Abstract
Towards an understanding of the interacting nature of the CLAVATA (CLV) complex, we predicted the 3D structures of CLV3/ESR-related (CLE) peptides and the ectodomain of their potential receptor proteins/kinases, and docking models of these molecules. The results show that the ectodomain of CLV1 can form homodimers and that the 12-/13-amino-acid CLV3 peptide fits into the binding clefts of the CLV1 dimers. Our results also demonstrate that the receptor domain of CORYNE (CRN), a recently identified receptor-like kinase, binds tightly to the ectodomain of CLV2, and this likely leads to an increased possibility for docking with CLV1. Furthermore, our docking models reveal that two CRN-CLV2 ectodomain heterodimers are able to form a tetramer receptor complex. Peptides of CLV3, CLE14, CLE19, and CLE20 are also able to bind a potential CLV2-CRN heterodimer or heterotetramer complex. Using a cell-division reporter line, we found that synthetic 12-amino-acid CLE14 and CLE20 peptides inhibit, irreversibly, root growth by reducing cell division rates in the root apical meristem, resulting in a short-root phenotype. Intriguingly, we observed that exogenous application of cytokinin can partially rescue the short-root phenotype induced by over-expression of either CLE14 or CLE20 in planta. However, cytokinin treatment does not rescue the short-root phenotype caused by exogenous application of the synthetic CLE14/CLE20 peptides, suggesting a requirement for a condition provided only in living plants. These results therefore imply that the CLE14/CLE20 peptides may act through the CLV2-CRN receptor kinase, and that their availabilities and/or abundances may be affected by cytokinin activity in planta.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Local signaling to maintain stem cells in the Arabidopsis shoot apical meristem (SAM) involves the CLAVATA-WUSCHEL (CLV-WUS) signal transduction pathway. WUS, a homeodomain transcription factor, acts non-cell autonomously to promote stem cell fate (Laux et al. 1996). WUS induces expression of CLAVATA3 (CLV3), which encodes a small, extra-cellular signal ligand (Fletcher et al. 1999) that is present in stem cells of the central zone (CZ) of the SAM. It is proposed that CLV3 is secreted from the stem cells of the CZ and interacts with CLAVATA1 (CLV1), a leucine-rich repeat (LRR) receptor kinase (Clark et al. 1997), which may in turn form a heterodimer with CLAVATA2 (CLV2) in the CZ, thereby restricting WUS expression specifically to the organizing center (OC) of the SAM (Schoof et al. 2000; Müller et al. 2006). In support of this view, biochemical evidence for a direct in vitro binding of the CLV3 peptide and the ectodomain region of CLV1 receptor kinase has been reported (Ogawa et al. 2008; Ohyama et al. 2009).
Loss-of-function CLV1, CLV2 or CLV3 mutations result in expanded WUS expression, enlarged shoot and floral meristems, and flowers with extra organs (Clark et al. 1997). Conversely, over-expression of CLV3 causes loss of WUS expression and premature shoot and floral meristem termination, thereby suggesting a relationship between the CLV3 peptide and the CLV1/CLV2 receptors that limits the size of the WUS expression domain and thus the size of the meristem. The CLV3-WUS signaling pathway forms a spatial, negative feedback loop that allows the SAM to balance the stem cells in the CZ with the cells in the peripheral zone (PZ), by controlling the transition rates of the daughters of the stem cells in the CZ, into the cells in the PZ (for a review, see Williams and Fletcher 2005). In the Arabidopsis root apical meristem (RAM) the 4 cells comprising the quiescent center (QC) are considered equivalent to the OC, and the cells encircling and in direct contact with the QC function as the stem cells. The size and the number of stem cells in the RAM of Arabidopsis appear generally constant (van den Berg et al. 1995; 1997). The structure of the RAM and SAM remains constant, although the size and the number of stem cells of the SAM varied over changed conditions and developmental stages (Geier et al. 2008; Brand et al. 2001; Weigel and Jürgens 2002). These results suggest that continuous processes of cell division and differentiation of daughter cells are well regulated.
Evidence also points to the operation of CLV-WUS-like pathways in the RAM of root tips. A WUS-related homeobox gene, termed WOX5, has been identified in the QC of the RAM in Arabidopsis. Functional equivalence between WUS and WOX5 has been suggested based on the fact that each can substitute reciprocally when ectopically expressed in the other’s expression domain (the OC or QC) (Sarkar et al. 2007). Loss-of-function of WOX5 causes differentiation of distal stem cells (columella initials) in the RAM. Conversely, gain-of-function of WOX5 blocks differentiation of daughter cells of the distal stem cells, into columella cells (Sarkar et al. 2007). Striking correspondences between WUS and WOX5 in the SAM and RAM imply that a CLV-like feedback loop may act to restrict WOX5 and thus negatively regulate meristem activity in the RAM. Using RT–PCR analysis, several CLE (CLV3/ESR-related) genes have been found to express in the root (Sharma et al. 2003). Over-expressing some CLE genes, including CLV3, triggers premature termination of the RAM (Strabala et al. 2006). Exogenous applications of a synthetic 12-amino-acid peptide derived from some CLE motifs have also been shown to cause a short-root phenotype (Ito et al. 2006). Furthermore, it has been recently reported that CLE40, which expresses in the differentiating daughters of distal root stem cells, restricts WOX5 expression and promotes differentiation of stem cells in the RAM (Stahl et al. 2009). Taken together these results thus suggest that a WUS-CLV-like pathway may function in the RAM of Arabidopsis root tips.
Our previous results have shown that over-expression of CLE14 or CLE20 in Arabidopsis triggers early termination of the RAM in a CLV1-independent and CLV2-dependent manner (Meng et al. 2010a). Cockcroft et al. (2000) suggested that cell division controlled meristem activity and the overall growth rate in plants (Cockcroft et al. 2000). One of the proposed functions of the CLV-WUS signaling loop in the SAM is to limit cell division rates of the stem cells in the CZ of the SAM. Reddy et al. (2004) and Reddy and Meyerowitz (2005) reported that CLV3 signaling in the SAM regulates cell fate specification and inhibits the cell division in the SAM (Reddy et al. 2004; Reddy and Meyerowitz 2005). However, there is thus far no detailed study describing the temporal distribution of cell division in the RAM of the roots showing a short-root phenotype induced by over-expression of the CLE genes.
