Abstract
Traditional histopathology, characterized by manual quantifications and assessments, faces challenges such as low-throughput and inter-observer variability that hinder the introduction of precision medicine in pathology diagnostics and research. The advent of digital pathology allowed the introduction of computational pathology, a discipline that leverages computational methods, especially based on deep learning (DL) techniques, to analyze histopathology specimens. A growing body of research shows impressive performances of DL-based models in pathology for a multitude of tasks, such as mutation prediction, large-scale pathomics analyses, or prognosis prediction. New approaches integrate multimodal data sources and increasingly rely on multi-purpose foundation models. This review provides an introductory overview of advancements in computational pathology and discusses their implications for the future of histopathology in research and diagnostics.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
The way histopathology analyses are carried out remained similar for close to two centuries. This classic way typically involves manually assessing specific patterns of injury (e.g., diffuse infiltration of neutrophilic granulocytes) or counting histological objects (e.g., mitoses). However, these manual quantifications and assessments can be cumbersome, low-throughput, error-prone, and subject to inter-observer variability—a hindrance to developing both diagnostics and research further.
The introduction of digital pathology is a transformative development for overcoming these challenges and developing pathology further towards precision medicine. Digital pathology has enabled the introduction of computational methods to pathology, a discipline termed computational pathology [1]. Broadly speaking, computational pathology encompasses computational methods used to analyze patient specimens for diseases. A main branch and topic of this review is computational histopathology that focuses on computational methods applied to digital histopathology images.
Benefits in computational histopathology mainly arise from using artificial intelligence (AI), especially deep learning (DL)–based techniques. DL is a subspecialty of machine learning (ML, a subspecialty of AI) that makes use of artificial neural networks (ANNs) [63]. ANNs, in short, are multilayer functions that progressively transform input data to produce a desired output. During training, the internal parameters of an ANN are automatically updated in an iterative process to produce outputs that are increasingly similar to the ground truth (i.e., the desired output) [63]. This is typically done using a training algorithm that takes actions to minimize a loss function. This paradigm allows training ANNs to map a multitude of input–output relationships, meaning ANNs can be trained to perform a task without being explicitly programmed for it. While that is exciting, it is important to keep in mind that DL model predictions are not of a causal nature, but are correlations based on the relations between input and output that were established in the DL model during training.
Typical applications of ANNs in pathology include classification (assigning a categorical label to data, e.g., hepatocellular carcinoma to a histopathology image) [23], segmentation (pixel-level classification; partitioning an image into discrete groups of pixels, typically corresponding to objects, e.g., glomeruli and tubules) [45], and regression (assigning a continuous value to an image, e.g., gene expression) [28].
Significant progress has been achieved using DL-based solutions in histopathology, e.g., inferring molecular alterations with high precision directly from whole slide images (WSI) [55], predicting the origin of cancers of unknown primary [68], predicting therapy response in colorectal cancer [33], or identifying morphometric biomarkers [3, 45].
In this review, we highlight a selection of studies applying deep learning to digital histopathology to achieve a variety of goals. We focus on major applications, i.e., histomorphometry, classification, and regression (Table 1 provides a selection of major studies on these applications in computational pathology).
Histomorphometry
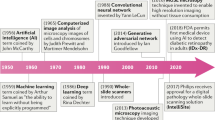
The hypothesis that form follows function has been extensively investigated in medical specialties such as anatomy or physiology. In principle, investigating the shape of a structure of interest can lead to a deeper understanding of its specific function. Tissue analysis based upon the form of tissue compartments or structures to derive novel insights into their functionality has been practiced for over 40 years [21]. However, these measurement and quantification tasks were mostly done manually, e.g., using digital planimeters [34] or drawing tubes mounted on microscopes [70]. Thus, these techniques were limited by their time-consumptive nature, technical capabilities, and inter-observer variability [22]. Computational pathology algorithms have evolved rapidly over the recent years, especially fueled by the digitalization of tissue slides (Fig. 1). Multiple commercial and open source software for image analysis have implemented histomorphometry workflows, e.g., MATLAB [99] or ImageJ—Fiji [25]. Whether these workflows are based on readily available plugins, self-coded macros, or in-built deep learning algorithms, they all perform the task of semantic segmentation to delineate structures of interest.
Digital pathology ecosystem encompassed in standard tissue analysis workflows. Regular tissue processing is followed by digitalization of tissue slides into whole slide images (WSIs). WSIs are the main data resource for digital pathology ecosystems in which they are stored, associated with other input data in the laboratory information system (LIS), and analyzed by machine or deep learning algorithms. Eventually, clinicians are provided with a bundle of extensive resources including the digitized tissue slide for informed decision making. ML, machine learning; DL, deep learning
Semantic segmentation
Histopathology segmentation tasks represent the precise delineation of complex tissue structures. Segmentation models produce pixel-level image masks for desired structures and compartments in WSIs. These masks allow for the extraction of hand-crafted features by further algorithms. Features represent different attributes of the structure of interest and can range from simple and explainable distance or area measures to complex readouts of texture (e.g., entropy, contrast, homogeneity) or image moments which range beyond the capabilities of the human eye. Extracted features can then be associated with clinical data such as laboratory values [67], be implemented in disease or outcome prediction models [117], and in downstream bioinformatics analyses [45] (Fig. 2). Feature importance for specific tasks can be computationally investigated, potentially allowing to identify novel associations between form and function.
Histomorphometry techniques for different organ systems including analysis applications. Convolutional neural networks (CNNs) are applied to image patches of organ histology for segmentation of regions of interest. These image masks are then used for calculation of morphometric features which can be implemented in downstream analyses. CNN, convolutional neural network
The quality and meaningfulness of features are inherently dependent on precise segmentation masks. To generate precise segmentation results, models need to learn based on a ground truth, i.e., the human prespecification on how structures of interest should be outlined, which often remains a laborious task. Therefore, an inherent challenge for segmentation in high-throughput settings, where manual control is not feasible, is continuous quality control of the segmentation output. Methods such as reverse classification accuracy [87] or anomaly detection [36] can help to automatically assess segmentation accuracy. As segmentation models require manual oversight as well as handcrafted annotations enhancing the learning process, they have to be adapted to each specific research setting. However, this also allows for considerable flexibility, as segmentation models can be applied to various resolutions and imaging modalities, such as bright-field, fluorescence, or electron microscopy. Depending on the respective task, this enables the segmentation of various levels of tissue architecture such as functional units, compartments, extracellular space, or nuclei.
Kidney
Kidney microanatomy consists of many diverse and complex structures such as glomeruli, different sections of tubules, the interstitium, and vessels. In recent years, considerable efforts have been made to accurately segment these structures in human and animal specimens for different histological stains, laying the groundwork for histomorphometry analysis [13, 44, 45, 50]. In diabetic nephropathy, one of the most prevalent causes of chronic kidney disease, segmentation models could accurately assess glomerulosclerosis as well as interstitial fibrosis and tubular atrophy (IFTA), which represent important hallmarks of chronic kidney disease [39]. The model’s quantification of IFTA reached high agreement with experienced renal pathologists (intra-class correlation coefficient of 0.94 when including the ANN as an observer) while being more time-efficient compared to human manual annotations.
Furthermore, detailed histomorphometry analysis can reveal alterations of tissue architecture that were not captured by functional and laboratory parameters. Klinkhammer et al. implemented two common murine chronic kidney disease models to analyze kidney disease recovery [59]. Interestingly, while kidney function measured by glomerular filtration rate normalized back to baseline 2 weeks after recovery, histomorphometry analysis of tubular architecture in more than half a million tubules revealed persisting dilation and atrophy of tubules leading to further nephron loss.
Liver
In liver histology, histomorphometry is mainly implemented for quantification of steatosis, i.e., fat accumulation in liver cells, and fibrosis. Both are important histopathological features of tissue remodeling pathways in steatohepatitis leading towards liver cirrhosis. Accurately quantifying liver steatosis and fibrosis allows for monitoring of disease progression and even therapy response for novel drugs. In a reanalysis of three randomized clinical trials including core-needle biopsy samples from 3662 patients with non-alcoholic steatohepatitis (NASH), Taylor-Weiner et al. developed a novel score summarizing fibrosis patterns on patient level termed the Deep Learning Treatment Assessment (DELTA) liver fibrosis score which accurately captures changes in tissue remodeling after anti-fibrotic therapy [105]. Broad implementation of the DELTA liver fibrosis score could facilitate standardized and reproducible assessment of treatment response in future trials regarding anti-fibrotic agents for liver disease. Liver steatosis quantification can also be implemented in liver transplant workflows, allowing for better organ allocation. Sun et al. developed a deep learning-based segmentation model for quantifying steatosis in frozen liver donor sections which could outperform the estimations of an on-service pathologist thus potentially leading to 9% fewer unnecessarily discarded liver transplants [103].
