Abstract
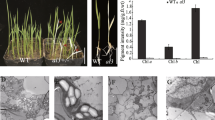
ClpP4 is a nuclear-encoded plastid protein that functions as a proteolytic subunit of the ATP-dependent Clp protease of higher plants. Given the lack of viable clpP4 knockout mutants, antisense clpP4 repression lines were prepared to study the functional importance of ClpP4 in Arabidopsis thaliana. Screening of transformants revealed viable lines with up to 90% loss of wild type levels of ClpP4 protein, while those with > 90% were severely bleached and strongly retarded in vegetative growth, failing to reach reproductive maturity. Of the viable antisense plants, repression of clpP4 expression produced a pleiotropic phenotype, of which slow growth and leaf variegation were most prominent. Chlorosis was most severe in younger leaves, with the affected regions localized around the mid-vein and exhibiting impaired chloroplast development and mesophyll cell differentiation. Chlorosis lessened during leaf expansion until all had regained the wild type appearance upon maturity. This change in phenotype correlated with the developmental expression of ClpP4 in the wild type, in which ClpP4 was less abundant in mature leaves due to post-transcriptional/translational regulation. Repression of ClpP4 caused a concomitant down-regulation of other nuclear-encoded ClpP paralogs in the antisense lines, but no change in other chloroplast-localized Clp proteins. Greening of the young chlorotic antisense plants upon maturation was accelerated by increased light, either by longer photoperiod or by higher growth irradiance; conditions that both raised levels of ClpP4 in wild type leaves. In contrast, shift to low growth irradiance decreased the relative amount of ClpP4 in wild type leaves, and caused newly developed leaves of fully greened antisense lines to regain the chlorotic phenotype.







Similar content being viewed by others
Abbreviations
- CaMV:
-
Cauliflower mosaic virus
- HSP:
-
Heat shock protein
- J PSII :
-
PSII electron transport
- Kan:
-
Kanamycin
- NPQ:
-
Nonphotochemical quenching
- q P :
-
Photochemical quenching
- ΦPSII :
-
Photosynthetic yield of PSII
References
Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K, Clarke AK (2001) Chloroplast and mitochondrial proteases in Arabidopsis: a proposed nomenclature. Plant Physiol 125:1912–1918
Adam Z, Zaltsman A, Sinvany-Villalobo G, Sakamoto W (2005) FtsH proteases in chloroplasts and cyanobacteria. Physiol Plant 123:386–390
Chen M, Jensen M, Rodermel S (1999) The yellow variegated mutant of Arabidopsis is plastid autonomous and delayed in chloroplast development. J Hered 90:207–214
Chen M, Choi Y, Voytas DF, Rodermel S (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J 22: 303–313
Clarke AK, MacDonald TM, Sjögren LLE (2005) The ATP-dependent Clp protease in chloroplasts of higher plants. Physiol Plant 123:406–412
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B (2002) AAA + proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett 529:6–10
Ganeteg U, Strand Å, Gustafsson P, Jansson S (2001) The properties of the chlorophyll a/b-binding proteins Lhca2 and Lhca3 studied in vivo using antisense inhibition. Plant Physiol 127:150–158
Grimaud R, Kessel M, Beuron F, Stevens AC (1998) Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem 273:12476–12481
Halperin T, Zheng B, Itzhaki H, Clarke AK, Adam Z (2001) Plant mitochondria contain proteolytic and regulatory subunits of the ATP-dependent Clp protease. Plant Mol Biol 45:461–468
Hendrickson L, Furbank RT Chow WS (2005) A simple alternative approach to assessing the fate of absorbed light energy using chlorophyll fluorescence. Photosynth Res 82:73–81
Huang C, Wang S, Chen L, Lemieux L, Otis C, Turmel M, Liu X-Q (1994) The Chlamydomonas chloroplast clpP gene contains translated large insertion sequences and is essential for cell growth. Mol Gen Genet 244:151–159
Huesgen PF, Schuhmann H, Adamska I (2005) The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol Plant 123:413–420
Kang SG, Maurizi MR, Thompson M, Mueser T, Ahvazi B (2004) Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP. J Struct Biol 148:338–352
Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425:86–89
Majeran W, Wollman FA, Vallon O (2000) Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12:137–149