Another approach for understanding the functional nature of the CLV-CLE pathway involves an appreciation of protein structure. A common structural element of many plant cell-surface receptors is the extra-cellular LRR domain that is generally thought to mediate ligand perception and to perform ligand binding (Kobe and Kajava 2001). Although biochemical evidence for a direct in vitro binding of the CLV3 peptide ligand with the CLV1 receptor has been reported (Ogawa et al. 2008; Ohyama et al. 2009), there is no information about the molecular location(s) at which interaction between CLV1 and CLV3 actually occurs. Moreover, the current results regarding the CLV3 peptide are controversial. The mature CLV3 peptide was first reported as a 12-amino-acid dodecapeptide, in which two of three proline residues were modified to hydroxyproline (Kondo et al. 2006). However, more recently it was reported that the mature CLV3 peptide was actually a 13-amino-acid arabinosylated glycopeptide (Ohyama et al. 2009). Additionally, it was shown that post-translational arabinosylation of CLV3 is critical both for its biological activity in the SAM and for high-affinity binding to CLV1 (Ohyama et al. 2009). By using firefly luciferase complementation imaging (LCI), and co-immunoprecipitation assays in both Arabidopsis mesophyll protoplasts and Nicotiana benthamiana leaves, Zhu et al. (2010) demonstrated that CLV2 did not interact with CLV1. Instead, it was shown that CLV2 directly interacts with CRN [a receptor-like protein kinase, also known as SOL2, isolated as a suppressor of root-specific over-expression of CLE19 (Müller et al. 2008; Miwa et al. 2008)] in the absence of CLV3, whereas CLV1 could interact with the CLV2–CRN heterodimer (Zhu et al. 2010). However, to date there is no structural evidence, including 3D, available on the interaction properties for any CLE peptides or CLV/CLV-like receptors, thus limiting our understanding of the functional nature of the CLV-CLE pathways in plants.
To determine the interaction properties of the CLV complex we undertook computational approaches in order to predict 3D structures and interaction models of the 12-/13-amino-acid CLV3 peptides, and the extra-cellular LRR domains of its receptors, CLV1 and CLV2. Over-expression of CLE14, CLE19, or CLE20 has been show to cause strong, short-root phenotypes in Arabidopsis, in a CLV1-independent, but CLV2-dependent manner (Meng et al. 2010a; Fiers et al. 2005). Therefore, 3D structures of the 12-amino-acid CLE14, CLE19, and CLE20 and their potential interaction models with CLV2 and CRN were also predicted.
Finally, using a GFP translational-fusion reporter system to explore the functional nature of the CLE14 and CLE20 pathway(s), we report that CLE 14 and CLE20 express in specific cells of the root. Using a synthetic 12-amino-acid peptide derived from either CLE14 or CLE20 motifs, and a cell-division marker line, BJ3 (Colon-Carmona et al. 1999), we subsequently demonstrate that both CLE14 and CLE20 peptides inhibit, irreversibly, root growth by reducing cell division rates in the RAM. Finally, we show that exogenous application of cytokinin is able to partially rescue the short-root phenotype induced by over-expression of CLE14/CLE20 in planta. However, cytokinin treatment does not rescue the short-root phenotype caused by exogenous application of the synthetic 12-amino-acid CLE14/CLE20 peptides. Our results provide new evidence and insights towards understanding the molecular basis of the CLE peptide-signaling pathway in the regulation of both plant meristem activity and in Arabidopsis development.
Materials and methods
Three-dimensional structure and dock modeling
Signal peptide cleavage sites and transmembrane domains were predicted using SignalP-HMM (Nielsen and Krogh 1998) and TMHMM (Krogh et al. 2001), respectively. The three-dimensional protein structures and docking models were predicted as described by Meng et al. (2010b). Briefly, the 3D structure of the 12/13-amino-acid CLV3, the 12-amino-acid CLE peptides, and the 29-amino-acid extra-cellular receptor domain of the CRN receptor-like kinase, were predicted using the ab initio method implemented in the I-TASSER algorithm, while the 3D structure models of the extra-cellular LRR region of CLV1 and CLV2 were inferred using the homology threading simulation of the I-TASSER algorithm (http://zhang.bioinformatics.ku.edu/I-TASSER/) (Zhang 2008). Superimposition analysis of the 3D models of CLV1/CLV2 and its templates was done using 3-dimensional structural superposition (3D-SS) (http://cluster.physics.iisc.ernet.in/3dss/severalinput.html) (Russell et al. 1992). Conserved amino acids at the protein surface were determined using ConSurf (Landau et al. 2005) (http://consurf.tau.ac.il/overview.html). The docking modeling of the CLV complex was conducted using PatchDock and FireDock (Schneidman-Duhovny et al. 2005; Mashiach et al. 2008).
Generating the CLE14 and CLE20 promoter GFP translational-fusion constructs
The expression patterns of CLE14 and CLE20 were analyzed by translational fusion of GFP to the full-length CLE gene driven by its promoter in the construct pCLE14::CLE14-GFP, or pCLE20::CLE20-GFP. For CLE14, a 2,490-bp fragment immediately upstream from the stop codon (TGA) of the CLE14 coding sequence was amplified by PCR from the genomic DNA of the wild-type Arabidopsis Col plants using the primers: SacpCLE14 5F: C GAG CTC AAT AAT AAC ATC ATT GAT CTC; BamCLE14 3R: GC GGA TCC CG TTT GTT GTG AAG CGG GTT AGG AC. The cloning sites, SacI and BamHI, were added at the 5′ end of the 5′ forward and 3′ reverse primers, respectively. The PCR fragment was cloned into pEZS-NL and fused to GFP at the C-terminus at the SacI and BamHI sites. Similarly, for CLE20, a 2,276-bp fragment immediately upstream of the stop codon (TGA) of the CLE20 coding region was amplified by PCR from the genomic DNA of Arabidopsis using the primers: pCLE20-5F: AAC ATG CGA AGA AGC TCG AGA; BamCLE20 3R: GC GGA TCC CG TCG TTT GTT GTG CAA AGG to GFP. The cloning site BamHI was added at the 5′ end of the 3′ reverse primer. The PCR fragment was cloned into pEZS-NL and fused to GFP at the C-terminus at the SacI and BamHI site. Subsequently, a SacI and NotI fragment containing the cloned CLE promoter and the full-length coding sequence of the CLE gene, GFP, and the OCS 3′ terminator was removed from the recombinant pEZS-CL plasmid and inserted into the binary vector pART27 for the stable transformation assay.
Plant transformation
The cloned CLE genes in the binary vector pART27 were introduced into Agrobacterium tumefaciens strain GV3101 using the freeze–thaw method (Höfgen and Willmitzer 1988). Transgenic plants were generated via the floral-dip method (Clough and Bent 1998).
Plant material and growth condition
Arabidopsis seeds were sterilized with 50% bleach (3% sodium hypochlorite) (CLOROX, Oakland, CA, USA) for 5–10 min, rinsed with sterilized ddH2O three times, followed by a cold-treatment at 4°C for 3 days. The T 0 seeds from the floral-dip transformation of the wild-type Col-0 plants were screened for the transgenic events on Petri dish plates containing: 1/2 MS, B5 vitamins, 1.5% sucrose, 50 μg L−1 of kanamycin, and 0.8% agarose, pH 5.7–5.8. Fifteen-day-old kanamycin-resistant seedlings were transferred to soil and grown in a greenhouse under a 16 h light, 8 h dark cycle at 23°C.