Heart and vasculature
Cardiomyocyte morphology is a key factor for understanding the pathophysiology of heart failure. Segmentation of cardiac myocytes in a murine model of transaortic constriction (TAC) as well as in autopsy samples of humans with aortic (valve) stenosis revealed similar increases in cardiomyocyte area and decreases of capillary contacts per area of cardiomyocytes signaling myocyte hypertrophy and microvascular rarefaction in pressure overload-induced heart failure [25]. Histomorphometry of cardiomyocyte hypertrophy on an electron microscopy scale in a guinea pig TAC model could also be linked to changes in intra- and transcellular electric conductance [35]. Fry et al. demonstrate how, although the topological arrangement of cells is retained in left ventricular hypertrophy, an increase in lateral cell-to-cell connections and intercalated disk space leads to altered three-dimensional cardiac action potential conduction velocity. Myocyte hypertrophy can not only lead to altered electrophysiology but also result in hemodynamic changes. In a multi-scale computational model of heart failure including cardiomyocyte histomorphometry, ventricular volume overload in pig hearts was directly associated with lengthening of cardiac myocytes leading to a decrease in measured ejection fraction during echocardiography [80].
Not only cardiac remodeling but also vascular remodeling in pulmonary hypertension is known to contribute to heart failure. Fayyaz et al. investigated vessel histomorphometry for pulmonary arteries, veins, and small indeterminate vessels in human autopsy or surgery specimens [31]. Interestingly, the severity of pulmonary hypertension and presence of heart failure were more determined by increases in intimal thickness of veins and small indeterminate vessels rather than the remodeling of arteries.
Soft tissue
Similar to the analysis of cardiac myocytes, skeletal muscle fibers can also be analyzed by histomorphometry allowing for characterization of different fiber types and muscle metabolism [99]. Quantifying the fiber size of human deltoid and pectoral muscle enables spatial analysis of location-specific muscle morphometry revealing different age-, sex-, and myopathy-related patterns of atrophy and hypertrophy [86].
In fatty tissue, segmentation of adipocytes has been widely established due to their rather simple shape and configuration. Osman et al. developed a MATLAB algorithm for high-throughput batch quantification of white fat morphology in high-fat diet-fed mice [77]. They precisely demonstrate how adipocytes significantly increase in size in high-fat diet-fed mice, although this increase is overestimated when using manual annotation methods. The authors propose that this is due to changes in adipocyte shape which cannot be accurately assessed by manual annotation methods.
Blood clots
In recent years, interest has sparked in analysis of cerebral blood clots retrieved during mechanical thrombectomy which can provide diagnostic insights into clot etiology as well as patient outcome [10]. This field can be enhanced by automated histomorphometric composition analysis. Automated quantitative analysis of the relative fractions of clot components (red or white blood cells, platelets, and fibrin) could differentiate between clot compositions of cardioembolic and noncardioembolic etiology in a retrospective study of 145 stroke patients. Further analysis of cryptogenic stroke thrombi revealed similar composition characteristics to cardioembolic thrombi, but distinct from noncardioembolic thrombi, supporting the hypothesis that many cryptogenic strokes are in fact cardioembolic [9].
Subcellular structures
Next to analysis of compartments or structures, histomorphometry can also expand to the level of subcellular organelles, mainly the segmentation and quantification of nuclei. Histomorphometry of nuclei is especially interesting for cancerous tissue. Nuclear features such as chromatin clumping have been identified as predictive of patient risk, overall survival, and recurrence in various cancer types such as breast [3, 113] or lung cancer [109].
Also considerable efforts have been made to enable segmentation of cellular organelles in electron microscopy. Heinrich et al. were able to segment over 30 different classes of cellular organelles, e.g., endoplasmic reticulum, microtubules, or ribosomes on nanometer level in reconstructed three-dimensional electron microscopy images [43]. This opens up a whole new resolution of histomorphometry analysis providing insights into spatial interactions and organelle architecture.
Pathomics
Due to the wide range of different features to characterize segmented tissue, feature extraction can generate large data frames containing thousands of segmented structures and all extracted features for each respective structure. These large data frames resemble datasets generated by molecular omics workflows, such as single cell transcriptomics and can be used for dimensionality reduction or trajectory inference analysis (Fig. 2). Hence, the term of pathomics has been proposed to characterize the data mining of histopathology images [14, 32, 42, 45]. Pathomics enables precise characterization of whole tissue architectures, but can also be applied to cancer specimens. Mining of pathomics data from cancer cells and their microenvironment allows for precise quantification of stromal, nuclear, or immune features thus enabling new prognostic biomarkers for breast [3] or colon cancer [32]. Pathomics is inherently spatial due to the traceable location of all segmented structures and could provide a missing link on how molecular processes influence tissue architecture and organ function [14].
Downstream analysis of datasets can be augmented by various readily available machine learning techniques. By linking pathomics with clinical and histopathological context, combined datasets can be applied to multivariate analyses for discovery of novel associations or developing for clinical practice. Established machine learning applications include support vector machines which find an optimal hyperplane that best captures the maximum differences of provided groups of interest. Depending on the dimensionality of the datasets, the hyperplane resembles a line (two-dimensional), a plane (three-dimensional), or more abstract geometric shapes (n-dimensional). Furthermore, tree-based models such as random forests, boosted trees, and classification and regression trees as well as more traditional statistical models such as linear, logistic, ridge, and lasso regression can be applied to pathomics data. For discovery of novel associations, k-means or hierarchical clustering algorithms can group statistically similar data points together and are already widely known in other omics fields. Pathomics has already been combined with other established omics such as genomics [3] or transcriptomics [104] in large-scale multi-omics studies and multiple applications and platforms exist for analyzing pathomics data [46, 82]. In the future, further bioinformatics methods have to be implemented to link multi-omics datasets to relevant patient-level outcomes [52].
Classification
Formulating a diagnosis in digital pathology can be framed as a classification task, i.e., assigning one or several categorical label(s) to WSIs. Consequently, many studies in computational histopathology have focused on end-to-end classification tasks [55, 68, 96]. In most studies, the ground truth used for training ANNs consists of WSI-label pairs, meaning that one or several WSI are associated with a categorical label, e.g., “Clear Cell Renal Cell Carcinoma” (Fig. 3). However, due to hardware constraints and data size, WSIs are typically tessellated into smaller image tiles. Because of that, not every image tile contains histological features that are diagnostic for the case label, e.g., when only a small part of the slide contains cancerous tissue.
Deep learning–based classification of histopathology and approaches to interpreting the basis for classification. A Whole slide images (WSIs) are tessellated into image tiles. In a multiple instance setting, typically, a pre-trained deep learning model is used as a feature extractor that transforms each image tile into a high level feature vector which is used by another model to formulate a prediction. B These predictions are usually opaque, but techniques exist to make them interpretable (explainable artificial intelligence (XAI)). Among the most popular XAI techniques in pathology are saliency maps, trust scores, prototype examples, and concept attribution. We thank Yu-Chia Lan, M.Sc., for providing a saliency map visualization for this figure
An often used approach to tackle this challenge is called multiple instance learning (MIL). In short, not every image tile in MIL inherits the case label, but the entire collection of tiles making up a WSI gets assigned a label. In a binary classification setup, this means that the entire collection of tiles is negative, if all tiles in the collection are negative, and positive, if at least one tile in the collection is positive [17]. This allows for efficient processing of WSIs in weakly supervised settings (e.g., one label per slide), which is beneficial for dataset generation, since weak labels are inherently easier to retrieve than strong labels. MIL and other classification techniques have been investigated for a multitude of pathology classification tasks in several organs. Although there are some exceptions [58, 93], most classification studies in computational pathology are concerned with cancer specimens. Due to the sheer number of studies published in the field, we had to limit ourselves to a selection of studies applying DL-based tools to major cancer types and pan-cancer approaches.
Deep learning–based classification of lung cancer
With an estimated 2.2 million new cases in 2020, carcinomas of the lung are among the most common types of cancer in both men and women and are the leading cause of cancer deaths [102]. Classification of histopathological images as normal or lung carcinoma (any type) was shown to be feasible using ANNs with AUROCs above 0.973 in a study using 4,704 WSI of lung histopathology [54]. This study investigated both a fully supervised and a weakly supervised approach, showing that the weakly supervised approach consistently showed better performance than the fully supervised approach [54]. Another study investigated prediction of mutations in the following genes: KRAS, TP53, EGFR, STK11, FAT1, and SETBP1 with AUROCs higher than 0.733 [23]. STK11 mutations could be predicted with the highest accuracy (AUROC 0.85). In addition, the authors investigated ANNs for distinguishing between lung squamous cell carcinomas and adenocarcinomas, a task for which they achieved high accuracy (AUROC of 0.97) [23]. Lung adenocarcinomas can present with different growth patterns. A lightweight (i.e., less complex) ANN was shown to be trainable to distinguish solid, cribriform, micropapillary, and acinar growth patterns of lung adenocarcinomas [37]. However, the primary use case of such a model would likely be delineating areas with different growth patterns, but the resolution of the classification heatmaps here is not on pixel level, but on tile level, which would lead to an inaccurate delineation, a limitation that could be circumvented using a segmentation model.
