolishing isochorismate synthase 1 activity (Wildermuth et al. 2001; Strawn et al. 2007). Analysis of the contribution of SA to the growth habit of esd4 mutants indicates an independence of SA, as judged by both crosses (Fig. 5; data not shown). We therefore concluded that esd4 and siz1, two mutations in nuclear-localized components of the sumoylation machinery, influence plant growth by distinct mechanisms, because growth regulation by ESD4 is SA-independent.
Discussion
In this work, we investigated a protease from the Arabidopsis sumoylation cascade. Arabidopsis encodes more than 65 reading frames with the C48 cysteine protease fold, which encompasses the SUMO-specific proteases (Novatchkova et al. 2004; van der Hoorn 2008). Only a limited number of them have been tested for activity towards SUMO (Murtas et al. 2003; Chosed et al. 2006; Colby et al. 2006). ESD4, the first characterized SUMO protease, has an important role in flower initiation and plant growth (Reeves et al. 2002; Murtas et al. 2003).
We investigated the closest relative of SUMO protease ESD4, ELS1 (termed AtULP1a by Kurepa et al. 2003), and show that it is an active SUMO protease (Fig. 4), confirming previous results by Chosed et al. (2006). Alignment of the ESD4 and ELS1 protein sequences indicates that similarity extends beyond the carboxyl-terminal catalytic domain towards the amino terminus (Fig. 1, Suppl. Fig. S1). ELS1 is ubiquitously expressed (Fig. 2). Its localization was determined by expression of ELS1–GFP and GFP–ELS1 fusion proteins in Arabidopsis and N. benthamiana (Fig. 3). Despite the overall similarity between ELS1 and ESD4, ELS1 localization is distinct from that of ESD4, which localizes to the nuclear periphery (Murtas et al. 2003; Xu et al. 2007). In contrast, ELS1 is mostly cytoplasmic, possibly associated with endo-membranes. While there was occasionally a weak signal detectable in the nucleolus, the protein was absent from all other parts of the nucleus, as opposed to its abundant presence in the cytoplasm. Expression in the homologous host, and the transient heterologous expression data are therefore strong indicators that ELS1 and ESD4 operate in different cellular compartments.
Another indicator of the functional difference between ELS1 and ESD4 is that ELS1 loss of function has only mild phenotypic consequences, whereas esd4 mutant plants are severely dwarfed (Fig. 5). ESD4 and ELS1 open reading frames were also expressed in S. cerevisiae cells with mutation in either of the two yeast SUMO proteases, ULP1 or ULP2. Whereas both ESD4 and ELS1 belong to the same structural group as yeast Ulp1 (Fig. 1a), only ELS1 can complement the essential function of Ulp1 in growth promotion (Fig. 7). It may seem surprising that ESD4 cannot complement, because both ESD4 in the plant, and ULP1 in yeast, localize to the nuclear periphery, whereas ELS1 is non-nuclear in the plant expression systems, and is therefore not likely to localize to the nuclear periphery upon expression in yeast. Interestingly, the requirement for Ulp1 can be bypassed by expression of mature SUMO in yeast, suggesting that an essential function of Ulp1 is its participation in SUMO maturation (Li and Hochstrasser 1999; Schwienhorst et al. 2000). If ELS1 can, in yeast, assist in generation of mature SUMO by proteolytic removal of the carboxyl-terminal extension of the primary translation product, it may complement the ulp1-ts mutant independent of its localization. If this explanation holds true, it follows that ESD4 is less able to assist in yeast SUMO maturation, although it can carry out this reaction in vitro (Murtas et al. 2003). Conversely, loss of ULP2, the yeast SUMO protease residing in the nucleoplasm, can be complemented by expression of ESD4 at low levels (Fig. 7). Neither ESD4 at high expression levels, nor ELS1 can complement the ulp2Δ mutation. The lack of complementation at high ESD4 expression levels may be attributed to a toxicity effect, similar to the finding that high level expression of full length or truncated versions of yeast Ulp1 is toxic to yeast cells (Li and Hochstrasser 2003; Panse et al. 2003). These results might indicate that in A. thaliana the major role of ESD4 is SUMO recycling from substrates while that of ELS1 is SUMO maturation.