The cell-division marker line BJ3 in the Arabidopsis Columbia ecotype (Col-0), which contains a pcyclinB1::cyclinB1-GFP construct, was provided by Dr. Peter Doerner (University of Edinburgh, UK). The construct represents a GFP translational fusion to an Arabidopsis mitotic cyclinB1 gene expressed under its own promoter. CyclinB1 is expressed in cells in the G2–M transition and can be used as a marker for cells entering or undergoing division (Colon-Carmona et al. 1999). Sterilized and cold-treated seeds, as described above were then sown and grown in Petri plates (radius 10 mm) containing control medium (1/2 MS + B5 vitamins + 1.5% sucrose + 0.8% agarose), or a treatment medium, i.e., the control medium plus 0.1 or 0.5 μM of a synthetic 12-amino-acid peptide derived from the CLE14 or CLE20 motif. The plates were incubated vertically in a growth chamber under 23 ± 1°C and a 16-h light cycle.
Analysis of root growth and cell division in the BJ3 seedlings
Root growth of the BJ3 seedlings was monitored by measuring the root length in mm, and by counting the numbers of cells undergoing cell division by examining GFP signals in the cells of the RAM using a fluorescence microscope.
Examination of GFP signals by fluorescence and confocal microscopy
Homozygous transgenic T3 plants which stably express the pCLE14::CLE14-GFP or the pCLE20::CLE20-GFP cassette were examined for GFP signals at different developmental stages using a Leica DM LB fluorescence microscope (Meyer Instruments, Inc., Houston, TX, USA), or a ZEISS LSM 510 confocal microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA), with excitation (nm)/emission (nm) light at 488/510. The same method was used to observe the GFP signals in the RAM of root tips of the BJ3 seedlings.
Histochemical GUS assays
GUS staining assays of the reporter marker lines were performed as described (Jefferson et al. 1987). The 7-day-old seedlings were incubated in the GUS staining buffer at 37°C for 2–6 h and subsequently fixed in 70% ethanol. Images were captured using a Leica DM LB compound microscope (Meyer Instruments, Inc.).
Results
Three-dimensional structures of the CLV components and interaction models of CLV complex
Using ab initio modeling in the I-TASSER algorithm, the 3D structural models of the CLV3 and CLE peptides, and the 29-amino-acid extra-cellular domain of CRN receptor-like kinase were obtained. The extra-cellular domain of the CLV1 or CLV2 receptor-like protein/kinase has also been predicted using homology threading simulation implemented in the I-TASSER algorithm. All of the structural models of the CLV components are statistically reliable with a score function (C score and TM score) significantly above the cutoff threshold (Table 1). C score, which is in the range [−5, 2], is a confidence score for estimating the quality of the predicted models by I-TASSER. A C score of a higher value indicates a model with a high confidence. TM score and RMSD measure how close the model is to the native structure, and both are estimated based on the C score. A TM score is in the range (0, 1]. A TM value >0.5 implies that the model is of correct topology. The score function cutoff is a C score of −1.5 and a TM score of 0.5. The false-positive and false-negative rates are 0.05 and 0.09, respectively, for a model with the score function at the cutoff (Zhang and Skolnick 2004).
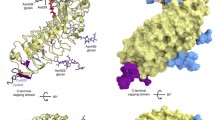
The 3D structure models show that the 12-amino-acid CLV3 peptide shares a roughly similar arch-shaped molecular structure with the CLE1, CLE2, and CLE6 peptides (Fig. 1a–d). CLE1 to CLE7 proteins, which belong to cluster group 2 (Oelkers et al. 2008), have been shown to cause a premature termination of the SAM when over-expressed in Arabidopsis (Strabala et al. 2006; Meng et al. 2010a). CLE1 and CLE6 can also provide a nearly full rescue in the SAM of clv3 mutant phenotypes (Ni and Clark 2006; Meng et al. 2010a). On the other hand, the 13-amino-acid CLV3 peptide has a much more curved molecular structure than that of the 12-amino-acid CLV3 peptide (compare Fig. 1e to a), and resembles those from the 12-amino-acid CLE14, CLE19, and CLE20 peptides (Fig. 1e–h). CLV3, CLE14, CLE19, and CLE20 have been found to trigger a short-root phenotype when over-expressed or when applied exogenously in the culture media in Arabidopsis (Fiers et al. 2005; Meng et al. 2010a). Analysis of the conserved residues at the molecular surfaces, using Consurf (http://consurf.tau.ac.il; Landau et al. 2005), reveals that the residues in the C-terminus of the CLE peptides are generally more conserved than those in the N-terminus, while the middle region of the CLE peptides located at the bottom of the peptide molecules is also relatively conserved, suggesting that the middle and C-terminal regions of CLE peptides may be important in performing receptor selection and binding of these peptide ligands (Fig. 1a–h, conservation of residues from variable to conserved are denoted by the color code from green to dark red).
Three-dimensional structural models of the CLE peptides and the ectodomains of CLV1/CLV2/CRN and the interaction models of these molecules. a–h 3D models of the CLE peptide ligands. a 12-amino-acid CLV3 (CLV3 12aa). 12-amino-acid CLE1 (b), CLE2 (c), and CLE6 (d). e 13-amino-acid CLV3 (CLV3 13aa). 12-amino-acid CLE14 (f), CLE19 (g), and CLE20 (h). i The 3D model of the 616-amino-acid extra-cellular LRR domain of CLV1 displayed in cartoon mode. j Superimposition of the 3D model of CLV1 (white) and the top three templates of CLV1: yellow (PDB ID code: 1ziwA), red (PDB ID code: 1o6sA), and green (PDB ID code: 1ogqA) using 3D-SS (http://cluster.physics.iisc.ernet.in/3dss/severalinput.html) (Russell et al. 1992). k The docking model for the homodimer of the 616-amino-acid extra-cellular LRR domain of CLV1. l The docking model showing the interaction between the homodimer of the 616-amino-acid extra-cellular LRR domain of CLV1 and the CLV3 peptides displayed in stick-and-ball mode. m–p The docking models of the CLV1 with the CLV3 peptides. m One 12-amino-acid CLV3 peptide (CLV3 12aa) binds to the deep binding cleft of CLV1 spanning from the concave to the convex sides. n One 13-amino-acid CLV3 peptide (CLV3 13aa) inserts into the deep binding cleft of CLV1 from the convex side of the CLV1 molecule. o One 12-amino-acid CLV3 peptide binds to and one 13-amino-acid CLV3 peptide insert into the deep binding cleft of CLV1, respectively. p Multiple 12-/13-amino-acid CLV3 peptides bind to the clefts of CLV1. q 3D model for the homodimer of the 29-amino-acid ectodomain of CRN receptor-like kinase displayed in cartoon mode and coated with solvent surface (1.4 Angstrom probe, rendered as discrete dots). r 3D structure of the 659-amino-acid ectodomain of CLV2 and its docking model with the 