Deep learning–based classification of prostate cancer
With an estimated 1.41 million new cases in 2020, prostate cancer is one of the most common types of cancer worldwide and a significant burden to global health [118]. Diagnostic criteria of prostate carcinomas are well defined, and its grading scheme—the Gleason grades—is defined by tissue architecture [48]. The combination of case abundance and well-defined histopathology makes prostate cancer an ideal entity for developing computational pathology tools. Automated Gleason grading was demonstrated in a large study including 5759 biopsies from 1243 patients [16]. The model showed excellent agreement with the ground truth (quadratic Cohen’s Kappa of 0.918), indicating its usefulness for clinical diagnostics [16].
High accuracy of prostate cancer and perineural invasion detection was achieved in another study [78]. Importantly, this was the first study reporting identification of a prostate cancer case by a DL system that was missed by a pathologist [78]. In the large Prostate cANcer graDe Assessment (PANDA) challenge that included approximately 11,000 WSI, many teams achieved accuracies on par or better than pathologists (e.g., sensitivity for tumor detection of 99.7% with specificity of 92.9%), mostly using end-to-end techniques with case level information only [15]. Given these impressive performances, it is not surprising that there are several commercial DL-based tools to be used in prostate cancer histopathology diagnostics that were already shown to help increase sensitivity and specificity of diagnostics [84].
Deep learning–based classification of breast cancer
Breast cancer is the most common invasive cancer in women with approximately 2.3 million new cases in 2020 [5]. Although the primary goal in computational breast pathology has been to detect and outline tumors both in mammary tissue and lymph nodes [26], histopathology classification studies using DL-based tools were performed relatively early as well [4, 112].
Automatic classification of lobular versus ductal invasive carcinoma of the breast with an accuracy of 94% was achieved with a neural network on tissue microarray cores [24]. Similarly, another study achieved an area under the receiver operating characteristics curve (AUROC) of > 95% in independent cohorts for breast cancer subtype classification using ANNs on WSI [27].
DeepGrade, an ensemble of multiple neural networks, could be used to re-stratify breast cancer cases classified as G2 (Nottingham grading system), corresponding to intermediate differentiation [111]. The DeepGrade-based stratification (Grade 2 high/low) was an independent predictor of patient survival, indicating usefulness of such stratification [111].
Point mutations in RB1, CDH1, NOTCH2, and TP53 (AUROCs > 0.729) could be directly inferred from histology using a classification DL network [83]. Another study showed that using an ANN, a germline BRCA1/2 mutation could be predicted directly from digital histopathology [110]. Although these molecular predictors are of interest, the accuracy is not high enough to use them instead of molecular investigations.
Deep learning–based classification of colorectal cancer
Predicting microsatellite instability (MSI) from WSI of colorectal cancer specimens using DL was one of the early landmark studies in computational pathology [57]. In the meantime, this was reproduced multiple times using a variety of approaches. DL-based MSI prediction tools have reached clinical grade performance (sensitivity of 0.99 and negative predictive value of 0.99), allowing the use of such models as screening tools before performing molecular analyses [108].
In addition to deficiencies in mismatch repair genes, other mutations were also shown to be predictable from WSIs of colorectal cancer specimens, including APC, KRAS, PIK3CA, SMAD4, and TP53 [49]. However, the predictive performance in this study was not high enough (AUROC 0.693–0.809) to justify using the model in clinical or research practice for this specific task.
The consensus molecular subtype (CMS) of colorectal cancer is a gene expression–based subtyping system with biological interpretability and prognostic implications [41]. The CMS was predicted directly from WSIs with good accuracy (AUROC 0.84) using an ANN [97]. Cox-proportional hazards analyses revealed similar predictive performances for the molecular and image-based CMS classification [97]. Thus, such an image-based classification could help cut costs for molecular analyses while at the same time providing similar predictive performances.
A multimodal DL-based approach using different immunostains (CD4, CD8, CD20, CD68) was used to predict the relapse free survival status after 3 years in a multicenter cohort of colorectal cancer patients (n > 1000) with good accuracy [33]. Data from these stains were integrated into a score termed AImmunscore by the authors, which proved to be a strong and independent predictor of prognosis in colorectal cancer patients [33].
Multi-cancer classification studies
In addition to studies focusing on DL applications for one type of cancer, several studies investigated DL approaches for multiple cancer types. Cancers of unknown primary (CUP), i.e., manifestations of cancerous tissue the origin of which is not known, are a major diagnostic challenge. In a landmark study, Lu et al. developed a DL model to simultaneously predict whether a tumor is primary or metastatic and predict its site of origin [68]. The model reached a top-1 accuracy of 0.83 and a top-3 accuracy (meaning the correct class is in the three predictions with the highest probability score) of 0.96, which is excellent [68]. Such a model could potentially have a major assistive impact for pathologists. An in-depth analysis of the usefulness of predictions in a prospective setting when pathologists cannot guess the site of origin from morphology would be highly interesting.
In an elegant study, an ImageNet-pretrained Xception model that was fine-tuned on colorectal cancer histopathology was used to extract features from WSI tiles of lung adenocarcinomas, head and neck squamous cell carcinomas, and colorectal carcinomas [20]. The tiles were then classified into four groups of clusters using unsupervised clustering (K-means) on a subset and nearest neighbor analysis on the remaining tiles. Tiles from different clusters correspond to different tissue areas and, interestingly, had different predictive values for mutation prediction, allowing the authors to guess which tissue parts are most informative about mutations [20]. This allows for the development of a more interpretable approach to mutation prediction than classic end-to-end approaches.
Approaches to interpretability of end-to-end classification models in computational pathology
Predictions made by a DL-based system are typically opaque, i.e., the basis for the prediction is unclear to the user. The field of explainable AI (XAI) aims to develop methods that help interpret and consequently understand the basis of model predictions (for an in-depth review on XAI in pathology, see [81]). Major XAI approaches include saliency maps, trust scores, prototypes, and concept attribution (Fig. 3).
Saliency maps are typically displayed as overlays indicating an estimation of the relevance of an area in the image to the model prediction. A major computational approach for producing saliency maps are gradient weighted class activation maps (Grad-CAM) [91]. Using these heatmap-like visualizations, a user can evaluate whether highlighted areas include morphologically typical variations for the predicted class (Fig. 3). However, when a task is performed for which no typical morphological variations are described (e.g., predicting certain mutations), evaluation of saliency maps can be challenging. Indeed, evaluation of saliency maps is prone to several sources of bias, for example, positive confirmation bias of pathologist expectation [29].
Trust scores help evaluate how confident a model prediction is. Given high model accuracy, this can be helpful for clinical implementation, e.g., to design a strategy for triaging cases which potentially do not require review by a pathologist, or cases that require review or to assist physicians in weighing the model prediction in their decision-making. However, confidence scores can be high for wrong predictions, which limits their validity. This is impressively demonstrated using adversarial attacks [38], where sometimes subvisual changes introduced to an image completely change the predicted class.
Prototypes are representations of instances (e.g., an image tile) that are “archetypal,” i.e., highly representative for the model classes. Often, the most predictive tiles from the patients with the highest prediction scores are depicted (e.g., in [58, 74]). However, synthetic generation of prototypical instances potentially allows more flexible interpretation of prototypes [66].
Concept attribution methods aim to estimate the relevance of manually defined features or concepts for model prediction [72]. These can range from local instance features, such as the circularity of a nucleus to more broad and potentially less understandable features, such as the image texture. It is necessary to select these concepts up front, which is a significant source of bias. However, novel unsupervised methods based on latent space deconvolution might provide a less biased approach to finding relevant concepts [40], although to our knowledge and at the time of writing this review, this has yet to be evaluated in histopathology.
These XAI techniques are important components of computational pathology techniques. They help evaluate bases for predictions and might be used to uncover new statistical correlations between images and a desired output. Importantly, they can help uncover spurious correlations or batch effects, e.g., when an ANN focuses on the background, not the tissue, while delivering good results. As such, they are helpful tools to build trust in model predictions.
Deep learning regression in pathology
Time-to-event regression
Regression techniques in digital pathology often encompass time-to-event regression, i.e., analyzing the length of time until the occurrence of a predefined endpoint. The most prominent applications are in cancer histopathology, especially predicting overall survival [56, 94, 98, 115, 116]. The implemented models are often based on end-to-end image regression and can predict the desired endpoint directly from the image input, thus not being dependent on the extraction of hand-crafted morphometric features. However, similar approaches include classification of patients into risk groups which are then implemented as a covariate in a Cox regression model [95]. Other than predicting overall survival, there have been multiple studies on predicting cancer recurrence in breast cancer [95] or hepatocellular carcinoma [88].