Maurizi MR, Singh SK, Thompson MW, Kessel M, Ginsburg A (1998) Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and ClpP. Biochemistry 37:7778–7786
Moore T, Keegstra K (1993) Characterization of a cDNA clone encoding a chloroplast-targeted Clp homologue. Plant Mol Biol 21:525–537
Nakabayashi K, Ito M, Kiosue T, Shinozaki K, Watanabe A (1999) Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol 40:504–514
Nielsen E, Akita M, Davila-Aponte J, Keegstra K (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16:935–946
Ortega J, Lee HS, Maurizi MR, Steven AC (2002) Alternating translocation of protein substrates from both ends of ClpXP protease. EMBO J 21:4938–4949
Peltier JB, Ytterberg J, Liberles DA, Roepstorff P, van Wijk KJ (2001) Identification of a 350 kDa ClpP and protease complex with 10 different Clp isoforms in chloroplasts of Arabidopsis thaliana. J Biol Chem 276:16318–16327
Peltier J, Ripoll DR, Friso G, Rudella A, Cai Y, Ytterberg J, Giacomelli L, Pillardy P, van Wijk KJ (2004) Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J Biol Chem 279:4768–4781
Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975:384–394
Porankiewicz J, Clarke AK (1997) Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. Strain PCC 7942. J Bacteriol 179:5111–5117
Sakamoto W, Tamura T, Hanba-Tomita Y, Sodmergen, Murata M (2002) The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Gene Cell 7:769–780
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Press, Cold Spring Harbor
Schirmer EC, Glover JR, Singer MA, Lindquist S (1996) HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci 21:289–296
Shanklin J, Dewitt ND, Flanagan JM (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7:1713–1722
Shikanai T, Shimizu K, Ueda K, Nishimura Y, Kuroiwa T, Hashimoto T (2001) The chloroplast clpP gene encoding a proteolytic subunit of ATP-dependent protease and is indispensable for chloroplast development in tobacco. Plant Cell Physiol 42:264–273
Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR (2000) Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA 97:8898–8903
Sjögren LLE, MacDonald TM, Sutinen S, Clarke AK (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol 136:4114–4126
Soikkeli S (1980) Ultrastructure of the mesophyll in Scots pine and Norway spruce: seasonal variation and molarity of fixative buffer. Protoplasma 103:241–252
Sokolenko A (2005) SppA peptidases: family diversity from heterotrophic bacteria to photoautotrophic eukaryotes. Physiol Plant 123:391–398
Sokolenko A, Lerbs-Mache S, Altschmied L, Herrmann RG (1998) Clp protease complexes and their diversity in chloroplasts. Planta 207:286–295
Spremulli L (2000) Protein synthesis, assembly, and degradation. In: Buchanan B, Gruissem W, Jones R (eds) Biochemistry and biology of plants. American Society of Plant Physiologists, pp 412–456
Strand Å, Zrenner R, Trevanion S, Stitt M, Gustafsson P, Gardeström P (2000) Decreased expression of two key enzymes in the sucrose biosynthesis pathway, cytosolic fructose-1,6-bisphosphate and sucrose phosphate synthase, has remarkably different consequences for photosynthetic carbon metabolism in transgenic Arabidopsis thaliana. Plant J 23:759–770
Sutinen S (1987) Cytology of Norway spruce needles I. Changes during ageing. Eur J For Path 17:65–73
Vierstra RD (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32:275–302
Wang J, Hartling JA, Flanagan JM (1997) The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell 91:447–456
Weaver LM, Froehlich JE, Amasino RM (1999) Chloroplast-targeted ERD1 protein declines but its mRNA increases during senescence in Arabidopsis. Plant Physiol 119:1209–1216
Zheng B, Halperin T, Hruskova-Heidingsfeldova O, Adam Z, Clarke AK (2002) Characterization of chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol Plant 114:92–101
Acknowledgements
This work was supported by grants to A.K.C. from The Swedish Research Council for Environment, Agricultural Science and Spatial Planning (Formas) and the Swedish Foundation for International Cooperation in Research and Higher Education (STINT). T.M.M. would like to acknowledge NSERC for an oversea postgraduate scholarship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, B., MacDonald, T.M., Sutinen, S. et al. A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana . Planta 224, 1103–1115 (2006). https://doi.org/10.1007/s00425-006-0292-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0292-2