29-amino-acid ectodomain of CRN receptor-like kinase. s Superimposition of 3D model of CLV2 (yellow) and the top three templates of CLV2: white (PDB ID code: 2z64), red (PDB ID code: 1ziwA), and green (PDB ID code: 1ogqA) using 3-dimensional structural superposition (3D-SS). t Tetramer model of CLV2 (two) and CRN (two) interacting complex in their ectodomain regions. u The docking models of the CLV2 with CRN and the CLV3. v The docking models of the CLV2 with the CRN and CLV1. w–y The docking models of the CLV2 with CRN and the CLE14 (w)/CLE19 (x)/CLE20 (y) peptides. Conservation of residues from variable to conserved was analyzed using Consurf (http://consurf.tau.ac.il/overview.html) (Landau et al. 2005), and are denoted by the color code from green to dark red (a–h, i, m–p, r). The yellow arrows point to the binding clefts of the CLV1 (i) and also point to the binding clefts of the CLV2 (r); The black arrows point to the 12-amino-acid CLV3 peptide (CLV3 12aa) (l–p, u), and also point to 12-amino-acid CLE14/CLE19/CLE20 (w–y); the blue arrows point to the 13-amino-acid CLV3 peptide (CLV3 13aa) (l–p, u); the white arrows point to the 29-amino-acid ectodomain of CRN kinase (r, t–y). All atoms are shown as 100% of the van der Waals when displayed in stick-and-ball mode (a–h, k–p, t–y). All the atoms from N-terminus to C-terminus are shown color coded from cold color [blue or green (k–l); blue (q, t–y)] to hot color [green or red (k–l); red (q, t–y)]
On the other hand, both the CLV1 and CLV2 have a horseshoe-shaped molecular structure with an interior, parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvents and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein’s hydrophobic core and is sterically tightly packed with leucine residues (Fig. 1i, r). Moreover, the 3D models show that there is a deep cleft and two small clefts at the middle and the N-terminal regions, respectively, of the ectodomain of CLV1 molecule (Fig. 1i, yellow arrows), whereas the CLV2 has two clefts located at the regions near both the C- and N-terminus of CLV2 (Fig. 1r, yellow arrows). These clefts in the extra-cellular domain of CLV1/CLV2 may facilitate its peptide binding.
The docking modeling demonstrates that the extra-cellular receptor domain of CLV1 is able to form a homodimer (Fig. 1k), and that the CLV1 homodimers dock with both 12- and 13-amino-acid CLV3 peptides (Fig. 1l). At a finer level of resolution, the docking models reveal that the 12-amino-acid CLV3 peptide fits into the binding clefts of CLV1, spanning from the convex to concave sides (Fig. 1m, o, black arrows), while the 13-amino-acid CLV3 peptide inserts into the deep cleft of the CLV1 from the convex side of the molecule (Fig. 1n, o, blue arrows). Moreover, the docking models show that the ectodomain of one CLV1 molecule can dock with multiple CLV3 peptides (Fig. 1p). On the other hand, the 29-amino-acid ectodomain of CRN has a helix-loop-helix 3D structure and is able to form homodimers (Fig. 1q). The docking models also demonstrate that the ectodomain region of the CRN binds tightly to the C-terminus of the extra-cellular receptor region of CLV2 (Fig. 1r), and that two of each CLV2 and CRN ectodomains can interact to form a tetramer receptor complex (Fig. 1t), which can dock with 12- or 13-amino-acid CLV3 peptide (Fig. 1u). Additionally, by binding to CLV2 the ectodomain of CRN appears to facilitate the interaction of CLV2 and CLV1 (Fig. 1v). Finally, the docking models suggest potential interactions between the 12-amino-acid CLE14/CLE19/CLE20 peptides and the CLV2-CRN heterodimer or heterotetramer complex (Fig. 1w–y). The synthetic CLE1 to CLE7 peptides do not affect root growth when used at a low concentration (Ito et al. 2006; Kinoshita et al. 2007), but at a high concentration, e.g., ≥10 μM, these CLE peptides are able to inhibit root growth (Whitford et al. 2008). Therefore, this suggests that the CLE1-to-CLE7 peptides may also be able to bind to the receptor(s) in the root, e.g., CLV2, although they are not the optimal ligands for the root receptor(s). The average global binding free energies from the ten best solutions of the docking models, however, suggest that the CLV2-CRN hetero-receptor does not favor a binding with the CLE1/CLE2/CLE6. The orders of the average global binding free energies for an interaction between the CLV2-CRN hetero-receptor, and with different CLE peptides are: CLV3-13 aa (−33.889) < CLE19 (−25.531) < CLV3-12 aa (−24.08) < CLE14 (−12.413) < CLE20 (−11.204) < CLE2 (−7.145) < CLE1 (3.77) < CLE6 (15.863). Additionally, the 12-amino-acid CLE14/CLE19/CLE20 peptides appear able to bind tightly to the CLV2-CRN heterotetramer complex (Fig. 1w–y).
CLE14 and CLE20 express in specific cells of roots
Using promoter and GFP translational fusion reporter constructs we show that CLE14 and CLE20 express in specific cells of the root. CLE14 expresses specifically in the epidermis of roots, including the root cap and root hairs (Fig. 2a–c), On the other hand, CLE20 expression is found initially at the sites of lateral root initiation (Fig. 2d), extending into the protoxylem of the vascular tissues of the mature primary roots (Fig. 2e–g), and into the meristem region of the young lateral roots (Fig. 2h).
CLE14 (a–c) and CLE20 (d–h) express in specific cells of roots in Arabidopsis plants analyzed by translational fusion of GFP to the CLE gene, driven by the CLE promoter. CLE14 expresses in the root epidermal and cap cells (a, b), or in root hairs in mature regions of the root (c) of a 7-day-old transgenic seedling. d GFP signals from the lateral root initiation sites of 7-day-old transgenic pCLE20::CLE20-GFP seedlings. e GFP signals from the protoxylem of the vascular tissues of the mature primary roots of a 10-day-old transgenic seedling. f, g Cross-sections of a primary root of 10-day-old transgenic pCLE20::CLE20-GFP seedlings. h A young lateral root of 10-day-old transgenic pCLE20::CLE20-GFP seedlings. b, e, f, h Viewed only for GFP. g Differential interference contrast (DIC) bright field image. a, c, d Overlay of DIC and GFP images. Arrows point to epidermis (b), a root hair (c), a lateral root initiation site (d), protoxylem of the vascular tissues of the mature roots (e–g), and the RAM in a young lateral root (h). Bar = 1 mm (h). Bar = 50 μm for all others
CLE14 and CLE20 peptides irreversibly inhibit root growth by reducing cell division rates in the RAM
The cell division marker line BJ3 harbors a translational fusion of GFP to the cyclinB1 gene driven by the cyclinB1 promoter (pCyclinB1::CyclinB1-GFP) construct in Arabidopsis Columbia ecotype Col-0 (personal communication). The GFP fusion gene is expressed upon entry into G2, which allows one to visually monitor the progression of the cell cycle in root meristems (Colon-Carmona et al. 1999).