Linear and logistic regression
Nevertheless, regression is not limited to time-to-event analysis, but also includes prediction of quantitative parameters. This can be especially interesting when deep learning models are able to predict quantitative clinical characteristics or pathophysiological alterations from histology. Recent studies have hypothesized that this approach holds advantages over classification workflows due to the continuous nature of measurements. Classification approaches often lead to dichotomization of the desired output value which results in a significant loss of predictive performance. Kather et al. demonstrate that a regression model for predicting homologous recombination deficiency, i.e., a biomarker for genome instability, outperformed traditional classification approaches in five of seven cancer types with similar predictive performance for the other two subtypes [28].
Similar to approaches in cancer pathology, multiple instance learning can be leveraged to accurately predict continuous brain age from post-mortem hippocampal sections [71]. The predicted brain age (HistoAge) unveiled novel determinants of age-related functional decline such as increased model attention to the C2 hippocampal subfield. HistoAge also showed significant associations with clinical features of cognitive impairment that were not found based on epigenetic methylation analysis.
Additionally, liver histology analysis could accurately predict the hepatic venous pressure gradient, an important physiological parameter for the estimation of portal hypertension in liver disease [12]. The proposed analysis method of liver biopsies from patients with compensated liver cirrhosis could quantify important hemodynamic changes which otherwise can only be determined by invasive interventional radiography.
Routine implementation in research and diagnostics
To implement computational pathology approaches in routine biomedical research and clinical pathology, several aspects must be considered. The most important aspects for implementation in both research and diagnostics likely are accuracy and reproducibility of model predictions [60] which resemble the common quality criteria for biomedical tests and research. However, ease of use is similarly important for distribution of deep learning applications in biomedical research, since many biomedical researchers lack the specific bioinformatics expertise needed to deploy trained models without dedicated available software.
Open source software that enables researchers to train and deploy deep learning models on their own with minimal or no coding expertise required are thus very valuable. Examples of such softwares include QuPath [6], which, e.g., has a StarDist [89] and a Cellpose [101] extension that can be used to perform DL-based cell segmentation. Another prominent example is napari [2], which has over 100 extensions [47] and can be used to access ImageJ features [92], which is highly interesting due to the broad use of ImageJ [90] in the biomedical community. A few other examples include CellProfiler [62], ilastik [100], or Orbit Image Analysis [76]. Given such tools, researchers can design their own deep learning–based biomedical image analysis pipelines without the need to code, potentially even using open source software assisting with downstream analysis of the generated data, such as CellProfiler Analyst [51] or Trigon [46].
To enable comparability of study results, standardization is needed. This is however largely lacking for studies performing pathomics analyses [14]. For the analysis of bone density and structure, histomorphometry is already a well-established application. Here, validated protocols and recognized definitions of structures have already been published [30]. Especially for renal osteodystrophy and osteoporosis, bone histomorphometry has been widely recognized as a valuable method for evaluating skeletal remodeling in clinical trials [8, 85]. Further efforts have to be made for reaching a consensus analysis workflow for other tissues as well to enable similar successes.
Before clinical implementation, a number of development and validation stages should ideally be completed (Fig. 4). Currently, the major hurdles for clinical diagnostic use and implementation are limited digitalization of pathology institutes and the lack of prospective evidence, although some studies emerge that investigate deep learning models in pathology prospectively [7]. Prospective evidence is needed to robustly prove a benefit of using deep learning models both for pathology diagnostics and for clinicians receiving new information (e.g., prognostic information generated by a model). That being said, another significant hurdle is the lack of reimbursement for using deep learning models in diagnostic pathology in many countries, which means, costs associated with buying software must be amortized through gaining efficiency. One reason for the lack of reimbursement might be that designing a reimbursement strategy for using image analysis tools in clinical medicine is challenging. Still, reimbursement concepts are increasingly emerging [79].
Workflow for implementation of computational pathology algorithms into clinical routine practice. Accurately formulating a clinical question, generating meaningful input data, and selecting an appropriate model architecture for the desired task are crucial for then developing a precise and robust algorithm. Computational pathology algorithms should ideally be validated in external independent cohorts and further evaluated in randomized controlled trials to demonstrate their impact on patient outcomes. After successful implementation in clinical workflows, algorithms need to be continuously monitored and adapted to the collected real-world data which provides important long-term longitudinal information to further improve model performance
Foundation models in computational pathology
Medical deep learning and also computational pathology show a trend towards using foundation models, i.e., models that are pre-trained on a wide range of data that can be adapted to perform many different downstream tasks (for an in-depth report on foundation models, see [11]). Probably the most well-known example of a foundation model is ChatGPT, based on a generative pre-trained transformer model [75]. In medicine, foundation models have been used primarily for non-imaging data [114], with a prominent example being the AlphaFold model [53].
In computational pathology, foundation models are only starting to appear. This is likely due to the fact that collecting a large and diverse enough dataset of WSI still poses a challenge. Still, some foundation models already exist in computational pathology. Virchow is a model that was trained on 1.5 million WSI of H&E stained tissue sections [107]. This model can be used to develop a cancer detection model with a very high accuracy (AUROC of 0.949 across 17 cancer types). While that is impressive, the Virchow model is not openly available, limiting its useability for the biomedical community. UNI is another foundation model for computational pathology [19]. UNI was shown to surpass the previous state of the art in several computational pathology tasks, e.g., tumor lymphocyte detection, assessed in the ChampKit benchmark [19]. A major advantage of UNI is that it is available for research purposes (for modes of access, see [19]), which will accelerate computational pathology research. CONCH is a visual-language foundation model for computational pathology developed by the same group that developed UNI [19]. CONCH was developed using 1.17 million text-image pairs. The intuition behind additionally using text is that image descriptions contain key information that might be hard to extract for a machine learning model automatically only from the image. The fact that pathologists can extrapolate from a few examples of image-text pairs might be seen as supportive for that intuition. CONCH outperformed several previous approaches across many tasks, such as image classification or segmentation [19]. CONCH is available for research purposes as well.
Outlook
Computational pathology transformed, transforms, and will further transform the way pathology diagnostics and research is being done. We assume that the current trend towards generalist foundation models will continue. These models will likely foster the development of more accurate specialist models that might even come with a text interface. Importantly, generalist multimodal models could be useful for integrating the vast amount of multimodal medical data produced everyday and perform a number of tasks. An interesting new paradigm is generalist medical AI (GMAI), i.e., multimodal foundation models that can perform many different medical specialist tasks without explicit training [73]. GMAI models would enable interactive procedures, in which physicians, e.g., perform analysis of a WSI image together with a GMAI model by querying the model using text- or voice-based input and receiving text-, voice-, or image-based outputs with explanations. While there currently is no GMAI model available, such a development does not seem far off given the current pace of research in foundation models. However, given the large number of parameters in these models, the computational overhead and consequently energy consumption of these models is potentially enormous and should be considered before large-scale implementation [106].
Conclusion
The evolution of histopathology from manual, error-prone analyses to the advent of digital pathology and the introduction of computational pathology, particularly computational histopathology, has revolutionized the way pathology specimens are analyzed. The use of AI, specifically deep learning–based techniques, further enhances transformation of pathology, e.g., enabling automated high-throughput classification, segmentation, and regression as well as multimodal data integration with high precision, a significant advance towards precision medicine and decoding the pathologic basis of diseases.
Data availability
No datasets were generated or analysed during the current study.