The double mutant els1 esd4 was also constructed. As shown in Fig. 5, the double mutant looks similar to the esd4 single mutant. This result is consistent with the other observations, pointing to distinct roles. The purpose of this experiment was to reveal more subtle interactions between the two SUMO proteases. For instance, the two SUMO-specific proteases of baker’s yeast, Ulp1 and Ulp2, do not co-localize, yet a double mutant in both genes does not show the sum of both single mutant phenotypes, but is actually less severely affected than one of the single mutants (Li and Hochstrasser 2000). As ELS1 and ESD4 showed functional complementarity in the yeast assay, a similar role might be observed in the Arabidopsis double mutant. We found that the growth habit of esd4 plants was unaffected by the simultaneous presence of the els1-1 mutation, but that one of the germ lines (presumably the male germ line) displayed higher viability as esd4 els1 double mutant, than as esd4 single mutant, leading to a segregation distortion of the els1 marker in the esd4 homozygous background. The pattern of SUMO1 conjugates was also similar between the esd4-2 single, and the esd4-2 els1-1 double mutant. Neither mutation influences the pattern of SUMO3 conjugates (Fig. 6).
We also investigated whether the dwarf phenotype of esd4 mutants is due to increased levels of salicylic acid, because decreased growth of Arabidopsis mutants can be due to induction of (biotic or abiotic) stress responses, and sumoylation has already been linked to stress response regulation. It was also recently found that decreased growth of mutants in another gene of the sumoylation cascade, the SIZ1 SUMO ligase, can be suppressed by decreasing endogenous SA levels. The SA levels in esd4 mutants therefore impinge on the question whether the increase in SUMO conjugates in the esd4 mutant affects the same processes as the decrease caused by a siz1 mutation. Preliminary characterization of esd4 siz1 double mutant plants indicated that this double mutant has the growth characteristics of esd4 single mutants, suggesting that the two mutations do not mitigate each others’ effects (R. Hermkes and A. Bachmair, unpublished results).
A possible role of altered SA levels in the esd4 mutant was assessed by expression of a nahG gene, which degrades SA to catechol, thereby decreasing the SA concentration. However, it was suggested that independent confirmation of results from nahG experiments is necessary, because catechol has biological activity and may therefore influence the outcome (van Wees and Glazebrook 2003). Therefore, we investigated SA dependence of growth by generation of both esd4-2 nahG plants, and esd4-2 sid2-1 plants. The SID2 gene is essential for SA biosynthesis (Wildermuth et al. 2001; Strawn et al. 2007), and its absence drastically reduces SA levels (without production of catechol). The finding that esd4-2 growth retardation is not suppressed by decreasing SA levels (Fig. 5, panels f–j) implies that excess SA is not the cause of the growth defects of esd4 mutants. Therefore, although both esd4 and siz1 mutants affect the same pathway and show reduced growth, the molecular mechanisms underlying this phenotype are likely to be distinct.
In summary, mutations in sumoylation components ESD4 and ELS1, as well as in SIZ1, have unique consequences for plants, even though these genes operate in the same pathway. ELS1 may be redundant with other de-sumoylating proteases because of the relatively weak and accession-dependent phenotype of the els1 mutation, but the closest homolog, ESD4, has little or no functional overlap with ELS1.