Root growth and mitotic activities in the RAM of the BJ3 seedlings, as assessed by the root length (Fig. 3a–b) and the number of cells undergoing cell division (Fig. 3c–d), respectively, were analyzed at different ages in roots treated with the CLE14/CLE20 peptide. Cells undergoing cell division were identified by observing GFP signals using a fluorescence microscope (Fig. 3d). Data were collected over 5 days of observation and measurement of BJ3 seedlings grown in the control or plus peptide-treatment media, beginning at day 2 and ending at day 7 after planting (indicated as “DPG”: day post germination, Fig. 3a–d). The results show that exogenous application of 0.5 μM of either the synthetic 12-amino-acid CLE14 or CLE20 motif peptide inhibits root growth as characterized by a strong reduction in the root lengths of the BJ3 seedlings (Fig. 3a–b). Moreover, the numbers of cells undergoing cell division in the RAM of the BJ3 seedlings also dramatically decreased in comparison to seedlings grown on control medium (Fig. 3c–d).
Exogenous application of the CLE14 or CLE20 peptide irreversibly inhibits root growth by reducing cell division rates in the RAM, as analyzed using the cell division marker line BJ3. Root length (a) or the numbers of the cells undergoing cell division (c) in 2- to 7-day-old BJ3 seedlings grown in the control medium (MS in red), or in the MS medium containing 0.5 μM CLE14 (in blue) or the CLE20 peptide (in green) (indicated as DPG, day post germination, i.e., days after sown in medium). b 7-day-old seedlings grown in the control medium (MS) or in the MS medium containing 0.5 μM of the CLE14 or the CLE20 peptide. d Root tips of 7-day-old BJ3 seedlings showing the cells undergoing cell division, which is visualized by the GFP signals from the pCyclinB1::CyclinB1-GFP construct in the BJ3 marker line. Root length (e–i) and the number of the cells undergoing cell division (j–n) in the 7-day-old BJ3 seedlings, which have been treated with the CLE peptide for a period between 0.0 and 7.0 days at the age of 0.0 (e, j), 1.5 (f, k), 2.0 (g, l), 2.5 (h, m), or 3.0 DPG (i, n) followed by a recovery in the control medium for a period of time to reach a total 7 days growth time. Bar = 5 mm (b). Bar = 50 μm (d)
Additionally, part of our efforts to understand CLE peptide functions in roots involved a detailed analysis of the mitotic activities in the RAM of the 7-day-old BJ3 seedlings after varying days of peptide treatment, followed by recovery on control (no peptide-containing) media. To avoid potential carry-over effects, the lowest (0.1 μM) and second lowest (0.5 μM) effective concentrations of CLE14 or CLE20 were used in this study. Peptide treatments were begun 0.0, 1.5, 2.0, 2.5, and 3.0 days after sowing (DPG), with treatments lasting between 0.0 (control) to 7.0 days, followed by varying times of recovery on the control medium (MS) to reach a total 7 days growth time. Root length and number of the cells undergoing cell division in the RAM of these 7-day-old seedlings were then analyzed (Fig. 3e–n).
The results demonstrate that root growth (Fig. 3e–i) and mitotic activity in the RAM (Fig. 3j–n), as indicated by root length and the number of cells undergoing cell division, respectively, generally cannot be restored after removal of the CLE peptides. For example, root length and the numbers of cells undergoing cell-division in the RAM are significantly reduced in 7-day-old BJ3 seedlings which have been treated with the CLE peptide only, for 1 day at the age of either 2.0, 2.5, or 3.0 DPG followed by a recovery in the control medium for 4.0, 3.5, and 3.0 days, respectively, in comparison to controls (treatment time 0.0) (Fig. 3g–i, l–n). The numbers of cells undergoing cell division in the RAM of the BJ3 seedlings which are treated for 2.0 days at the age of 2.5 and 3.0 DPG, followed by a recovery of 2.5 and 2 days, respectively, are nearly the same as those treated for 4.5 and 4.0 day without recovery (Fig. 3m, n), suggesting a permanent loss of cell division capability in the RAM of these plants. Our results from this study thus show a progressive loss of cell division capability with prolonged peptide treatment time and, in general, the losses are permanent. BJ3 seedlings at different ages also appear to be differentially sensitive to the CLE peptide treatments. Seedlings at ages between 2.0 and 3.0 DPG are sensitive to the CLE treatment (Fig. 3e–n). [Note that the number of the cells undergoing cell division in the RAM of the seedlings grown in the control (MS) medium increases rapidly at an age between 2 and 3.5 DPG (Fig. 3b, in red).]
CLE14 and CLE20 peptides irreversibly inhibit cell division, but do not change QC identity in the RAM
Over-expression of CLE14 or CLE20 in Arabidopsis has been found to trigger early consumption of the RAM (Meng et al. 2010a). Additionally, over-expression of CLE14 and CLE20 in Arabidopsis wild-type Col plants resulted in a significantly earlier differentiation of root hairs and in vascular tissue extending into the region adjacent (proximal) to the QC, resulting in extremely short roots (Fig. 4a–c).
Over-expressing CLE14/CLE20 caused early cell differentiation (a–c), whereas exogenous treatment of CLE14/CLE20 peptide does not result in a loss of QC identity in the RAM of the 7-day-old Arabidopsis seedlings (d–l). a Non-transgenic wild-type Col-0 seedling (WT). b Transgenic seedling over-expressing CLE14 (CLE14-ox), and c, over-expressing CLE20 (CLE20-ox). d–f Root tips of QC25 enhancer trap line seedlings. g–i Root tips of seedlings harboring the pWOX5::GUS construct. j–l Root tips of seedlings expressing the construct of pDR5::GUS. d, g, and j Seedlings grown on control MS medium (MS). e, h, and k Seedlings grown in MS medium containing 0.5 μM CLE14 peptide (MS + CLE14). f, i, and l Seedlings grown on the MS medium containing 0.5 μM CLE20 peptide (MS + CLE20). Bar = 50 μm. Arrows (b, c) point to a root hair (b) or the vascular tissue (c), respectively
Three QC marker lines were used to evaluate QC identity in roots treated with the CLE peptides: QC25, an enhancer trap reporter for QC identity (Sabatini et al. 2003); WOX5, a WUS-related homeobox gene which expresses exclusively in the QC (Sarkar et al. 2007); and DR5, a highly active synthetic auxin response element (AuxRE) that expresses in response to auxin, with maximal expression at the distal region of the QC (Ulmasov et al. 1997). The results, however, show that the CLE peptide treatment does not eliminate the identity of the QC, as is evident by the expected expression patterns in the QC for each of these QC markers (Fig. 4d–l).