References
Abels E, Pantanowitz L, Aeffner F, Zarella MD, van der Laak J, Bui MM, Vemuri VN, Parwani AV, Gibbs J, Agosto-Arroyo E, Beck AH, Kozlowski C (2019) Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol 249:286–294
Ahlers J, Althviz Moré D, Amsalem O, Anderson A, Bokota G, Boone P, Bragantini J, Buckley G, Burt A, Bussonnier M, Can Solak A, Caporal C, Doncila Pop D, Evans K, Freeman J, Gaifas L, Gohlke C, Gunalan K, Har-Gil H, Harfouche M, Harrington KIS, Hilsenstein V, Hutchings K, Lambert T, Lauer J, Lichtner G, Liu Z, Liu L, Lowe A, Marconato L, Martin S, McGovern A, Migas L, Miller N, Muñoz H, Müller J-H, Nauroth-Kreß C, Nunez-Iglesias J, Pape C, Pevey K, Peña-Castellanos G, Pierré A, Rodríguez-Guerra J, Ross D, Royer L, Russell CT, Selzer G, Smith P, Sobolewski P, Sofiiuk K, Sofroniew N, Stansby D, Sweet A, Vierdag W-M, Wadhwa P, Weber Mendonça M, Windhager J, Winston P, Yamauchi K (2023) napari: a multi-dimensional image viewer for Python. Zenodo
Amgad M, Hodge JM, Elsebaie MAT, Bodelon C, Puvanesarajah S, Gutman DA, Siziopikou KP, Goldstein JA, Gaudet MM, Teras LR, Cooper LAD (2023) A population-level digital histologic biomarker for enhanced prognosis of invasive breast cancer. Nat Med 1–13
Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, Polónia A, Campilho A (2017) Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 12:e0177544
Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S, Soerjomataram I (2022) Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast 66:15–23
Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW (2017) QuPath: open source software for digital pathology image analysis. Sci Rep 7:16878
Baxi V, Edwards R, Montalto M, Saha S (2022) Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol 35:23–32
Behets GJ, Spasovski G, Sterling LR, Goodman WG, Spiegel DM, De Broe ME, D’Haese PC (2015) Bone histomorphometry before and after long-term treatment with cinacalcet in dialysis patients with secondary hyperparathyroidism. Kidney Int 87:846–856
Boeckh-Behrens T, Kleine JF, Zimmer C, Neff F, Scheipl F, Pelisek J, Schirmer L, Nguyen K, Karatas D, Poppert H (2016) Thrombus histology suggests cardioembolic cause in cryptogenic stroke. Stroke 47:1864–1871
Boeckh-Behrens T, Schubert M, Förschler A, Prothmann S, Kreiser K, Zimmer C, Riegger J, Bauer J, Neff F, Kehl V, Pelisek J, Schirmer L, Mehr M, Poppert H (2016) The impact of histological clot composition in embolic stroke. Clin Neuroradiol 26:189–197
Bommasani R, Hudson DA, Adeli E, Altman R, Arora S, von Arx S, Bernstein MS, Bohg J, Bosselut A, Brunskill E, Brynjolfsson E, Buch S, Card D, Castellon R, Chatterji N, Chen A, Creel K, Davis JQ, Demszky D, Donahue C, Doumbouya M, Durmus E, Ermon S, Etchemendy J, Ethayarajh K, Fei-Fei L, Finn C, Gale T, Gillespie L, Goel K, Goodman N, Grossman S, Guha N, Hashimoto T, Henderson P, Hewitt J, Ho DE, Hong J, Hsu K, Huang J, Icard T, Jain S, Jurafsky D, Kalluri P, Karamcheti S, Keeling G, Khani F, Khattab O, Koh PW, Krass M, Krishna R, Kuditipudi R, Kumar A, Ladhak F, Lee M, Lee T, Leskovec J, Levent I, Li XL, Li X, Ma T, Malik A, Manning CD, Mirchandani S, Mitchell E, Munyikwa Z, Nair S, Narayan A, Narayanan D, Newman B, Nie A, Niebles JC, Nilforoshan H, Nyarko J, Ogut G, Orr L, Papadimitriou I, Park JS, Piech C, Portelance E, Potts C, Raghunathan A, Reich R, Ren H, Rong F, Roohani Y, Ruiz C, Ryan J, Ré C, Sadigh D, Sagawa S, Santhanam K, Shih A, Srinivasan K, Tamkin A, Taori R, Thomas AW, Tramèr F, Wang RE, Wang W, Wu B, Wu J, Wu Y, Xie SM, Yasunaga M, You J, Zaharia M, Zhang M, Zhang T, Zhang X, Zhang Y, Zheng L, Zhou K, Liang P (2021) On the opportunities and risks of foundation models. arXiv [cs.LG]
Bosch J, Chung C, Carrasco-Zevallos OM, Harrison SA, Abdelmalek MF, Shiffman ML, Rockey DC, Shanis Z, Juyal D, Pokkalla H, Le QH, Resnick M, Montalto M, Beck AH, Wapinski I, Han L, Jia C, Goodman Z, Afdhal N, Myers RP, Sanyal AJ (2021) A machine learning approach to liver histological evaluation predicts clinically significant portal hypertension in NASH cirrhosis. Hepatology 74:3146–3160
Bouteldja N, Klinkhammer BM, Bülow RD, Droste P, Otten SW, Freifrau von Stillfried S, Moellmann J, Sheehan SM, Korstanje R, Menzel S, Bankhead P, Mietsch M, Drummer C, Lehrke M, Kramann R, Floege J, Boor P, Merhof D (2021) Deep learning-based segmentation and quantification in experimental kidney histopathology. J Am Soc Nephrol 32:52–68
Bülow RD, Hölscher DL, Costa IG, Boor P (2023) Extending the landscape of omics technologies by pathomics. NPJ Syst Biol Appl 9:38
Bulten W, Kartasalo K, Chen P-HC, Ström P, Pinckaers H, Nagpal K, Cai Y, Steiner DF, van Boven H, Vink R, Hulsbergen-van de Kaa C, van der Laak J, Amin MB, Evans AJ, van der Kwast T, Allan R, Humphrey PA, Grönberg H, Samaratunga H, Delahunt B, Tsuzuki T, Häkkinen T, Egevad L, Demkin M, Dane S, Tan F, Valkonen M, Corrado GS, Peng L, Mermel CH, Ruusuvuori P, Litjens G, Eklund M, PANDA challenge consortium (2022) Artificial intelligence for diagnosis and Gleason grading of prostate cancer: the PANDA challenge. Nat Med 28:154–163
Bulten W, Pinckaers H, van Boven H, Vink R, de Bel T, van Ginneken B, van der Laak J, Hulsbergen-van de Kaa C, Litjens G (2020) Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol 21:233–241
Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ (2019) Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 25:1301–1309
Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, Mao Q, Yu H, Cai X (2020) Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol 4:14
Chen RJ, Ding T, Lu MY, Williamson DFK, Jaume G, Song AH, Chen B, Zhang A, Shao D, Shaban M, Williams M, Oldenburg L, Weishaupt LL, Wang JJ, Vaidya A, Le LP, Gerber G, Sahai S, Williams W, Mahmood F (2024) Towards a general-purpose foundation model for computational pathology. Nat Med 30:850–862
Chen Z, Li X, Yang M, Zhang H, Xu XS (2023) Optimization of deep learning models for the prediction of gene mutations using unsupervised clustering. Hip Int 9:3–17
Collan Y (1984) Morphometry in pathology: another look at diagnostic histopathology. Pathol Res Pract 179:189–192
Collan Y, Montironi R, Mariuzzi GM, Torkkeli T, Marinelli F, Pesonen E, Collina G, Kosma VM, Jantunen E, Kosunen O (1986) Observer variation in interactive computerized morphometry. Appl Pathol 4:9–14
Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A (2018) Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 24:1559–1567
Couture HD, Williams LA, Geradts J, Nyante SJ, Butler EN, Marron JS, Perou CM, Troester MA, Niethammer M (2018) Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 4:30
Droste P, Wong DWL, Hohl M, von Stillfried S, Klinkhammer BM, Boor P (2023) Semiautomated pipeline for quantitative analysis of heart histopathology. J Transl Med 21:666
Duggento A, Conti A, Mauriello A, Guerrisi M, Toschi N (2021) Deep computational pathology in breast cancer. Semin Cancer Biol 72:226–237
Ektefaie Y, Yuan W, Dillon DA, Lin NU, Golden JA, Kohane IS, Yu K-H (2021) Integrative multiomics-histopathology analysis for breast cancer classification. NPJ Breast Cancer 7:147
El Nahhas OSM, Loeffler CML, Carrero ZI, van Treeck M, Kolbinger FR, Hewitt KJ, Muti HS, Graziani M, Zeng Q, Calderaro J, Ortiz-Brüchle N, Yuan T, Hoffmeister M, Brenner H, Brobeil A, Reis-Filho JS, Kather JN (2024) Regression-based deep-learning predicts molecular biomarkers from pathology slides. Nat Commun 15:1253
Evans T, Retzlaff CO, Geißler C, Kargl M, Plass M, Müller H, Kiehl T-R, Zerbe N, Holzinger A (2022) The explainability paradox: challenges for xAI in digital pathology. Future Gener Comput Syst 133:281–296
Evenepoel P, Behets GJS, Laurent MR, D’Haese PC (2017) Update on the role of bone biopsy in the management of patients with CKD-MBD. J Nephrol 30:645–652