Abbreviations
- SUMO:
-
Small ubiquitin-related modifier
- SAE:
-
SUMO activating enzyme
- SCE:
-
SUMO conjugating enzyme
- ESD4:
-
EARLY IN SHORT DAYS4
- ELS1:
-
ESD4 LIKE SUMO PROTEASE
References
Anckar J, Sistonen L (2007) SUMO: getting it on. Biochem Soc Trans 35:1409–1413
Bucher E, Sijen T, de Haan P, Goldbach R, Prins M (2003) Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J Virol 77:1329–1336
Budhiraja R, Hermkes R, Müller S, Schmidt J, Colby T, Coupland G, Bachmair A (2009) Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on de-sumoylation. Plant Physiol 149:1529–1540
Bylebyl GR, Belichenko I, Johnson ES (2003) The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem 45:44113–44120
Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua N-H (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19:2952–2966
Chosed R, Mukherjee S, Lois LM, Orth K (2006) Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis sumoylation. Biochem J 398:521–529
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Colby T, Matthäi A, Boeckelmann A, Stuible H-P (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142:318–332
Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20:2894–2908
Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8:947–956
Hay RT (2007) SUMO-specific proteases: a twist in the tail. Trends Cell Biol 17:370–376
Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I (2006) Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281:16117–16127
Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim W-Y, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through effects on FLC chromatin structure. Plant J 53:530–540
Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73:355–382
Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung D-Y, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. J Biol Chem 278:6862–6872
Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, Kim D, Lee SY, Salt DE, Mengiste T, Gong Q, Ma S, Bohnert HJ, Kwak SS, Bressan RA, Hasegawa PM, Yun DJ (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49:79–90
Li S-J, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398:246–251
Li S-J, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20:2367–2377
Li S-J, Hochstrasser M (2003) The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J Cell Biol 160:1069–1081
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Jun DJ, Hasegawa PM (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102:7760–7765
Miura K, Jin JB, Hasegawa P (2007a) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10:1–8
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007b) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19:1403–1414
Mumberg D, Müller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22:5767–5768
Murtas G, Reeves PH, Fu Y-F, Bancroft I, Dean C, Coupland G (2003) A nuclear protease required for flowering time regulation in Arabidopsis reduces the abundance of small ubiquitin-related modifier conjugates. Plant Cell 15:2308–2319
Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A (2004) SUMO conjugation in plants. Planta 220:1–8
Panse VG, Kuster B, Gerstberger T, Hurt E (2003) Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nature Cell Biol 5:21–27
Panse VG, Kressler D, Pauli A, Petfalski E, Gnädig M, Tollervey D, Hurt E (2006) Formation and nuclear export of preribosomes are functionally linked to the small ubiquitin-related modifier pathway. Traffic 7:1311–1321
Reeves PH, Murtas G, Dash S, Coupland G (2002) early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129:5349–5361
Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res 30:94–97
Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007) Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145:119–134
Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM (2009) Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 149:1196–1204
Schwienhorst I, Johnson ES, Dohmen RJ (2000) SUMO conjugation and deconjugation. Mol Gen Genet 263:771–786
Strawn MA, Marr SK, Inoue K, Inada N, Zubieta C, Wildermuth MC (2007) Arabidopsis isochorismate synthase function in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J Biol Chem 282:5919–5933
Tang Z, Hecker CM, Scheschonka A, Betz H (2008) Protein interactions in the sumoylation cascade: lessons from X-ray structures. FEBS J 275:3003–3015
Ulrich HD (2008) The fast-growing business of SUMO chains. Mol Cell 32:301–305
Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282:3416–3417
van den Burg HA, Kini RK, Schuurink RC, Takken FL (2010) Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22:1998–2016
van der Hoorn RAL (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59:191–223
van Wees SCM, Glazebrook J (2003) Loss of non-host resistance of Arabidopsis nahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J 33:733–742
Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 414:562–565
Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I (2007) NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19:1537–1548
Yeh ETH (2009) SUMOylation and de-SUMOylation: wrestling with life’s processes. J Biol Chem 284:8223–8227
Yin X-J, Volk S, Ljung K, Dolezal K, Mehlmer N, Ditengou F, Hanano S, Davis SJ, Schmelzer E, Sandberg G, Teige M, Palme K, Pickart C, Bachmair A (2007) Ubiquitin lysine 63 chain-forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19:1898–1911
Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM (2006) SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142:1548–1558
Acknowledgments
We wish to thank Michaela Lehnen, Kerstin Luxa and Rainer Franzen for technical assistance, Maret Kalda for photography, Dr. Hailong An (MPIPZ) and Dr. Franziska Turck (MPIPZ) for vectors, Michael Bartsch (MPIPZ) and Dr. Jane Parker (MPIPZ) for plant material and the consortia from SALK and Génoplante for mutant lines. We thank Dr. Kristina Uzunova (University of Cologne, Germany) for yeast strains and Dr. Ralf Kölling (University of Hohenheim, Germany) for plasmids. This work was supported by the Max Planck Society, and by the German Research Foundation (SFB 635 to G.C. and J.D., BA1158/3-1 and SPP1365 to A.B.), by the Austrian Science Foundation (FWF grant P21215 to A.B.) and by pre-doctoral fellowships from the International Max Planck Research School to R.B. and R.H.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
R. Hermkes and Y.-F. Fu are joint first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hermkes, R., Fu, YF., Nürrenberg, K. et al. Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233, 63–73 (2011). https://doi.org/10.1007/s00425-010-1281-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1281-z