The short-root phenotype triggered by CLE14 and CLE20 peptide treatments is not rescued by exogenous application of IAA or 6-BA
Indole-3-acetic acid (IAA), the most common plant auxin, and the cytokinin, 6-BA, were employed to study the combined effects of plant hormones and CLE peptides on root growth and mitotic activity in the RAM. The results show that the short-root phenotype (Fig. 5a, c) and the loss of the mitotic activity in the RAM (Fig. 5b), persist in all the seedlings treated with the CLE peptides plus auxin or cytokinin at various concentrations. The short-root phenotype induced by the CLE14 and CLE20 peptide treatments cannot be overcome by exogenous applications of IAA or 6-BA (Fig. 5).
The short-root phenotype caused by treatments with either the CLE14 or CLE20 peptides in vitro cannot be rescued by combined application of various concentrations of the phytohormones, IAA or 6-BA. Root lengths in mm (a), and the numbers of the cells undergoing cell division in the RAM (b) of 7-day-old BJ3 seedlings grown in a control medium (MS), or on medium containing 0.1/0.5 μM of CLE14 or CLE20, plus a specified concentration of IAA or 6-BA. c Morphological images of the 7-day-old BJ3 seedlings grown on a control medium (MS), or on a medium containing 0.5 μM of CLE14 or CLE20, plus a specified concentration of IAA or 6-BA. The concentrations of IAA and 6-BA are in nanomoles (nM) (a–c). Bar = 5 mm (c)
Cytokinin 6-BA can partially rescue the short-root phenotypes caused by over-expression of CLE14 or CLE20 in Arabidopsis plants
Although the cytokinin 6-BA does not rescue the short-root phenotype induced by the exogenous application of CLE peptides, 6-BA partially releases the short-root phenotype caused by over-expression of CLE14 or CLE20 in planta (Fig. 6). Twenty-day-old seedlings that over-express CLE14 or CLE20 have a much longer root when grown in a medium containing 0.1 or 0.5 μM of 6-BA compared to controls on medium lacking 6-BA (Fig. 6a, b). On the other hand, plants over-expressing CLE14 and CLE20 also appear to be less sensitive to the 6-BA. Whereas treatments of 6-BA restore root growth in plants over-expressing CLE14 or CLE20, treatments of 0.1 or 0.5 μM of 6-BA significantly inhibits root growth in plants over-expressing CLE1 (Fig. 6c), and in wild-type Arabidopsis plants (Fig. 6d). [Note that over-expression of CLE1 does not affect root growth (Fig. 6c, top panel with no 6-BA) (Meng et al. 2010a).] These results will be considered subsequently in regard to the view that cytokinin is believed to promote cell differentiation in the RAM, thus suppressing root growth, leading to a short-root phenotype (Dello Ioio et al. 2007).
Discussion
The potential binding clefts of CLV1 dock with both the 12- and 13-amino-acid CLV3 peptides
The molecular nature of the mature CLV3 peptide is controversial. By in situ matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analyses, the endogenous, mature CLV3 peptide was first reported as a 12-amino-acid dodecapeptide, in which two of three proline residues were modified to hydroxyproline (RTVPhSGPhDPLHH) (Kondo et al. 2006). However, using Nano-LC-MS and Nano-LC-MS/MS analysis of the apoplastic CLV3 peptide which accumulated in the medium, Ohyama et al. (2009) recently showed that the mature CLV3 peptide was in fact a 13-amino-acid arabinosylated glycopeptide. An additional histidine position at the C-terminal, immediately outside CLE motif (H13), is included in the 13-amino-acid CLV3 peptide, and Hyp7 (hydroxyproline at position 7) is post-translationally modified with linear β-1,2-linked tri-arabinoside to result in the peptide, RTVPSGPh-tri-l-arabinoseDPLHHH. A direct in vitro interaction of the 12-/13-amino-acid CLV3 peptide and CLV1 has been reported (Ogawa et al. 2008; Ohyama et al. 2009), and the 13-amino-acid arabinosylated CLV3 peptide has been reported to have a higher binding affinity to CLV1 compared to the 12-amino-acid CLV3 peptide (Ohyama et al. 2009). The 12-amino-acid CLE2 and CLE9 peptides have also been found able to interact with CLV1 (Ogawa et al. 2008; Ohyama et al. 2009). However, to date there is no information available on the molecular structures and on the interaction site(s) at which the binding of CLV1 and CLV3 actually occurs.
To further our understanding of the interacting nature of the CLAVATA (CLV) complex, we predicted 3D structures and interaction models of the 12-/13-amino-acid CLV3 peptides and the extra-cellular LRR domains of its receptors, CLV1 and CLV2, using computational approaches. Our results show that there is a single deep and two small binding clefts in the regions near the middle and the N-terminus of the ectodomain, respectively, of the CLV1 molecule (Fig. 1i, yellow arrows). These potential binding clefts of the CLV1 protein appear able to dock with both of the 12- and 13-amino-acid CLV3 peptides (Fig. 1l–o). Our results also show that the ectodomain of one CLV1 molecule may dock with multiple CLV3 peptides (Fig. 1p). CLV1 can self-interact forming a homodimer (Fig. 1k), which can further dock with both 12- and 13-amino-acid CLV3 peptides (Fig. 1l). This result is in an agreement with that reported by Bleckmann et al. (2010) in which the CLV1 is able to form homodimers independently of CLV3 (Bleckmann et al. 2010).
Based on our dock modeling results, and the biochemical evidence from Kondo et al. (2006), Ogawa et al. (2008), and Ohyama et al. (2009) for CLV1 and CLV3 molecules, we propose that the endogenous, mature CLV3 peptide may exist in planta as both a 12- and 13-amino-acid peptide, and that both the 12- and 13-amino-acid CLV3 peptides may be able to interact with a specific binding cleft in the ectodomain of CLV1. It is, however, worth noting that the majority CLE members do not have an additional residue outside the CLE motif. Their protein sequences terminate at the last residue (H/N) of the C-terminal CLE motif. Although some CLE members do have a short extension at the C-terminus of the CLE motif, as in CLV3, the immediately adjacent residue to the C-terminal CLE motif is highly variable. Therefore, it is reasonable to believe that the endogenous, mature CLE peptides are, in general, 12-amino-acid peptides.