Fayyaz AU, Edwards WD, Maleszewski JJ, Konik EA, DuBrock HM, Borlaug BA, Frantz RP, Jenkins SM, Redfield MM (2018) Global pulmonary vascular remodeling in pulmonary hypertension associated with heart failure and preserved or reduced ejection fraction. Circulation 137:1796–1810
Feng L, Liu Z, Li C, Li Z, Lou X, Shao L, Wang Y, Huang Y, Chen H, Pang X, Liu S, He F, Zheng J, Meng X, Xie P, Yang G, Ding Y, Wei M, Yun J, Hung M-C, Zhou W, Wahl DR, Lan P, Tian J, Wan X (2022) Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study. Lancet Digit Health 4:e8–e17
Foersch S, Glasner C, Woerl A-C, Eckstein M, Wagner D-C, Schulz S, Kellers F, Fernandez A, Tserea K, Kloth M, Hartmann A, Heintz A, Weichert W, Roth W, Geppert C, Kather JN, Jesinghaus M (2023) Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat Med 29:430–439
Fogo A, Hawkins EP, Berry PL, Glick AD, Chiang ML, MacDonell RC Jr, Ichikawa I (1990) Glomerular hypertrophy in minimal change disease predicts subsequent progression to focal glomerular sclerosis. Kidney Int 38:115–123
Fry CH, Gray RP, Dhillon PS, Jabr RI, Dupont E, Patel PM, Peters NS (2014) Architectural correlates of myocardial conduction: changes to the topography of cellular coupling, intracellular conductance, and action potential propagation with hypertrophy in Guinea-pig ventricular myocardium. Circ Arrhythm Electrophysiol 7:1198–1204
Galati F, Zuluaga MA (2021) Efficient model monitoring for quality control in cardiac image segmentation. In: Functional imaging and modeling of the heart. Springer International Publishing, pp 101–111
Gertych A, Swiderska-Chadaj Z, Ma Z, Ing N, Markiewicz T, Cierniak S, Salemi H, Guzman S, Walts AE, Knudsen BS (2019) Convolutional neural networks can accurately distinguish four histologic growth patterns of lung adenocarcinoma in digital slides. Sci Rep 9:1483
Ghaffari Laleh N, Truhn D, Veldhuizen GP, Han T, van Treeck M, Buelow RD, Langer R, Dislich B, Boor P, Schulz V, Kather JN (2022) Adversarial attacks and adversarial robustness in computational pathology. Nat Commun 13:5711
Ginley B, Jen K-Y, Han SS, Rodrigues L, Jain S, Fogo AB, Zuckerman J, Walavalkar V, Miecznikowski JC, Wen Y, Yen F, Yun D, Moon KC, Rosenberg A, Parikh C, Sarder P (2021) Automated computational detection of interstitial fibrosis, tubular atrophy, and glomerulosclerosis. J Am Soc Nephrol 32:837–850
Graziani M, Mahony LO, Nguyen A-P, Müller H, Andrearczyk V (2023) Uncovering unique concept vectors through latent space decomposition. arXiv [cs.LG]
Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E, Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S (2015) The consensus molecular subtypes of colorectal cancer. Nat Med 21:1350–1356
Gupta R, Kurc T, Sharma A, Almeida JS, Saltz J (2019) The emergence of pathomics. Curr Pathobiol Rep 7:73–84
Heinrich L, Bennett D, Ackerman D, Park W, Bogovic J, Eckstein N, Petruncio A, Clements J, Pang S, Xu CS, Funke J, Korff W, Hess HF, Lippincott-Schwartz J, Saalfeld S, Weigel AV, COSEM Project Team (2021) Whole-cell organelle segmentation in volume electron microscopy. Nature 599:141–146
Hermsen M, de Bel T, den Boer M, Steenbergen EJ, Kers J, Florquin S, Roelofs JJTH, Stegall MD, Alexander MP, Smith BH, Smeets B, Hilbrands LB, van der Laak JAWM (2019) Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol 30:1968–1979
Hölscher DL, Bouteldja N, Joodaki M, Russo ML, Lan Y-C, Sadr AV, Cheng M, Tesar V, Stillfried SV, Klinkhammer BM, Barratt J, Floege J, Roberts ISD, Coppo R, Costa IG, Bülow RD, Boor P (2023) Next-generation morphometry for pathomics-data mining in histopathology. Nat Commun 14:470
Hölscher DL, Goedertier M, Klinkhammer BM, Droste P, Costa IG, Boor P, Bülow RD (2024) tRigon: an R package and Shiny app for integrative (path-)omics data analysis. BMC Bioinformatics 25:98
Home. https://www.napari-hub.org/. Accessed 10 Apr 2024
Humphrey PA (2017) Histopathology of prostate cancer. Cold Spring Harb Perspect Med 7. https://doi.org/10.1101/cshperspect.a030411
Jang H-J, Lee A, Kang J, Song IH, Lee SH (2020) Prediction of clinically actionable genetic alterations from colorectal cancer histopathology images using deep learning. World J Gastroenterol 26:6207–6223
Jayapandian CP, Chen Y, Janowczyk AR, Palmer MB, Cassol CA, Sekulic M, Hodgin JB, Zee J, Hewitt SM, O’Toole J, Toro P, Sedor JR, Barisoni L, Madabhushi A, Nephrotic Syndrome Study Network (NEPTUNE) (2021) Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int 99:86–101
Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE (2008) Cell Profiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9:482
Joodaki M, Shaigan M, Parra V, Bülow RD, Kuppe C, Hölscher DL, Cheng M, Nagai JS, Goedertier M, Bouteldja N, Tesar V, Barratt J, Roberts IS, Coppo R, Kramann R, Boor P, Costa IG (2023) Detection of PatIent-Level distances from single cell genomics and pathomics data with Optimal Transport (PILOT). Mol Syst Biolhttps://doi.org/10.1038/s44320-023-00003-8
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589
Kanavati F, Toyokawa G, Momosaki S, Rambeau M, Kozuma Y, Shoji F, Yamazaki K, Takeo S, Iizuka O, Tsuneki M (2020) Weakly-supervised learning for lung carcinoma classification using deep learning. Sci Rep 10:9297
Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, Krause J, Niehues JM, Sommer KAJ, Bankhead P, Kooreman LFS, Schulte JJ, Cipriani NA, Buelow RD, Boor P, Ortiz-Brüchle N-N, Hanby AM, Speirs V, Kochanny S, Patnaik A, Srisuwananukorn A, Brenner H, Hoffmeister M, van den Brandt PA, Jäger D, Trautwein C, Pearson AT, Luedde T (2020) Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer 1:789–799
Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis C-A, Gaiser T, Marx A, Valous NA, Ferber D, Jansen L, Reyes-Aldasoro CC, Zörnig I, Jäger D, Brenner H, Chang-Claude J, Hoffmeister M, Halama N (2019) Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med 16:e1002730
Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T (2019) Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 25:1054–1056
Kers J, Bülow RD, Klinkhammer BM, Breimer GE, Fontana F, Abiola AA, Hofstraat R, Corthals GL, Peters-Sengers H, Djudjaj S, von Stillfried S, Hölscher DL, Pieters TT, van Zuilen AD, Bemelman FJ, Nurmohamed AS, Naesens M, Roelofs JJTH, Florquin S, Floege J, Nguyen TQ, Kather JN, Boor P (2022) Deep learning-based classification of kidney transplant pathology: a retrospective, multicentre, proof-of-concept study. Lancet Digit Health 4:e18–e26
Klinkhammer BM, Buchtler S, Djudjaj S, Bouteldja N, Palsson R, Edvardsson VO, Thorsteinsdottir M, Floege J, Mack M, Boor P (2022) Current kidney function parameters overestimate kidney tissue repair in reversible experimental kidney disease. Kidney Int 102:307–320
van der Laak J, Litjens G, Ciompi F (2021) Deep learning in histopathology: the path to the clinic. Nat Med 27:775–784
Laleh NG, Muti HS, Loeffler CML, Echle A, Saldanha OL, Mahmood F, Lu MY, Trautwein C, Langer R, Dislich B, Buelow RD, Grabsch HI, Brenner H, Chang-Claude J, Alwers E, Brinker TJ, Khader F, Truhn D, Gaisa NT, Boor P, Hoffmeister M, Schulz V, Kather JN (2022) Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology. Med Image Anal 102474
Lamprecht MR, Sabatini DM, Carpenter AE (2007) Cell Profiler: free, versatile software for automated biological image analysis. Biotechniques 42:71–75
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Leo P, Lee G, Shih NNC, Elliott R, Feldman MD, Madabhushi A (2016) Evaluating stability of histomorphometric features across scanner and staining variations: prostate cancer diagnosis from whole slide images. J Med Invest 3:047502
Liao H, Long Y, Han R, Wang W, Xu L, Liao M, Zhang Z, Wu Z, Shang X, Li X, Peng J, Yuan K, Zeng Y (2020) Deep learning-based classification and mutation prediction from histopathological images of hepatocellular carcinoma. Clin Transl Med 10:e102
Li O, Liu H, Chen C, Rudin C (2018) Deep learning for case-based reasoning through prototypes: a neural network that explains its predictions. AAAI 32. https://doi.org/10.1609/aaai.v32i1.11771
Lucarelli N, Ginley B, Zee J, Mimar S, Paul AS, Jain S, Han SS, Rodrigues L, Ozrazgat-Baslanti T, Wong ML, Nadkarni G, Clapp WL, Jen K-Y, Sarder P (2023) Correlating deep learning-based automated reference kidney histomorphometry with patient demographics and creatinine. Kidney360 4:1726–1737
Lu MY, Chen TY, Williamson DFK, Zhao M, Shady M, Lipkova J, Mahmood F (2021) AI-based pathology predicts origins for cancers of unknown primary. Nature 594:106–110
Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M, Mahmood F (2021) Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Enghttps://doi.org/10.1038/s41551-020-00682-w
Malluche HH, Sherman D, Meyer W, Massry SG (1982) A new semiautomatic method for quantitative static and dynamic bone histology. Calcif Tissue Int 34:439–448
Marx GA, Kauffman J, McKenzie AT, Koenigsberg DG, McMillan CT, Morgello S, Karlovich E, Insausti R, Richardson TE, Walker JM, White CL 3rd, Babrowicz BM, Shen L, McKee AC, Stein TD, PART Working Group, Farrell K, Crary JF (2023) Histopathologic brain age estimation via multiple instance learning. Acta Neuropathol 146:785–802
Graziani M, Andrearczyk V, Marchand-Maillet S, Müller H (2020) Concept attribution: explaining CNN decisions to physicians. Comput Biol Med 123:103865
Moor M, Banerjee O, Abad ZSH, Krumholz HM, Leskovec J, Topol EJ, Rajpurkar P (2023) Foundation models for generalist medical artificial intelligence. Nature 616:259–265
Niehues JM, Quirke P, West NP, Grabsch HI, van Treeck M, Schirris Y, Veldhuizen GP, Hutchins GGA, Richman SD, Foersch S, Brinker TJ, Fukuoka J, Bychkov A, Uegami W, Truhn D, Brenner H, Brobeil A, Hoffmeister M, Kather JN (2023) Generalizable biomarker prediction from cancer pathology slides with self-supervised deep learning: a retrospective multi-centric study. Cell Rep Med 4:100980
OpenAI, Achiam J, Adler S, Agarwal S, Ahmad L, Akkaya I, Aleman FL, Almeida D, Altenschmidt J, Altman S, Anadkat S, Avila R, Babuschkin I, Balaji S, Balcom V, Baltescu P, Bao H, Bavarian M, Belgum J, Bello I, Berdine J, Bernadett-Shapiro G, Berner C, Bogdonoff L, Boiko O, Boyd M, Brakman A-L, Brockman G, Brooks T, Brundage M, Button K, Cai T, Campbell R, Cann A, Carey B, Carlson C, Carmichael R, Chan B, Chang C, Chantzis F, Chen D, Chen S, Chen R, Chen J, Chen M, Chess B, Cho C, Chu C, Chung HW, Cummings D, Currier J, Dai Y, Decareaux C, Degry T, Deutsch N, Deville D, Dhar A, Dohan D, Dowling S, Dunning S, Ecoffet A, Eleti A, Eloundou T, Farhi D, Fedus L, Felix N, Fishman SP, Forte J, Fulford I, Gao L, Georges E, Gibson C, Goel V, Gogineni T, Goh G, Gontijo-Lopes R, Gordon J, Grafstein M, Gray S, Greene R, Gross J, Gu SS, Guo Y, Hallacy C, Han J, Harris J, He Y, Heaton M, Heidecke J, Hesse C, Hickey A, Hickey W, Hoeschele P, Houghton B, Hsu K, Hu S, Hu X, Huizinga J, Jain S, Jain S, Jang J, Jiang A, Jiang R, Jin H, Jin D, Jomoto S, Jonn B, Jun H, Kaftan T, Kaiser Ł, Kamali A, Kanitscheider I, Keskar NS, Khan T, Kilpatrick L, Kim JW, Kim C, Kim Y, Kirchner JH, Kiros J, Knight M, Kokotajlo D, Kondraciuk Ł, Kondrich A, Konstantinidis A, Kosic K, Krueger G, Kuo V, Lampe M, Lan I, Lee T, Leike J, Leung J, Levy D, Li CM, Lim R, Lin M, Lin S, Litwin M, Lopez T, Lowe R, Lue P, Makanju A, Malfacini K, Manning S, Markov T, Markovski Y, Martin B, Mayer K, Mayne A, McGrew B, McKinney SM, McLeavey C, McMillan P, McNeil J, Medina D, Mehta A, Menick J, Metz L, Mishchenko A, Mishkin P, Monaco V, Morikawa E, Mossing D, Mu T, Murati M, Murk O, Mély D, Nair A, Nakano R, Nayak R, Neelakantan A, Ngo R, Noh H, Ouyang L, O’Keefe C, Pachocki J, Paino A, Palermo J, Pantuliano A, Parascandolo G, Parish J, Parparita E, Passos A, Pavlov M, Peng A, Perelman A, de Avila Belbute Peres F, Petrov M, de Oliveira Pinto HP, Michael, Pokorny, Pokrass M, Pong VH, Powell T, Power A, Power B, Proehl E, Puri R, Radford A, Rae J, Ramesh A, Raymond C, Real F, Rimbach K, Ross C, Rotsted B, Roussez H, Ryder N, Saltarelli M, Sanders T, Santurkar S, Sastry G, Schmidt H, Schnurr D, Schulman J, Selsam D, Sheppard K, Sherbakov T, Shieh J, Shoker S, Shyam P, Sidor S, Sigler E, Simens M, Sitkin J, Slama K, Sohl I, Sokolowsky B, Song Y, Staudacher N, Such FP, Summers N, Sutskever I, Tang J, Tezak N, Thompson MB, Tillet P, Tootoonchian A, Tseng E, Tuggle P, Turley N, Tworek J, Uribe JFC, Vallone A, Vijayvergiya A, Voss C, Wainwright C, Wang JJ, Wang A, Wang B, Ward J, Wei J, Weinmann CJ, Welihinda A, Welinder P, Weng J, Weng L, Wiethoff M, Willner D, Winter C, Wolrich S, Wong H, Workman L, Wu S, Wu J, Wu M, Xiao K, Xu T, Yoo S, Yu K, Yuan Q, Zaremba W, Zellers R, Zhang C, Zhang M, Zhao S, Zheng T, Zhuang J, Zhuk W, Zoph B (2023) GPT-4 technical report. arXiv [cs.CL]
Orbit image analysis. https://www.orbit.bio/. Accessed 10 Apr 2024
Osman OS, Selway JL, Kępczyńska MA, Stocker CJ, O’Dowd JF, Cawthorne MA, Arch JR, Jassim S, Langlands K (2013) A novel automated image analysis method for accurate adipocyte quantification. Adipocyte 2:160–164
Pantanowitz L, Quiroga-Garza GM, Bien L, Heled R, Laifenfeld D, Linhart C, Sandbank J, Albrecht Shach A, Shalev V, Vecsler M, Michelow P, Hazelhurst S, Dhir R (2020) An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: a blinded clinical validation and deployment study. The Lancet Digital Health 2:e407–e416
Parikh RB, Helmchen LA (2022) Paying for artificial intelligence in medicine. NPJ Digit Med 5:63
Peirlinck M, Sahli Costabal F, Sack KL, Choy JS, Kassab GS, Guccione JM, De Beule M, Segers P, Kuhl E (2019) Using machine learning to characterize heart failure across the scales. Biomech Model Mechanobiol 18:1987–2001
Plass M, Kargl M, Kiehl T-R, Regitnig P, Geißler C, Evans T, Zerbe N, Carvalho R, Holzinger A, Müller H (2023) Explainability and causability in digital pathology. Hip Int 9:251–260
Pocock J, Graham S, Vu QD, Jahanifar M, Deshpande S, Hadjigeorghiou G, Shephard A, Bashir RMS, Bilal M, Lu W, Epstein D, Minhas F, Rajpoot NM, Raza SEA (2022) TIAToolbox as an end-to-end library for advanced tissue image analytics. Commun Med 2:120
Qu H, Zhou M, Yan Z, Wang H, Rustgi VK, Zhang S, Gevaert O, Metaxas DN (2021) Genetic mutation and biological pathway prediction based on whole slide images in breast carcinoma using deep learning. NPJ Precis Oncol 5:87
Raciti P, Sue J, Retamero JA, Ceballos R, Godrich R, Kunz JD, Casson A, Thiagarajan D, Ebrahimzadeh Z, Viret J, Lee D, Schüffler PJ, DeMuth G, Gulturk E, Kanan C, Rothrock B, Reis-Filho J, Klimstra DS, Reuter V, Fuchs TJ (2023) Clinical validation of artificial intelligence-augmented pathology diagnosis demonstrates significant gains in diagnostic accuracy in prostate cancer detection. Arch Pathol Lab Med 147:1178–1185
Recker R, Dempster D, Langdahl B, Giezek H, Clark S, Ellis G, de Villiers T, Valter I, Zerbini CA, Cohn D, Santora A, Duong LT (2020) Effects of odanacatib on bone structure and quality in postmenopausal women with osteoporosis: 5-year data from the phase 3 long-term odanacatib fracture trial (LOFT) and its extension. J Bone Miner Res 35:1289–1299
Reyes-Fernandez PC, Periou B, Decrouy X, Relaix F, Authier FJ (2019) Automated image-analysis method for the quantification of fiber morphometry and fiber type population in human skeletal muscle. Skelet Muscle 9:15
Robinson R, Valindria VV, Bai W, Oktay O, Kainz B, Suzuki H, Sanghvi MM, Aung N, Paiva JM, Zemrak F, Fung K, Lukaschuk E, Lee AM, Carapella V, Kim YJ, Piechnik SK, Neubauer S, Petersen SE, Page C, Matthews PM, Rueckert D, Glocker B (2019) Automated quality control in image segmentation: application to the UK Biobank cardiovascular magnetic resonance imaging study. J Cardiovasc Magn Reson 21:18
Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky J-M, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J (2020) Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology 72:2000–2013