Dock modeling suggests a physical interaction among CLAVATA2, CORYNE, and CLV3/CLE14/CLE20
Over-expression of CLE14, CLE19, or CLE20 has been shown to cause strong short-root phenotypes in a CLV1-independent, but CLV2-dependent manner in Arabidopsis (Meng et al. 2010a; Fiers et al. 2005). It has also been suggested that many of the extra-cellular domains derived from the CLV1-like LRR-RKs may be able to interact with a specific CLE peptide in vitro (Ogawa et al. 2008). In addition, genetic and biochemical evidence has suggested that the receptor-like kinase, CRN, may be involved in the CLV signaling pathway and may interact with CLV2. CRN was first isolated as a suppressor of the short-root phenotype resulting from over-expression of CLE19 (Müller et al. 2008; Miwa et al. 2008). The crn/clv2 double mutant shows a similar phenotype to that from each of the single mutants, whereas the crn/clv1 double mutant has more severe phenotypes than that from either of the single mutants. These results imply that CRN and CLV2 may act in the same pathway, whereas the CRN and CLV1 might act in parallel (Müller et al. 2008). Moreover, both sol2 and clv2 single mutants show a similar resistance to 26 different synthetic 12-amino-acid CLE peptides, including CLE14 and CLE20 peptides in roots, thereby suggesting that CRN and CLV2 are required for the CLE peptide functions, and that CRN may interact with CLV2 to regulate the RAM activity (Miwa et al. 2008; Müller et al. 2008). Finally, using firefly luciferase complementation imaging (LCI) and co-immunoprecipitation assays, Zhu et al. (2010) demonstrated that CLV2 did not interact with CLV1, but directly interacted with CRN in the absence of CLV3, whereas CLV1 could interact with the heterodimer of CLV2 and CRN (Zhu et al. 2010). Bleckmann et al. (2010) have convincingly demonstrated that CLV1 did not interact with CLV2, but interacted with the CRN/CLV2 heteromer. Both CRN and CLV1 could form a homodimer, and their self-interactions are CLV3 independent (Bleckmann et al. 2010). Bleckmann et al. (2010) further proposed that CLV1 homodimers and CLV2/CRN heterotetramers coexist at the plasma membrane of the stem cells in the SAM.
In contrast to CLV2, which encodes a LRR receptor protein lacking a functional kinase domain, CRN, a receptor-like kinase, has only a 29-amino-acid ectodomain. It is thus unlikely that either CLV2 or CRN, alone, can mediate cellular signal transduction, but together they may form a functional receptor kinase. Our docking models show that the ectodomain of CRN binds tightly to the C-terminus of the extra-cellular receptor region of CLV2 (Fig. 1r, white arrows). Moreover, the binding of the ectodomain of CRN with CLV2 appears to increase the possibility of interaction between CLV2 and CLV1 (Fig. 1v, white arrows). The ectodomain of CNR appears to fill the gap and support the interaction between CLV1 and CLV2 (Fig. 1v). Furthermore, our results also show that the potential heterotetramer complex of CLV2-CRN is able to dock with the 12- or 13-amino-acid CLV3 peptide (Fig. 1u), and also dock with the 12-amino-acid CLE14/CLE19/CLE20 peptides (Fig. 1w–y), suggesting a potential interaction between CLV2 and CRN, and these CLE peptides.
Endogenous CLE14 and CLE20 may regulate cell differentiation outside the meristem niche
CLE peptides have been shown to negatively regulate stem cell fates in the SAM and/or RAM (Strabala et al. 2006; Meng et al. 2010a). In contrast to CLV3 that expresses in the stem cells of the SAM, CLE40 expresses in differentiated cells and promotes differentiation of stem cells in the RAM by restricting WOX5 expression (Stahl et al. 2009). Using a GFP translational fusion reporter system we show that CLE14 and CLE20 express specifically in differentiating cells of roots. For example, CLE14 was found to express in the epidermis of roots, including the root cap and root hairs (Fig. 2a–c), whereas CLE20 expression is found in the protoxylem of the mature primary roots (Fig. 2e–g). Consistently, early differentiation of the root hairs and vascular tissue has been observed in Arabidopsis plants over-expressing CLE14 and CLE20, respectively (Fig. 4b, c). These results imply that CLE14 and CLE20 may promote cell (e.g., epidermis and vascular tissue) differentiation beyond the RAM in roots.
CLE14 and CLE20 peptides irreversibly inhibit root growth by reducing cell division rates, but do not change QC identity in the RAM
Our results demonstrate that the short-root phenotype caused by CLE14/CLE20 peptide treatment is due to a remarkable reduction in the cell division rate, but not to a reduction in cell elongation in the RAM of Arabidopsis roots (Fig. 3a–d). Fully differentiated plant cells generally cannot divide or produce cells of a different type unless they are caused to de-differentiate by specific, and strong stimuli, e.g., by an induction with proper concentrations of plant hormones (auxin and cytokinin). The identity and activity of the QC are crucial for the RAM maintenance, and the capability of a cell in the RAM to divide and to differentiate appears to depend on its distance from the QC (van den Berg et al. 1997; Sabatini et al. 2003). A failure in maintenance of the RAM activities and root growth therefore could be due to the loss of stem cell niche specification, which may be caused by the loss of the identities of, and/or activities in the QC (van den Berg et al. 1997; Sabatini et al. 2003), or only due to the temporal suppression of cell division in the RAM. In the first case, the loss of the RAM activity can be permanent, while, in the second case the RAM activity may be restored upon removal of the inhibition factors. It might be suggested that mitotic activity of the RAM should be able to resume following removal of the CLE peptides, assuming the specification of the QC remains. However, our results demonstrate that the growth and mitotic activity of the RAM generally cannot be restored after removal of the CLE peptides (Fig. 3e–n), suggesting a permanent loss of cell division capability in the RAM of these plants.
A loss of stem cell niche specification may be a consequence of a loss of either QC identity or a lack of the QC activity (van den Berg et al. 1997; Sabatini et al. 2003). In the first case, a loss of the expression of QC markers would be expected. However, the results from using three QC markers: QC25 (Sabatini et al. 2003), WOX5 (Sarkar et al. 2007), and DR5 (Ulmasov et al. 1997), show that CLE peptide treatment does not result in a loss of the QC identity in these Arabidopsis marker lines (Fig. 4d–l). These results would suggest that the permanent loss of meristem activity in the RAM induced by the CLE peptide treatment is not due to the loss of QC identity, but could be caused by lack of some other QC-related activity in the RAM.
Cytokinin may influence CLE14/CLE20 functions by affecting the processing of CLE peptides in vivo, resulting in a change in the availability and/or abundance of CLE14/CLE20 peptides in planta
Auxin participates in a range of developmental processes, including cell-fate specification, cell division, cell expansion, and initiation of root growth. Cytokinin is implicated in the control of cell division in the SAM, but promotes cell differentiation in the RAM (Meng et al. 2010c). Our results reveal that the inhibitive effects of the mature CLE peptide treatments on the roots are likely epistatic, or independent of auxin or cytokinin. The short-root phenotype induced by the exogenous application of the CLE14 or CLE20 peptide cannot be overcome by IAA or 6-BA treatments (Fig. 5). Although the cytokinin 6-BA does not rescue the root phenotype that is induced by exogenous application of CLE peptides, 6-BA partially releases the short-root phenotype caused by over-expression of CLE14 or CLE20 in planta (Fig. 6). These results therefore suggest that this rescue by cytokinin can only overcome the CLE14/CLE20 inhibitory effects that occur endogenously in plants. The results thus imply that cytokinin may influence CLE14/CLE20 functions by affecting the processing of CLE peptides in vivo, resulting in a change in the availability and/or abundance of CLE14/CLE20 peptides. Moreover, because plants over-expressing CLE14/CLE20 are resistant to 6-BA, with no additional inhibition to root length, this suggests that in these over-expressing lines that cytokinin is unable to promote cell differentiation in the RAM. This contrasts with the described short-root type phenotype reported for cytokinin-treated roots (Dello Ioio et al. 2007), and with our results showing that 0.1 or 0.5 μM of 6-BA significantly inhibits root growth in the plants over-expressing CLE1 (Fig. 6c), or in the wild-type Arabidopsis plants (Fig. 6d). [Note that over-expression of CLE1 does not affect root growth (Fig. 6c, top panel with no 6-BA; Meng et al. 2010a).] These results thus reveal an antagonistic relationship between CLE proteins and phytohormone cytokinin in vivo.