Schmidt U, Weigert M, Broaddus C, Myers G (2018) Cell detection with star-convex polygons. arXiv [cs.CV]
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2016) Grad-CAM: visual explanations from deep networks via gradient-based localization. arXiv [cs.CV]
Selzer GJ, Rueden CT, Hiner MC, Evans EL 3rd, Harrington KIS, Eliceiri KW (2023) napari-imagej: ImageJ ecosystem access from napari. Nat Methods 20:1443–1444
Seraphin TP, Luedde M, Roderburg C, van Treeck M, Scheider P, Buelow RD, Boor P, Loosen SH, Provaznik Z, Mendelsohn D, Berisha F, Magnussen C, Westermann D, Luedde T, Brochhausen C, Sossalla S, Kather JN (2023) Prediction of heart transplant rejection from routine pathology slides with self-supervised deep learning. Eur Heart J Digit Health 4:265–274
Shao W, Wang T, Huang Z, Han Z, Zhang J, Huang K (2021) Weakly supervised deep ordinal cox model for survival prediction from whole-slide pathological images. IEEE Trans Med Imaging 40:3739–3747
Shi Y, Olsson LT, Hoadley KA, Calhoun BC, Marron JS, Geradts J, Niethammer M, Troester MA (2023) Predicting early breast cancer recurrence from histopathological images in the Carolina Breast Cancer Study. NPJ Breast Cancer 9:92
Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN (2022) Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer 3:1026–1038
Sirinukunwattana K, Domingo E, Richman SD, Redmond KL, Blake A, Verrill C, Leedham SJ, Chatzipli A, Hardy C, Whalley CM, Wu C-H, Beggs AD, McDermott U, Dunne PD, Meade A, Walker SM, Murray GI, Samuel L, Seymour M, Tomlinson I, Quirke P, Maughan T, Rittscher J, Koelzer VH, S:CORT consortium, (2021) Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 70:544–554
Skrede O-J, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, Danielsen HE (2020) Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet 395:350–360
Smith LR, Barton ER (2014) SMASH – semi-automatic muscle analysis using segmentation of histology: a MATLAB application. Skelet Muscle 4:1–16
Sommer C, Straehle C, Köthe U, Hamprecht FA (2011) ilastik: interactive learning and segmentation toolkit. In: 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE, pp 230–233
Stringer C, Wang T, Michaelos M, Pachitariu M (2021) Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18:100–106
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249
Sun L, Marsh JN, Matlock MK, Chen L, Gaut JP, Brunt EM, Swamidass SJ, Liu T-C (2020) Deep learning quantification of percent steatosis in donor liver biopsy frozen sections. EBioMedicine 60:103029
Suppli MP, Rigbolt KTG, Veidal SS, Heebøll S, Eriksen PL, Demant M, Bagger JI, Nielsen JC, Oró D, Thrane SW, Lund A, Strandberg C, Kønig MJ, Vilsbøll T, Vrang N, Thomsen KL, Grønbæk H, Jelsing J, Hansen HH, Knop FK (2019) Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am J Physiol Gastrointest Liver Physiol 316:G462–G472
Taylor-Weiner A, Pokkalla H, Han L, Jia C, Huss R, Chung C, Elliott H, Glass B, Pethia K, Carrasco-Zevallos O, Shukla C, Khettry U, Najarian R, Taliano R, Subramanian GM, Myers RP, Wapinski I, Khosla A, Resnick M, Montalto MC, Anstee QM, Wong VW-S, Trauner M, Lawitz EJ, Harrison SA, Okanoue T, Romero-Gomez M, Goodman Z, Loomba R, Beck AH, Younossi ZM (2021) A machine learning approach enables quantitative measurement of liver histology and disease monitoring in NASH. Hepatology 74:133–147
Vafaei Sadr A, Bülow R, von Stillfried S, Schmitz NEJ, Pilva P, Hölscher DL, Ha PP, Schweiker M, Boor P (2024) Operational greenhouse-gas emissions of deep learning in digital pathology: a modelling study. Lancet Digit Health 6:e58–e69
Vorontsov E, Bozkurt A, Casson A, Shaikovski G, Zelechowski M, Liu S, Severson K, Zimmermann E, Hall J, Tenenholtz N, Fusi N, Mathieu P, van Eck A, Lee D, Viret J, Robert E, Wang YK, Kunz JD, Lee MCH, Bernhard J, Godrich RA, Oakley G, Millar E, Hanna M, Retamero J, Moye WA, Yousfi R, Kanan C, Klimstra D, Rothrock B, Fuchs TJ (2023) Virchow: a million-slide digital pathology foundation model. arXiv [eess.IV]
Wagner SJ, Reisenbüchler D, West NP, Niehues JM, Zhu J, Foersch S, Veldhuizen GP, Quirke P, Grabsch HI, van den Brandt PA, Hutchins GGA, Richman SD, Yuan T, Langer R, Jenniskens JCA, Offermans K, Mueller W, Gray R, Gruber SB, Greenson JK, Rennert G, Bonner JD, Schmolze D, Jonnagaddala J, Hawkins NJ, Ward RL, Morton D, Seymour M, Magill L, Nowak M, Hay J, Koelzer VH, Church DN, TransSCOT consortium, Matek C, Geppert C, Peng C, Zhi C, Ouyang X, James JA, Loughrey MB, Salto-Tellez M, Brenner H, Hoffmeister M, Truhn D, Schnabel JA, Boxberg M, Peng T, Kather JN, (2023) Transformer-based biomarker prediction from colorectal cancer histology: a large-scale multicentric study. Cancer Cell 41:1650-1661.e4
Wang X, Janowczyk A, Zhou Y, Thawani R, Fu P, Schalper K, Velcheti V, Madabhushi A (2017) Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci Rep 7:13543
Wang X, Zou C, Zhang Y, Li X, Wang C, Ke F, Chen J, Wang W, Wang D, Xu X, Xie L, Zhang Y (2021) Prediction of BRCA gene mutation in breast cancer based on deep learning and histopathology images. Front Genet 12:661109
Wang Y, Acs B, Robertson S, Liu B, Solorzano L, Wählby C, Hartman J, Rantalainen M (2022) Improved breast cancer histological grading using deep learning. Ann Oncol 33:89–98
Wei B, Han Z, He X, Yin Y (2017) Deep learning model based breast cancer histopathological image classification. In: 2017 IEEE 2nd International Conference on Cloud Computing and Big Data Analysis (ICCCBDA). IEEE, pp 348–353
Whitney J, Corredor G, Janowczyk A, Ganesan S, Doyle S, Tomaszewski J, Feldman M, Gilmore H, Madabhushi A (2018) Quantitative nuclear histomorphometry predicts oncotype DX risk categories for early stage ER+ breast cancer. BMC Cancer 18:610
Wornow M, Xu Y, Thapa R, Patel B, Steinberg E, Fleming S, Pfeffer MA, Fries J, Shah NH (2023) The shaky foundations of large language models and foundation models for electronic health records. NPJ Digit Med 6:135
Wulczyn E, Steiner DF, Xu Z, Sadhwani A, Wang H, Flament-Auvigne I, Mermel CH, Chen P-HC, Liu Y, Stumpe MC (2020) Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE 15:e0233678
Yao J, Zhu X, Jonnagaddala J, Hawkins N, Huang J (2020) Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med Image Anal 65:101789
Yi Z, Xi C, Menon MC, Cravedi P, Tedla F, Soto A, Sun Z, Liu K, Zhang J, Wei C, Chen M, Wang W, Veremis B, Garcia-Barros M, Kumar A, Haakinson D, Brody R, Azeloglu EU, Gallon L, O’Connell P, Naesens M, Shapiro R, Colvin RB, Ward S, Salem F, Zhang W (2023) A large-scale retrospective study enabled deep-learning based pathological assessment of frozen procurement kidney biopsies to predict graft loss and guide organ utilization. Kidney Inthttps://doi.org/10.1016/j.kint.2023.09.031
Zhang W, Cao G, Wu F, Wang Y, Liu Z, Hu H, Xu K (2023) Global burden of prostate cancer and association with socioeconomic status, 1990–2019: a systematic analysis from the global burden of disease study. J Epidemiol Glob Health 13:407–421
Acknowledgements
We thank Yu-Chia Lan, M.Sc., for providing a saliency map visualization for Fig. 3B.
Funding
Open Access funding enabled and organized by Projekt DEAL. RDB receives funding from the START-Program of the Faculty of Medicine, Rheinisch-Westfälische Technische Hochschule Aachen (grant number 148/21).
Author information
Authors and Affiliations
Contributions
DLH and RDB performed literature search and study selection. DLH created and RDB reviewed the figures. DLH and RDB wrote and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the special issue on Artificial Intelligence in Pflügers Archiv—European Journal of Physiology.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hölscher, D.L., Bülow, R.D. Decoding pathology: the role of computational pathology in research and diagnostics. Pflugers Arch - Eur J Physiol (2024). https://doi.org/10.1007/s00424-024-03002-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00424-024-03002-2