In summary, our 3D structure prediction and dock modeling results demonstrate that the mature CLV3 peptide may function as both 12- and 13-amino-acid peptides, and that both CLV3 peptides could interact with the homodimer of CLV1 or with the heterodimer/heterotetramer CLV2-CRN complex. Our results also suggest that 12-amino-acid CLE14/CLE20 peptides may function through the potential heterodimer/heterotetramer CLV2-CRN complex. Furthermore, we show that both CLE14 and CLE20 peptides inhibit, irreversibly, root growth by reducing cell division rates in the RAM, and we suggest that CLE14/CLE20 peptides may promote cell differentiation in roots outside of the RAM. The availabilities and/or abundances of CLE14/CLE20 may be affected post-translationally through the regulation of the processing of CLE peptides, and this may involve cytokinin activity in planta. Our results provide new evidence and insights towards understanding the molecular basis of the CLE peptide-signaling pathway in plant meristem activity and, more broadly, in Arabidopsis development.
Abbreviations
- ESR:
-
Embryo surrounding region
- WUS:
-
WUSCHEL
- CLV:
-
CLAVATA
- CLE:
-
CLAVATA3/ESR-related
- SAM:
-
Shoot apical meristem
- OC:
-
Organizing center
- CZ:
-
Central zone
- PZ:
-
Peripheral zone
- RAM:
-
Root apical meristem
- QC:
-
Quiescent center
- LRR:
-
Leucine-rich repeat
- LRR-RK:
-
Leucine-rich repeat receptor kinase
References
Bleckmann A, Peters SW, Seidel CAM, Simon R (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 152:166–176
Brand U, Hobe M, Simon R (2001) Functional domains in plant shoot meristems. Bioessays 23:134–141
Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 8:575–585
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cockcroft CE, den Boer BGW, Healy JMS, Murray JAH (2000) Cyclin D control of growth rate in plants. Nature 405:575–579
Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20:503–508
Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17:678–682
Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17:2542–2553
Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283:1911–1914
Geier F, Lohmann JU, Gerstung M, Maier AT, Timmer J, Fleck C (2008) A quantitative and dynamic model for plant stem cell regulation. PLoS One 3(10):e3553
Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9877
Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313:842–845
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Kinoshita A, Nakamura Y, Sasaki E, Kyozuka J, Fukuda H, Sawa S (2007) Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 48:1821–1825
Kobe B, Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 11:725–732
Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313:845–848
Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N (2005) ConSurf: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33 (Web Server issue):W299–W302
Laux T, Mayer KF, Berger J, Jurgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122:87–96
Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ (2008) FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res 36:229–232
Meng L, Ruth KC, Fletcher JC, Feldman L (2010a) The roles of different CLE domains in Arabidopsis CLE polypeptide activity and functional specificity. Mol Plant. doi:10.1093/mp/ssq021
Meng L, Wong J, Feldman L, Lemaux PG, Buchanan BB (2010b) A membrane-associated thioredoxin required for plant growth moves from cell-to-cell suggestive of a role in intercellular communication. Proc Natl Acad Sci USA 107:3900–3905
Meng L, Zhang S, Lemaux PG (2010c) Toward molecular understanding of in vitro and in planta shoot organogenesis. Crit Rev Plant Sci 29:108–122
Miwa H, Betsuyaku S, Iwamoto K, Kinoshita A, Fukuda H, Sawa S (2008) The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol 49:1752–1757
Müller R, Borghi L, Kwiatkowska D, Laufs P, Simon R (2006) Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18:1188–1198
Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20:934–946
Ni J, Clark SE (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140:726–733
Nielsen H, Krogh A (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. In: Proceedings of the sixth international conference on intelligent systems for molecular biology (ISMB 6). AAAI Press, Menlo Park, California, pp 122–130
Oelkers K, Goffard N, Weiller GF, Gresshoff PM, Mathesius U, Frickey T (2008) Bioinformatic analysis of the CLE signalling peptide family. BMC Plant Biol 8:1
Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319:294
Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5:578–580
Reddy GV, Meyerowitz EM (2005) Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310:663–667
Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM (2004) Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development 131:4225–4237
Russell RB, Breed J, Barton GJ (1992) Conservation analysis and secondary structure prediction of the SH2 family of phosphotyrosine binding domains. FEBS Lett 304:5–20
Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17:354–358
Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446:811–814
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl Acids Res 33:W363–W367
Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100:635–644
Sharma VK, Ramirez J, Fletcher JC (2003) The Arabidopsis CLV3-like (CLE) genes are expressed in diverse tissues and encode secreted proteins. Plant Mol Biol 51:415–425
Stahl Y, Wink RH, Ingram GC, Simon RA (2009) Signaling module controlling the stem cell niche in arabidopsis root meristems. Curr Biol 19:909–914
Strabala TJ, O’donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N (2006) Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol 140:1331–1344
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/lAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971
van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B (1995) Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378:62–65
van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390:287–289
Weigel D, Jürgens G (2002) Stem cells that make stems. Nature 415:751–754
Whitford R, Fernandez A, De Groodt R, Ortega E, Hilson P (2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc Natl Acad Sci USA 105:18625–18630
Williams L, Fletcher JC (2005) Stem cell regulation in the Arabidopsis shoot apical meristem. Curr Opin Plant Biol 8:582–586
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinform 9:40
Zhang Y, Skolnick J (2004) Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci USA 101:7594–7599
Zhu Y, Wang Y, Li R, Song X, Wang Q, Huang S, Jin JB, Liu CM, Lin J (2010) Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J 61:223–233
Acknowledgments
We thank Dr. David King for synthesis of the CLE peptides and Dr. Robert L. Fischer for thoughtful suggestions in the experimental design. We are grateful to Mengruo Yang for analyzing cell division and root growth. We thank Dr. Peter Doerner for providing the pcycB1::GFP and pcycB1::GUS transgenic lines. We thank Drs. S. Ruzin and D. Schichnes of the UCB Biological Imaging Facility for assistance. This research was supported by a National Science Foundation grant to Lewis J. Feldman and Jennifer C. Fletcher (Arabidopsis 2010 MCB-0313546). Ling Meng was partially supported by an award from the Agricultural and Environmental Chemistry Graduate Program at UC, Berkeley.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Meng, L., Feldman, L.J. CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232, 1061–1074 (2010). https://doi.org/10.1007/s00425-010-1236-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1236-4